Abstract
Transposable elements (TEs) represent ∼45% of the human genome and 50–90% of some grass genomes. While most elements contain inactivating mutations, others are reversibly inactivated (silenced) by epigenetic mechanisms, including cytosine methylation. Previous studies have shown that retrotransposons can influence the expression of adjacent host genes. In this study, the methylation patterns of TEs and their flanking sequences in different tissues were undertaken using a novel technique called transposon methylation display (TMD). TMD was successfully applied on a highly copied (∼1000 copies), newly amplified LTR retrotransposon family in rice called Dasheng. We determined that the methylation status of a subset of LTRs varies in leaves vs. roots. In addition, we determined that tissue-specific LTR methylation correlated with tissue-specific expression of the flanking rice gene. Genes showing tissue-specific expression were in opposite orientation relative to the LTR. Antisense transcripts were detected in the tissue where the sense transcripts from that gene were not detected. Comparative analysis of Dasheng LTR methylation in the two subspecies, japonica vs. indica revealed LTR-mediated differences in subspecies gene expression. Subspecies-specific expression was due either to polymorphic Dasheng insertion sites between the two subspecies or to subspecies-specific methylation of LTRs at the same locus accounted for observed differences in the expression of adjacent genes.
TRANSPOSABLE elements (TEs) are mobile DNA sequences that have the ability to “jump” to new locations in the genome. They are divided into two classes. Class I retrotransposons, or retroelements, move to a new genomic location via an RNA intermediate that is reverse-transcribed into an extrachromosomal cDNA that then integrates into a new location (Finnegan 1989). There are two types of retrotransposons: LTR retrotransposons, flanked by long terminal repeats (LTRs) that include the promoter and terminator regions and non-LTR elements (such as long interspersed nuclear elements, (LINEs) and short interspersed nuclear elements, (SINEs) that use an internal promoter or are transcribed from a flanking gene's promoter. Transcription of LTR retrotransposons initiates from a promoter in the 5′ LTR and terminates in the 3′ LTR. However, because the 5′ and 3′ LTRs are identical upon insertion, transcripts can initiate from the promoter in the 3′ LTR and read out into flanking host sequences. In addition, transcripts can read out from solo LTRs, which are produced by homologous recombination between LTRs. The replicative mode of transposition of retroelements allows them to reach a high copy number (up to 1 million copies). In some plants they represent up to 80% of the genome (Bennetzen and Kellogg 1997; Feschotte et al. 2002). On the other hand, class II elements, also called DNA elements or transposons, move directly by a cut and paste mechanism (Finnegan 1989).
Plants protect their genes by targeting TEs for methylation (Kumar and Bennetzen 1999; Zilberman and Henikoff 2004). As such, TEs in plant genomes are hypermethylated in comparison with host genes and are said to be epigenetically silenced (Bennetzen et al. 1994; Flavell 1994; Martienssen 1998; Rabinowicz et al. 2003; Khodosevich et al. 2004; Okahara et al. 2004; Lavie et al. 2005). Inactive TEs in plants are often heavily methylated (Bennetzen et al. 1994; Flavell 1994; Martienssen 1998) and can be reactivated in genetic backgrounds containing methylation-defective mutants or during tissue culture (Miura et al. 2001; Singer et al. 2001). For example, in Arabidopsis thaliana, a higher frequency of mutation in genetic backgrounds containing methylation-defective mutations, such as ddm1, has been associated with the transcriptional activation of endogenous TEs and the subsequent insertion of a member of at least one TE family into Arabidopsis genes (Miura et al. 2001; Singer et al. 2001). Moreover, methylation patterns of retroelements have been shown to change in a tissue-specific manner, such as the human endogenous retrovirus family K (HERV-K) (Khodosevich et al. 2004). In some plant species, methylation patterns also change dramatically during the formation of polyploids (Shaked et al. 2001; Madlung et al. 2002). In newly synthesized wheat polyploids, several cases of genomewide methylation changes have been documented where retrotransposons were found to be demethylated (Shaked et al. 2001). In the early stages after TE amplification, readout transcription from LTRs can impact the expression of dozens of host genes (Kashkush et al. 2003). In addition, the spectrum of affected genes depends on the methylation status of the LTRs, which can vary from generation to generation and/or in different subspecies. In addition, DNA methylation patterns were shown to change during the life of an organism (Finnegan 1989; Jaenisch and Bird 2003).
Our hypothesis is that unmethylated LTRs adjacent to host genes might impact host gene expression. Here, the methylation patterns of TEs and their flanking sequences were undertaken using transposon methylation display (TMD). TMD was applied to various diploid and polyploid rice species. A newly amplified LTR retrotransposon family called Dasheng with its ∼1000 insertions in the rice genome (Jiang et al. 2002a,b) was the focus of this study. The impact of Dasheng insertions on the expression of rice genes has been assessed by identifying Dasheng elements located near or into genes, then assaying LTR methylation status of these elements and the nearby gene expression. Furthermore, the availability of sequence drafts for japonica and indica permitted the identification of polymorphic Dasheng insertion sites in the two subspecies and facilitated the design of experiments to determine if the presence/absence of Dasheng led to differential flanking gene expression. Our data indicate that tissue-specific LTR methylation inversely correlated with the expression of adjacent genes. In addition, we show that TEs can be responsible in part for the gene expression differences between subspecies.
MATERIALS AND METHODS
Plant material:
Six diploid Oryza species: sativa ssp. japonica (genome AA), sativa ssp. indica (genome AA), nivara (genome AA), punctata (genome BB), eichingeri (genome CC), officinails (genome CC), and a tetraploid Oryza african (genome BBCC) were used in this study. Seed was supplied by National Small Grains Collection, United States Department of Agriculture (USDA) (http://www.ars-grin.gov/npgs/). For DNA and RNA extractions, roots and leaves were collected from seedlings 2 weeks after germination.
DNA analysis:
TMD:
This technique utilizes two methylation-sensitive enzymes HpaII and MspI that both recognize CCGG sites. While HpaII is sensitive to methylation of either cytosine (except when the external cytosine is hemi-methylated), MspI is sensitive only when the external cytosine is methylated. Methylation of CCGG will result in different cleavage by the isoschizomers and to the generation of polymorphic PCR fragments. Use of both enzymes insures that all combinations of cytosine methylation will be detected. Genomic DNA was extracted from leaves and roots using DNeasy plant kit (QIAGEN, Valencia CA) and digested (200–300 ng DNA) with either HpaII or MspI. Adapter ligation was carried out as previously described (Shaked et al. 2001). Preamplification PCR was carried out with a previously described (Shaked et al. 2001) primer complementary to the core of the HpaII–MspI (H/M) adapter sequence and with a primer whose 5′ end anneals to the Dasheng LTR and whose 3′ end primes DNA synthesis toward the adjacent genomic DNA (5′- TTTAGGTCTCGTGCGCTACC -3′). The 3′-LTR primer is located downstream of H/M sites in the LTR (see Figure 1A). Selective amplification PCR was carried out using the 3′-LTR primer and an adapter primer [with substituting primers any of the 16 (24) primers with different combinations of the last two nucleotides: 5′-ATCATGAGTCCTGCTCGGNN-3′]. Combinations of adapter primers with different selective bases and a specific 33P-end-labeled LTR primer were used to amplify different chimeric sequences (LTR/flanking). Reaction products were separated in denaturing 6% polyacrylamide gel (19:1 acrylamide:bisacrylamide, 7.5 m urea and 1× Tris-borate-EDTA buffer, pH 7.8) (43 cm) at 300 V for 1.5 hr and exposed to X-ray film. Bands were extracted and reamplified (using the same PCR conditions as in the preamplification reaction for TMD), cloned, and sequenced.
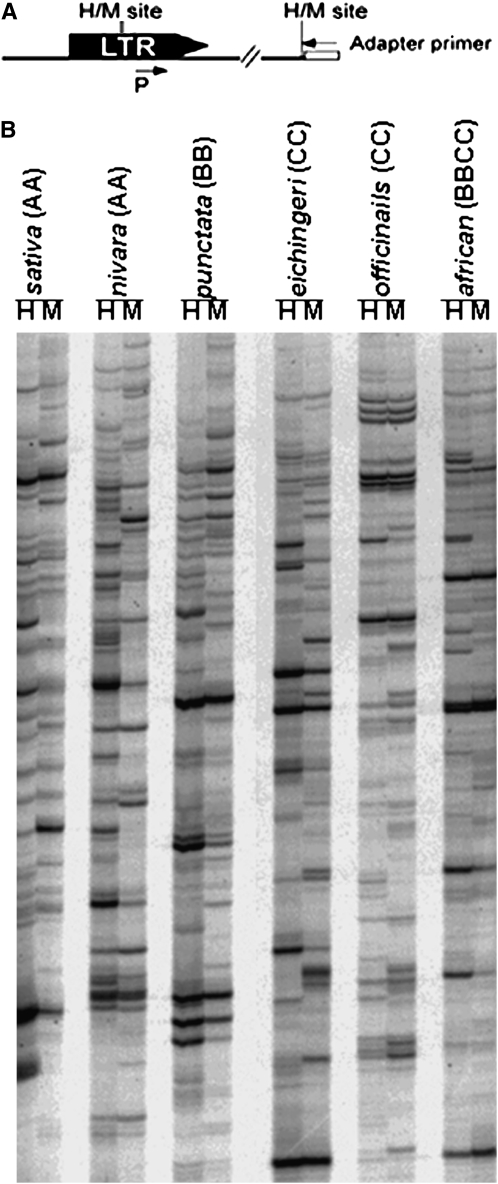
Figure 1.—
Transposon methylation display (TMD) in Oryza species. (A) LTR and flanking sequences are shown together with the cleavage sites of HpaII (H) and MspI (M) and the position of primers used in TMD reactions (see materials and methods). (B) Autoradiograph of transposon methylation display using P and the adapter primer +(AT).
Methylation status of individual LTRs:
Undigested genomic DNA or DNA digested with either HpaII or MspI served as template for PCR using one primer derived from flanking sequence and the other primer derived from Dasheng LTR sequence. Primers were designed to amplify fragments that contained CCGG sites only in the LTR and not in the flanking sequence. PCR products were visualized under UV light following electrophoresis through 1.7% agarose gels containing ethidium bromide. Using this procedure a positive PCR product from DNA digested with HpaII or MspI indicates that the LTR is methylated. Primer sequences are available upon request.
RNA analysis:
Total RNA was extracted from leaves and roots using Trizol Reagent (Invitrogen, Carlsbad, CA) (Chomczynski 1993). First strand cDNA synthesis was carried out as previously described (Sambrook et al. 1989). RT–PCR were carried out as previously described (Kashkush et al. 2002, 2003). A sense gene-specific oligonucleotide was used for the synthesis of the first strand cDNA. This primer can anneal only to an antisense transcript. The resulting cDNA was then used as template for the RT–PCR reaction using the same conditions as previously described by Kashkush et al. (2003). RACE reactions were carried out according to the manufacturer's instructions using the SMART RACE cDNA amplification kit (Clontech, Mountain View, CA). Primer sequences are available upon request.
Computer-assisted analysis:
Annotation of sequences flanking LTRs:
Annotation was done using the BLAST package 2.0 from the NCBI (http://www.ncbi.nlm.nih.gov/BLAST/), Institute for Genomic Research (http://tigrblast.tigr.org/tgi/) and Knowledge-based Oryza Molecular Biology Encyclopedia (http://cdna01.dna.affrc.go.jp/cDNA/).
Promoter prediction:
Public promoter prediction program NNPP version 2.2 (http://www.fruitfly.org/seq_tools/promoter.html) was used to analyze a consensus Dasheng LTR sequence generated from 20 LTRs of full-length Dasheng elements by BioEdit Sequence Alignment Editor software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html).
RESULTS
Methylation status of chimeric (Dasheng/flanking) sequences in diploid and polyploid rice species:
Methylation status of Dasheng loci was investigated by using TMD to detect methylation of chimeric DNA fragments (LTR/host flanking). TMD is a modification of the methylation-sensitive amplified polymorphism (MSAP) and transposon display techniques, which are, in turn, modifications of amplified fragment length polymorphism (AFLP) (Vos et al. 1995; Xiong et al. 1999; Casa et al. 2000; Shaked et al. 2001). TMD allows one to perform a systematic genomewide analysis of the methylation status of CCGG sites in DNA flanking TEs.
Genomic DNA from leaf tissue was first digested with either HpaII or MspI and the resultant restriction fragments were ligated to adapters and then used as template for PCR using primers designed to amplify Dasheng ends and flanking genomic DNA (Figure 1A). The high copy number of Dasheng elements necessitated the addition of two selective bases to the primer complementary to the adapter sequence to reduce the number of bands amplified in each TMD reaction (see materials and methods).
The methylation patterns of chimeric sequences (LTR/host flanking) were determined in six Oryza species (see materials and methods). A gel displaying the products of one TMD adapter primer (of 16 possible primers) is shown in Figure 1B. Each band has at one end Dasheng LTR sequence and, at the other end, an unmethylated HpaII or MspI site in flanking host DNA. A monomorphic band (present in both H and M lanes) in the TMD gel indicates that no methylation was detected at the CCGG site flanking the LTR. A polymorphic band between the H and M lanes indicates the presence of methylation at either internal cytosine (bands present in M lanes only) or at external cytosine (bands present in H lanes only) of the CCGG site flanking the LTR (see materials and methods for more details on the isoschizomers). Use of one TMD primer combination produced 60–80 (with an average of 68.3) clear bands when DNA from each one of the six species was used as template (Table 1). On average, ∼38% of the CCGG sites flanking Dasheng LTRs were methylated in leaf (Table 1). Note that the TMD experiments were repeated twice and identical patterns were seen when the same primer combinations were used (supplemental Figure 1 at http://www.genetics.org/supplemental/).
TABLE 1.
Number of TMD bands amplified using one selective primer combination in the six Oryza species
| Species | Total no. of bandsa | Total no. of methylated sitesb (%) |
|---|---|---|
| sativa ssp. japonica | 68 | 22 (32.3) |
| nivara | 80 | 32 (40.0) |
| punctata | 66 | 28 (42.4) |
| eichingeri | 72 | 28 (38.8) |
| officinails | 60 | 26 (43.0) |
| african | 64 | 20 (31.2) |
| Average | 68.3 | 26 (37.9) |
The primer combination used a Dasheng LTR primer (5′-TTTAGGTCTCGTGCGCTACC-3′) and an adapter primer +AT (see materials and methods). Bands that were monomorphic (identical in both lanes) were counted only once.
Bands were considered methylated if they showed polymorphism between the two isoschizomers.
A subset of 39 TMD bands were recovered from the acrylamide gel, reamplified and sequenced. While all contained Dasheng LTR sequences at one end, the other end contained host DNA flanking the LTR and corresponded to noncoding DNA, known genes, or no hits found in rice databases (supplemental Table 1 at http://www.genetics.org/supplemental/).
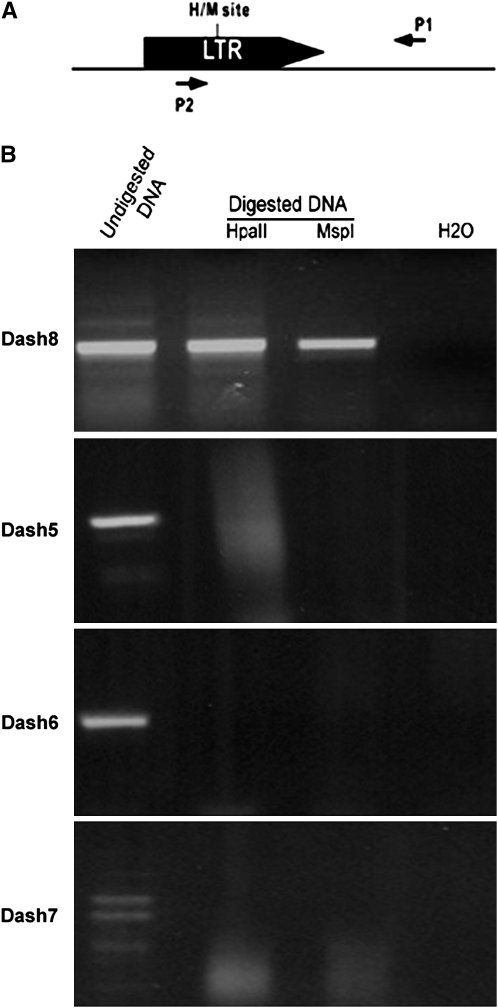
To test the methylation status of CCGG sites in the Dasheng LTRs, primers were generated from the DNA sequence flanking each LTR (P1 in Figure 2A) and used in PCR (undigested genomic DNA or DNA digested with either HpaII or MspI served as template) with a primer generated from the Dasheng LTR sequence upstream of the HpaII and MspI sites (P2 in Figure 2A). The presence of a band in both the HpaII and MspI lanes indicates that the CCGG sites in the LTR sequence are methylated (because the fragment was not digested, see materials and methods). For example, the LTR in Dash8 (supplemental Table 1) is methylated (Figure 2B), while the LTR in Dash5, -6, and -7 (supplemental Table 1) is unmethylated (Figure 2B). For each locus tested, the LTR methylation status was the same as that of the adjacent host sequence.
Figure 2.—
Methylation of CCGG sites in the LTR. (A) LTR and flanking sequences are shown together with the cleavage sites of the restriction enzymes (H, HpaII, M, MspI) in the LTR and the position of primers used in PCR analysis (see materials and methods). (B) PCR analysis (using primers P2 and P1) of LTR methylation in leaves using undigested genomic DNA or DNA digested with either HpaII or MspI as template. Dash8 pattern was used as a control for the digestion quality because the same DNA templates (Oryza sativa, see supplemental Table 1 at http://www.genetics.org/supplemental/) were used for Dash5, -6, -7, and -8. For a negative control, H2O was used as a template in PCR reaction using primers P1 and P2.
Tissue-specific Dasheng LTR methylation:
It is known that methylation patterns change in a tissue-specific manner (Khodosevich et al. 2004; Okahara et al. 2004; Lavie et al. 2005) and that TE methylation status has been shown to alter the methylation of flanking host sequences during development (Turker 2002; Khodosevich et al. 2004). To test the methylation status of Dasheng LTRs, a two-step procedure was devised whereby tissue-specific methylation changes were first detected in host sequences flanking Dasheng LTRs and then these particular LTRs were scrutinized for tissue-specific methylation and readout transcription.
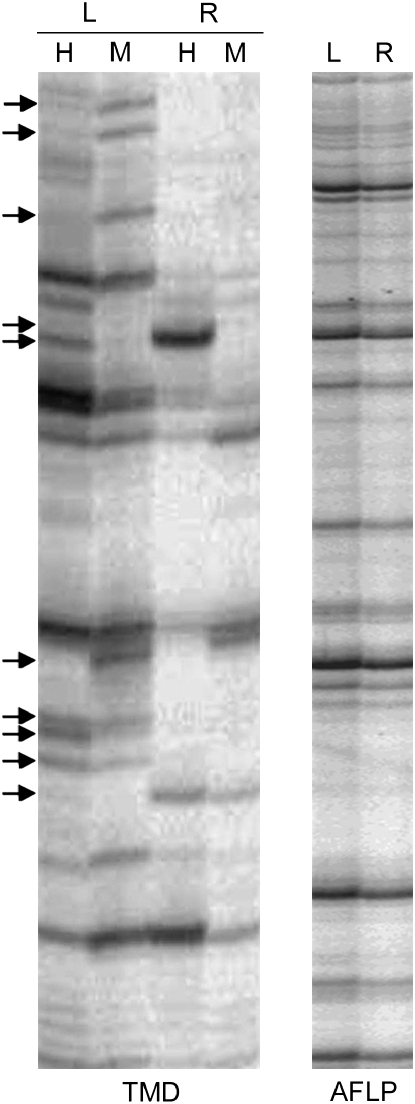
Changes in tissue-specific methylation of host sequences flanking Dasheng LTRs was investigated by using TMD to detect methylation of chimeric DNA fragments (LTR/host flanking) in leaves vs. roots. An example of a gel displaying the products of one TMD adapter primer is shown in Figure 3 . Each band has at one end Dasheng LTR sequence and, at the other end, an unmethylated HpaII or MspI site in flanking host DNA. If, for example, a site is methylated in leaves and not in roots, the TMD gel will have a band in the roots (R) lane but not in the leaves (L) lane. Use of 4 of the 16 possible TMD primer combinations (see materials and methods) led to the detection of 220 and 152 bands, respectively, when japonica or indica genomic DNA was used as template. Of these 372 bands, 78 (42 in japonica and 36 in indica) were tissue-specific (a band is present only in leaves or in roots, see arrows in Figure 3).
Figure 3.—
Transposon methylation display reveals tissue-specific methylation in DNA flanking LTRs in leaves (L) and roots (R). Autoradiograph of transposon methylation display using P and the adapter primer +(TT) (see Figure 1A). The arrows indicate sites that show tissue-specific methylation. A standard AFLP [primers reaction (using MseI and EcoRI restriction enzymes] was used for DNA quality control in leaves and roots (Vos et al. 1995).
A subset of 34 of these tissue-specific bands was chosen at random, recovered from the gel, reamplified, and sequenced (Table 2). While all contained Dasheng LTR sequences at one end, the other end contained host DNA (with ∼100% identity in sequence alignment) flanking the LTR and corresponded to (1) ESTs and predicted genes (15 fragments), (2) TEs (4 fragments), (3) genomic DNA fragments with no similarity to ESTs or predicted genes (2 fragments), and (4) sequences with no hits in the database (13 fragments) (Table 2). Some of the bands were detected in only one of the two subspecies (ND in Table 2), in part due to polymorphism in Dasheng insertion sites in japonica vs. indica. An example of this is the HAK4 gene (Dash14) where Dasheng has inserted adjacent to HAK4 in japonica but not in indica as was seen after comparing the two sequence drafts of the two subspecies. The TMD bands with no hits in the rice databases were detected in the japonica and/or indica genomes by PCR amplification (supplemental Figure 2 at http://www.genetics.org/supplemental/) thus confirming that they are unsequenced genomic regions and not experimental artifacts.
TABLE 2.
Characterization of TMD fragments
| Methylation statusb
|
||||||
|---|---|---|---|---|---|---|
| Subspecies distributiona |
japonica
|
indica
|
Database hits of flanking sequences (accession no.)c | |||
| Clone | Leaf | Root | Leaf | Root | ||
| Dash1 | j, i | − | + | − | + | EST (A078092) |
| Dash2 | ND | − | + | ND | ND | EST (TC229808) |
| Dash3 | j, i | − | + | − | + | Dasheng sequence |
| Dash4 | j, i | − | + | − | + | Dasheng sequence |
| Dash5 | j, i | − | + | − | + | No hits |
| Dash6 | j, i | − | + | − | + | No hits |
| Dash7 | ND | − | + | ND | ND | No hits |
| Dash8 | j, i | − | + | − | + | No hits |
| Dash9 | ND | − | + | ND | ND | No hits |
| Dash10 | j, i | + | − | + | − | Ribosomal protein L32 (XP_372959) |
| Dash11 | j, i | + | − | + | − | Cryptochrome 1b CRY1b (BAB70688) |
| Dash12 | j, i | + | − | + | − | Non-LTR sequence of putative retroelement (NP809988) |
| Dash13 | j, i | + | − | ND | ND | Putative transposase (NP810207) |
| Dash14 | j | − | + | ND | ND | Potassium transporter HAK4 (AAF36497.1) |
| Dash15 | j, i | + | − | + | − | Jasmonate inducible protein (AF395880) |
| Dash16 | j, i | + | − | + | − | Transcription factor IID (AAF65405) |
| Dash17 | j, i | + | − | + | − | No hits |
| Dash18 | ND | + | − | ND | ND | No hits |
| Dash19 | ND | + | − | ND | ND | No hits |
| Dash20 | ND | + | − | ND | ND | No hits |
| Dash21 | ND | ND | ND | − | + | No hits |
| Dash22 | ND | ND | ND | − | + | No hits |
| Dash23 | j, i | ND | ND | − | + | NADH-ubiquinone oxidoreductase (Q96068) |
| Dash24 | j, i | ND | ND | + | − | Unknown protein (NP_912736) |
| Dash25 | j, i | − | + | − | + | RING finger protein GEG (CAB45241) |
| Dash26 | j, i | − | + | − | + | Peptide transfer (AP003621) |
| Dash27 | j, i | ND | ND | + | − | Phosphoenolpyruvate carboxylase (CAA11414) |
| Dash28 | j, i | − | + | − | + | EST (TC115648) |
| Dash29 | j, i | − | + | − | + | EST (TC215714) |
| Dash30 | j, i | + | − | + | − | EST (TC115647) |
| Dash31 | j, i | + | − | + | − | Genomic DNA (OSJNBa0017B18) |
| Dash32 | j, i | + | − | + | − | Genomic DNA (OSJNBa0030C11) |
| Dash33 | j, i | − | + | − | + | No hits |
| Dash34 | ND | + | − | ND | ND | No hits |
j, i, Dasheng inserted in both subspecies; j or i, Dasheng inserted in japonica or in indica. ND, could not be detected because the sequence is not available in one of the sequence drafts (see text).
(−), unmethylated site; (+), methylated site. ND, not detected in one of the two subspecies.
No significant sequence hits in databases at e-value < e−10.
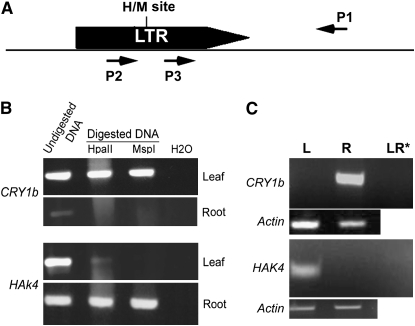
To test the methylation status of CCGG sites in the Dasheng LTRs that are adjacent to the 15 ESTs and predicted genes, primers were generated from the DNA sequence flanking each LTR (P1 in Figure 4A) and used in PCR (undigested genomic DNA or DNA digested with either HpaII or MspI served as template) with a primer generated from the Dasheng LTR sequence upstream of the HpaII and MspI sites (P2 in Figure 4A). The presence of a band in both the HpaII and MspI lanes indicates that the CCGG sites in the LTR sequence are methylated (because the fragment was not digested, see materials and methods). For example, the LTR in Dash11 (LTR/CRY1b) is methylated in leaves and unmethylated in roots in both japonica (Figure 4B) and indica (data not shown), while the LTR adjacent to HAK4 (Dash14) is methylated in roots but not in japonica leaves (Figure 4B). The HAK4 gene is unmethylated in both leaves and roots of indica (supplemental Figure 3 at http://www.genetics.org/supplemental/), where no Dasheng insertion close to the gene was identified.
Figure 4.—
LTR methylation at loci showed tissue-specific chimeric transcripts. (A) LTR and flanking sequences are shown together with the cleavage sites of the restriction enzymes (H, HpaII; M, MspI) in the LTR and the position of primers used in PCR analysis (see materials and methods). (B) PCR analysis (using primers P2 and P1) of LTR methylation in leaves (L) and roots (R) using undigested genomic DNA or DNA digested with either HpaII or MspI as template. The presence of a band in both the HpaII and MspI lanes indicates that the CCGG sites in the LTR sequence are methylated (because the fragment was not digested, see materials and methods). (C) RT–PCR analysis of chimeric transcripts in leaves and roots of japonica using a primer from the LTR (P3) and a second primer derived from the flanking sequence (P1). LR* is a negative control (RNA templates treated with RNase) and actin was used as a control for RNA quality.
Expression analysis of flanking rice genes:
To test whether the methylation status of Dasheng LTRs is associated with the expression of adjacent genes, we identified Dasheng elements located near or into genes using the publicly available rice sequence draft. Over 100 of the sequence-flanked Dasheng elements were annotated as genes (supplemental Table 2 at http://www.genetics.org/supplemental/). Flanking genes represent a broad spectrum of functional classes including products potentially involved in metabolism, disease resistance, and cell cycle regulation (Table 3). About half of these insertions were downstream of genes, 30% were upstream, and 20% were in introns. Eleven loci from the computer output of genomic insertion sites were selected for further analysis.
TABLE 3.
Genes flanking 28 Dasheng LTRs from in silico analysis
| Gene or protein product | Protein database accession no. | cDNA clone database accession no. | Intact element/solo LTR | Insertion sites and position of Dashenga | Subspecies distributionb |
|---|---|---|---|---|---|
| Amino acid transporter | XP_467580.1 | AK069154 | Intact | 3′-UTR | Both |
| NBS-LRR resistance protein | NP_911138.1 | AK103061 | Intact | 3′-UTR | Both |
| RNA helicase | XP_466992.1 | AK066504 | Solo | 811 bp, upstream | Both |
| UDP-glucose glucosyl-transferase | XP_450076.1 | AK059022 | Solo | 663 bp, downstream | Both |
| Plasmodium vivax circumsporozoite protein | Q9FTX1 | TC215254 | Intact | 765 bp, upstream | Both |
| LeOPT1-oligopeptide transporter | UP|Q94LI9 | TC236771 | Intact | 3′-UTR | Both |
| Cryptochrome 1b-CRY1b | BAB70688 | AB073547 | Solo | Intron | Both |
| rRNA methylase | XP_475694.1 | XM_475694 | Solo | Intron | Both |
| Phosphoenolpyruvate carboxylase | CAA11414 | CB642292 | Intact | 3′-UTR | Both |
| Cytochrome P450 monooxygenase | CAA06156.1 | AK110009 | Intact | 3′-UTR | japonica |
| Potassium transporter-HAK4 | AAF36497.1 | AK119564 | Intact | 3′-UTR | japonica |
| Protein kinase (ADK1) | XP_466811.1 | AK101388 | Intact | 198 bp, upstream | japonica |
| Auxin response-like protein | BAD45739 | TC236741 | Intact | 700 bp, downstream | japonica |
| Deoxynucleoside kinase | NP_565032.2 | AK063257 | Intact | 418 bp, upstream | japonica |
| Serine/threonine kinase | XP_462758.1 | AK072359 | Intact | 3′-UTR | japonica |
| Isopentenyl-diphosphate | AAT94033.1 | AK060336 | Intact | 757 bp, downstream | japonica |
| Cell division protein | Q39102 | TC234557 | Intact | 100 bp, downstream | japonica |
| Pyrophosphate-dependent phosphofructokinase α subunit | GP_29647436 | TC149299 | Intact | 100 bp, downstream | japonica |
| Phosphoribosylglycinamide formyltransferase | BAD08899 | TC221784 | Intact | 715 bp, downstream | japonica |
| Splicing factor U2AF | Q8GVW8 | TC223844 | Intact | 3′-UTR | japonica |
| Peptide transporter | AP003621 | AAT77837.1 | Intact | 763 bp, downstream | japonica |
| Male fertility protein | XP_478624.1 | XM_478624 | Intact | 3′-UTR | japonica |
| Cell wall invertase | AAR07091 | AY387483 | Solo | Intron | japonica |
| Sesquiterpene cyclase | XP_465466 | AK070116 | Solo | Intron | japonica |
| 2-dehydro-3-deoxyphosphooctonat aldolase | AAO72599.1 | AK072548 | Solo | 874 bp, downstream | indica |
| RPR1h | XP_481094.1 | AK100590 | Solo | Inton | indica |
| Homeobox protein related | NP937617 | AC079736.12 | Solo | 170 bp, upstream | indica |
| Early nodulin 75 precursor-like protein | GP|22775647 | CA755439 | Solo | 3′-UTR | indica |
Position of Dasheng element or solo LTRs relative to the gene.
Presence of Dasheng element in japonica vs. indica.
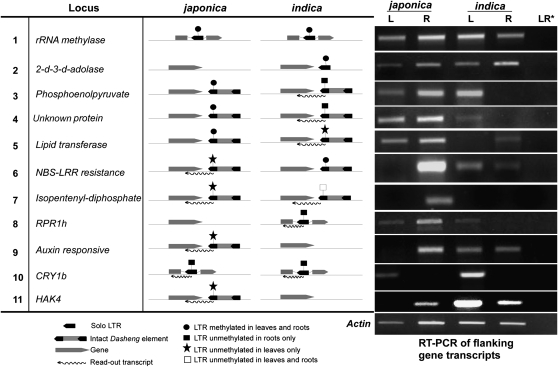
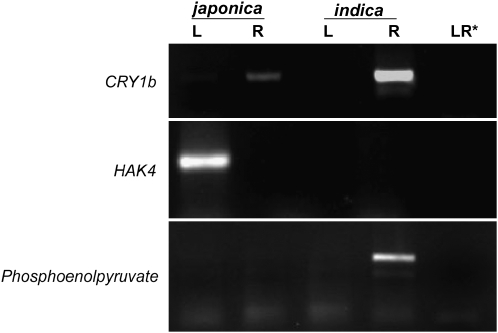
The methylation status of each LTR and the chimeric transcription (LTR/flanking) were determined. Transcripts were detected in the tissue where the LTRs were shown to be unmethylated. For example, the LTR in Dash11 (LTR/CRY1b), which is unmethylated in roots and methylated in leaves (Table 2, Figure 4B), is transcribed in roots but not in leaves (Figure 4C). Similarly, for Dash14 (LTR/HAK4) transcripts were detected in the tissue where the adjacent LTR is unmethylated (Table 2, Figure 4, B and C). The methylation status and chimeric transcripts in leaves vs. roots of each LTR are presented in Figure 5 (middle). The chimeric transcripts were initiated from the LTR as seen by 5′-RACE analysis (data not shown) for cases 4, 10, and 11 (see Figure 5). In addition, transcripts from flanking genes were analyzed by RT–PCR (qualitatively, presence or absence of a sense transcript, after 35 PCR cycles) using gene-specific primers in conjunction with cDNA synthesized from RNA isolated from the leaves (L) and roots (R) of japonica and indica rice (Figure 5, right).
Figure 5.—
Expression analysis of putative genes adjacent to Dasheng LTRs in leaves (L) and roots (R) of japonica and indica rice tissue. The relative position of Dasheng and flanking genes (including exons and introns) in japonica and/or indica is shown schematically. The LTR methylation status and the readout transcription (in the tissue where the LTR is unmethylated) are indicated (see key at the bottom). (Right) RT–PCR products (gene sense transcripts) after using gene specific primers are shown. LR* and actin are controls (see Figure 4 legend).
A consistent picture emerges from these data regarding the presence or absence of an LTR and its methylation status and the presence or absence of chimeric transcripts and flanking gene transcripts. For example, in both subspecies transcription of the rRNA methylase gene in the leaves and roots correlated with the absence of transcriptional readout from the methylated LTR in the intron. Similarly, a polymorphic LTR insertion that is methylated has no obvious impact on flanking gene expression (Figure 5, case 2). In these two cases, and perhaps in many others, the presence of a methylated LTR has no apparent impact on flanking gene expression. In contrast, for 9 of the 11 loci analyzed, tissue-specific methylation correlates with tissue-specific readout transcription and with tissue-specific expression of the flanking gene (Figure 5, cases 3–11). For example, detection of CRY1b transcripts in leaves but not in roots of both subspecies correlates with methylation of the intronic LTR in leaves and the detection of readout transcripts only in roots. Reduced tissue-specific methylation correlates inversely with expression of adjacent genes. Note that the absence of flanking gene transcripts in roots or leaves of japonica or indica was not due to cytosine methylation of the gene (supplemental Figure 4 at http://www.genetics.org/supplemental/).
Subspecies-specific patterns of gene expression were observed, as expected, for several polymorphic insertions. In three instances (Figure 5, cases 8, 9, and 11), rice gene transcripts were detected in leaves and roots in the subspecies without the insertion. However, in the subspecies with the insertion, flanking gene transcripts were only detected in the tissue where the LTR was methylated. For example, HAK4 transcripts were found in leaves and roots of indica rice which lacks the HAK4 insertion (Figure 5, case 11), but HAK4 transcripts were not detected in japonica leaves where the unmethylated Dasheng LTR in the 3′-UTR promotes readout transcription. In addition, in five instances (Figure 5, cases 3–7), subspecies differences in the expression of flanking genes were not due to polymorphic insertions but to subspecies-specific LTR methylation. For example, while Dasheng is located in the 3′-UTR of a gene encoding phosphoenolpyruvate in both japonica and indica (Figure 5, case 3), the 3′ LTR only promotes readout transcription in indica roots where the LTR is unmethylated.
Antisense transcription detection:
From Figure 5 it can be seen that genes showing tissue-specific expression were in opposite orientation relative to LTR readout transcription. The consistency of this result suggests that the inability to detect flanking gene transcripts was due to the formation of dsRNA between the readout and the gene transcript and the subsequent degradation of gene transcripts. This possibility was explored by analyzing three representative genes that showed tissue-specific expression (Figure 5, cases 3, 10, and 11). Antisense strands were detected for each gene by RT–PCR using sense primers to synthesize antisense cDNA from leaves (L) and roots (R) of japonica and indica (Figure 6). In all three cases, the antisense transcripts were detected in the tissue where sense transcripts from that gene were not detected.
Figure 6.—
Detection of antisense transcripts. RT–PCR analysis of antisense transcripts in leaves (L) and roots (R) of japonica and indica using gene specific primers. LR* is a negative control (RNA templates treated with RNase).
DISCUSSION
In this study, we pinpointed methylated LTRs and detected tissue-specific LTR methylation, and then we tested the impact of readout transcription from unmethylated LTRs on the expression of adjacent genes. Dasheng was the focus of this study because of its young age, high copy number, and the high level of polymorphism among Oryza species (Jiang et al. 2002b). Dasheng elements are classified as nonautonomous LTR retrotransposons because they do not encode any of the proteins necessary for retrotransposition (Jiang et al. 2002b). The transposition intermediate for both autonomous and nonautonomous LTR retrotransposons is a transcript that initiates from a promoter in the 5′ LTR and terminates in the 3′ LTR. Note that if active, the promoter in the 3′ LTR (or in a solo LTR) could initiate transcription into flanking host DNA. A promoter in the Dasheng LTR was predicted with a high degree of certainty by employing a consensus Dasheng LTR as the input sequence for a promoter identification program (supplemental Figure 5 at http://www.genetics.org/supplemental/). We hypothesized that unmethylated LTRs adjacent to host genes might impact host gene expression. We searched for and found evidence of Dasheng LTR-promoted transcripts among the rice ESTs and full-length cDNA collections (our unpublished data). The EST and cDNA libraries from which the Dasheng transcripts were identified were prepared from tissues of rice plants grown in normal conditions, suggesting that Dasheng LTRs do not need stress conditions to be transcribed. Several studies show that LTRs of retroelements in both human and plant genomes have retained their promoter activity (Domansky et al. 2000; Kashkush et al. 2003; Druker et al. 2004; Mack et al. 2004; Okahara et al. 2004; Steinhoff and Schulz 2004; Lavie et al. 2005). Promoters of HERVs LTRs were shown to be activated in various human tissues during development, and that this promoter activity is correlated with the LTR methylation (Okahara et al. 2004; Lavie et al. 2005).
Dasheng methylation:
The methylation patterns of Dasheng LTRs and flanking host sequences were undertaken using a novel technique called TMD. The technique permits the analysis of hundreds of TE loci in an unbiased manner in a relatively short period of time and requires small amounts of DNA. Band recovery from the TMD gel is almost possible for all visible bands. The technique can easily be applied to any mulicopy TE family including TEs in organisms with limited genomic sequence information. One limitation is that DNA methylation might vary from cell to cell in the same tissue and can cause the production of different band intensities between different tissues in TMD gel because TMD is a PCR-based technique. Hence, our analysis was done in a qualitative manner (polymorphic vs. monomorphic bands). Here, TMD was successfully applied in leaves of various Oryza species to determine the methylation status of chimeric sequences (Dasheng/flanking) (Figure 1). We show that Dasheng methylation can change during plant development because part of the LTRs was tissue specific (see Figures 3 and 4).
Impact of Dasheng insertion on gene expression:
This study provides several instances of correlation between tissue-specific LTR methylation and tissue-specific expression of adjacent genes (Figure 5). Although this work is based on correlative data, it suggests that tissue-specific expression of some rice genes might be regulated by adjacent TEs. Alteration of gene expression as a result of the outward promoter activity of adjacent TE elements was reported in mouse for the insertion of intracisternal A-particle (IAP) elements in the A (agouti) locus (Michaud et al. 1994) and in Drosophila for the insertion of Foldback-like transposon Kepler in the CG13617 locus (Puig et al. 2004). Changes in host gene expression during development as a result of readout transcription from an adjacent TE was first reported for the mutant hcf106 allele, which contains an insertion of the Mutator element near the start of transcription (Martienssen et al. 1990). Correlation between transposon readout activity (via methylation–demethylation) and gene expression was never shown on a genomewide scale. The genomewide readout transcription activity that was reported for Wis2-1A in newly synthesized allotetraploid wheat (Kashkush et al. 2003) is potentially associated with methylation alteration of the element because methylation patterns of DNA sequences including TEs were shown to alter as a result of polyploidization (Shaked et al. 2001; Madlung et al. 2002).
Subspecies-specific expression:
The availability of sequencing drafts for japonica and indica allowed for the analysis of polymorphic insertion sites and for how this phenomenon contributed to gene expression differences. Subspecies-specific expression was observed for 8 of the 11 genes tested (Figure 5). The source of the subspecies-specific expression was correlated with (1) polymorphic insertions of Dasheng, where the readout transcript from an unmethylated LTR in japonica or in indica correlated with the expression of the gene in a tissue-specific manner (Figure 5, cases 8, 9, and 11), and (2) subspecies-specific LTR methylation (Figure 5, cases 3–7). Our data suggest that TEs can be responsible in part for the gene expression differences between subspecies and because of this they increase the diversity between different subspecies.
In summary, the increasing availability of genomic sequences has permitted a better understanding of the relationship between hosts and their TEs. In this study, we focused on the impact of one young retrotransposon family that was recently amplified in the rice genome to reach ∼1000 copies. Here, we showed that this family itself has the potential to impact the expression of hundreds of rice genes. Availability of methylation-defective mutants in rice will be very useful to better understand the role of TEs on host gene regulation and to better understand the epigenetic regulation of TEs. Another question that can be addressed in future studies is the association, if any, between changes in gene expression caused by the readout transcripts from Dasheng and phenotype. Our study highlights some of the possible evolutionary roles of TEs on regulation of host genes during development and TEs contribution to the diversity in gene expression in natural populations as a major source of genetic variation.
Acknowledgments
The authors would like to thank Susan Wessler, Moshe Feldman, and Avi Levy for their support and helpful discussions. This work was supported by a Ben-Gurion University startup grant, by the European Molecular Biology Organization, and by the Vaadia-Binational Agricultural Research and Development Fund (for K.K).
References
- Bennetzen, J. L., and E. A. Kellogg, 1997. Do plants have a one-way ticket to genomic obesity? Plant Cell 9: 1509–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J. L., K. Schrick, P. S. Springer, W. E. Brown and P. Sanmiguel, 1994. Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome 37: 565–576. [DOI] [PubMed] [Google Scholar]
- Casa, A. M., C. Brouwer, A. Nagel, L. J. Wang, Q. Zhang et al., 2000. The MITE family Heartbreaker (Hbr): molecular markers in maize. Proc. Natl. Acad. Sci. USA 97: 10083–10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski, P., 1993. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 15: 532. [PubMed] [Google Scholar]
- Domansky, A. N., E. P. Kopantzev, E. V. Snezhkov, Y. B. Lebedev, C. Leib-Mosch et al., 2000. Solitary HERV-K LTRs possess bi-directional promoter activity and contain a negative regulatory element in the U5 region. FEBS Lett. 472: 191–195. [DOI] [PubMed] [Google Scholar]
- Druker, R., T. J. Bruxner, N. J. Lehrbach and E. Whitelaw, 2004. Complex patterns of transcription at the insertion site of a retrotransposon in the mouse. Nucleic Acids Res. 32: 5800–5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte, C., N. Jiang and S. R. Wessler, 2002. Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 3: 329–341. [DOI] [PubMed] [Google Scholar]
- Finnegan, D. J., 1989. Eukaryotic transposable elements and genome evolution. Trends Genet. 5: 103–107. [DOI] [PubMed] [Google Scholar]
- Flavell, R. B., 1994. Inactivation of gene-expression in plants as a consequence of specific sequence duplication. Proc. Natl. Acad. Sci. USA 91: 3490–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch, R., and A. Bird, 2003. Epigenetic regulation of gene ex pression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33: 245–254. [DOI] [PubMed] [Google Scholar]
- Jiang, N., Z. Bao, S. Temnykh, Z. Cheng, J. Jiang et al., 2002. a Dasheng: a recently amplified nonautonomous long terminal repeat element that is a major component of pericentromeric regions in rice. Genetics 161: 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, N., I. K. Jordan and S. R. Wessler, 2002. b Dasheng and RIRE2. A nonautonomous long terminal repeat element and its putative autonomous partner in the rice genome. Plant Physiol. 130: 1697–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush, K., M. Feldman and A. A. Levy, 2002. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160: 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush, K., M. Feldman and A. A. Levy, 2003. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33: 102–106. [DOI] [PubMed] [Google Scholar]
- Khodosevich, K., Y. Lebedev and E. D. Sverdlov, 2004. Large-scale determination of the methylation status of retrotransposons in different tissues using a methylation tags approach. Nucleic Acids Res. 32: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A., and J. L. Bennetzen, 1999. Plant retrotransposons. Annu. Rev. Genet. 33: 479–532. [DOI] [PubMed] [Google Scholar]
- Lavie, L., M. Kitova, E. Maldener, E. Meese and J. Mayer, 2005. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2). J. Virol. 79: 876–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack, M., K. Bender and P. M. Schneider, 2004. Detection of retroviral antisense transcripts and promoter activity of the HERV-K(C4) insertion in the MHC class III region. Immunogenetics 56: 321–332. [DOI] [PubMed] [Google Scholar]
- Madlung, A., R. W. Masuelli, B. Watson, S. H. Reynolds, J. Davison et al., 2002. Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 129: 733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen, R., 1998. Transposons, DNA methylation and gene control. Trends Genet. 14: 263–264. [DOI] [PubMed] [Google Scholar]
- Martienssen, R., A. Barkan, W. C. Taylor and M. Freeling, 1990. Somatically heritable switches in the DNA modification of Mu transposable elements monitored with a suppressible mutant in maize. Genes Dev. 4: 331–343. [DOI] [PubMed] [Google Scholar]
- Michaud, E. J., M. J. van Vugt, S. J. Bultman, H. O. Sweet, M. T. Davisson et al., 1994. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev. 8: 1463–1472. [DOI] [PubMed] [Google Scholar]
- Miura, A., S. Yonebayashi, K. Watanabe, T. Toyama, H. Shimada et al., 2001. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411: 212–214. [DOI] [PubMed] [Google Scholar]
- Okahara, G., S. Matsubara, T. Oda, J. Sugimoto, Y. Jinno et al., 2004. Expression analyses of human endogenous retroviruses (HERVs): tissue-specific and developmental stage-dependent expression of HERVs. Genomics 84: 982–990. [DOI] [PubMed] [Google Scholar]
- Puig, M., M. Caceres and A. Ruiz, 2004. Silencing of a gene adjacent to the breakpoint of a widespread Drosophila inversion by a transposon-induced antisense RNA. Proc. Natl. Acad. Sci. USA 101: 9013–9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz, P. D., L. E. Palmer, B. P. May, M. T. Hemann, S. W. Lowe et al., 2003. Genes and transposons are differentially methylated in plants, but not in mammals. Genome Res. 13: 2658–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shaked, H., K. Kashkush, H. Ozkan, M. Feldman and A. A. Levy, 2001. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13: 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, T., C. Yordan and R. A. Martienssen, 2001. Robertson's mutator transposons in A-thaliana are regulated by the chromatin-remodeling gene decrease in DNA methylation (DDM1). Genes Dev. 15: 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhoff, C., and W. A. Schulz, 2004. Transcriptional regulation of the human LINE-1 retrotransposon L1.2B. Mol. Genet. Genomics 270: 394–402. [DOI] [PubMed] [Google Scholar]
- Turker, M. S., 2002. Gene silencing in mammalian cells and the spread of DNA methylation. Oncogene 21: 5388–5393. [DOI] [PubMed] [Google Scholar]
- Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. Vandelee et al., 1995. AFLP - a new technique for DNA-fingerprinting. Nucleic Acids Res. 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L. Z., C. G. Xu, M. A. S. Maroof and Q. F. Zhang, 1999. Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol. Gen. Genet. 261: 439–446. [DOI] [PubMed] [Google Scholar]
- Zilberman, D., and S. Henikoff, 2004. Silencing of transposons in plant genomes: kick them when they're down. Genome Biol. 5: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]