Abstract
Despite its status as one of the world's major crops, linkage disequilibrium (LD) patterns have not been systematically characterized across the genome of Asian rice (Oryza sativa). Such information is critical to fully exploit the genome sequence for mapping complex traits using association techniques. Here we characterize LD in five 500-kb regions of the rice genome in three major cultivated rice varieties (indica, tropical japonica, and temperate japonica) and in the wild ancestor of Asian rice, Oryza rufipogon. Using unlinked SNPs to determine the amount of background linkage disequilibrium in each population, we find that the extent of LD is greatest in temperate japonica (probably >500 kb), followed by tropical japonica (∼150 kb) and indica (∼75 kb). LD extends over a shorter distance in O. rufipogon (≪40 kb) than in any of the O. sativa groups assayed here. The differences in the extent of LD among these groups are consistent with differences in outcrossing and recombination rate estimates. As well as heterogeneity between groups, our results suggest variation in LD patterns among genomic regions. We demonstrate the feasibility of genomewide association mapping in cultivated Asian rice using a modest number of SNPs.
DOMESTICATED Asian rice, Oryza sativa, is the world's the most widely cultivated crop species, serving as the staple for more than half of the world's population (Yu et al. 2002). Oryza sativa was domesticated during the Neolithic ∼10,000 years ago from Oryza rufipogon Griff, with at least two centers of domestication—one in the lower Yangtze Valley of China that gave rise to the japonica variety group and the other possibly in South Asia that gave rise to the indica variety group (Khush 1997; Crawford and Chen 1998; Garris et al. 2005). The temperate japonica variety group, which is grown widely in Japan, Korea, and northeast China, is closely related to the tropical japonica subpopulation that is grown on hillsides throughout Southeast Asia (Khush 1997; Garris et al. 2005). Domestication in rice also resulted in a change in the breeding system; O. sativa is autogamous, outcrossing at a rate of ∼1–2%, while O. rufipogon outcrosses at a much higher rate (∼7–56%) (Oka 1988; Gao et al. 2007).
The importance of rice as a major world crop has engendered interest in determining the levels and patterns of variation in its genome. Single nucleotide polymorphism (SNP) levels are generally low in the O. sativa and O. rufipogon genomes, with mean silent Watterson's θ levels of 2.92 and 5.42/kb for rice and its wild ancestor, respectively (Caicedo et al. 2007). SNP levels are highest in indica (θW = 2.21/kb) and lowest in temperate japonica (θW = 0.948/kb), with tropical japonica having intermediate polymorphism levels (θW = 1.70/kb). There appears to be an excess of high-frequency-derived SNPs in O. sativa but not in O. rufipogon, and modeling suggests that the patterns of polymorphism across the rice genome are best explained either by a bottleneck and strong selection on various domestication traits or by more complex demographic models that include bottlenecks, population subdivision, and migration (Caicedo et al. 2007).
Although the levels and patterns of SNPs in rice have begun to be characterized, there is still little information on linkage disequilibrium (LD) in this crop species and its wild relative. There has been intense interest in characterizing LD levels and patterns in this species, both to infer evolutionary forces (Rakshit et al. 2007) and to exploit this information for gene discovery (Garris et al. 2003). Several forces—including mutation, drift, population bottlenecks, population substructure, population admixture, levels of inbreeding, and selection—contribute to the emergence and maintenance of LD. The extent of LD is also dependent on the effective recombination rate, since LD between two loci is degraded by crossover between genes. In a population of constant size, if recombination rates do not vary across the genome, a strong correlation is expected between interlocus distance and LD, and loci in close proximity will remain in disequilibrium for longer periods than those located farther apart (Hartl and Clark 1997).
Patterns of linkage disequilibrium have been characterized in several crop species and their relatives. In maize (Zea mays ssp. mays), r2 decays within 0.3–2 kb, and this rapid decay may be due to this species being outcrossing (Remington et al. 2001; Tenaillon et al. 2001). In the related species sorghum (Sorghum bicolor), r2 >0.1 is observed up to 15–20 kb (Hamblin et al. 2005). High levels of marker association (r2 > 0.1) across a 212-kb region are observed in cultivated, elite varieties of the selfing crop barley (Hordeum vulgare ssp. vulgare), while in landrace accessions, high LD levels persist to ∼90 kb (Caldwell et al. 2006). LD in wild barley (H. vulgare ssp. spontaneum), a highly selfing species, decays intragenically, with a range of only a few kilobases (Morrell et al. 2005). Linkage disequilibrium in soybean (Glycine max), which is also a selfer, can extend from 90 kb to > 500 kb in landrace material, although the level is dependent on the population sample (Hyten et al. 2007).
There has been no large-scale assessment of LD in O. sativa, although the first study in rice found an LD decay of ∼100 kb around a disease resistance locus in the aus subpopulation (Garris et al. 2003) and a more recent study reported an LD decay of ∼50 kb in indica and of ∼5 kb in O. rufipogon (Rakshit et al. 2007). Here we determine the extent of LD in three major subpopulations of domesticated Asian rice (indica, tropical japonica, and temperate japonica) and its wild relative O. rufipogon. Using data from five ∼500-kb genomic regions in the rice genome, we are able to demonstrate differing extents of LD in these cultivated and wild groups and determine that a modest number of SNP markers can provide genomewide coverage for association studies.
MATERIALS AND METHODS
Plant material:
We used a sample set composed of 82 rice accessions, including 60 O. sativa strains and 21 cultivars of O. rufipogon collected in the wild (supplemental Table S1 at http://www.genetics.org/supplemental/) and used in previous studies of SNP and SSR variation (Garris et al. 2005; Olsen et al. 2006; Caicedo et al. 2007). We also included in our analysis the sequence from the whole-genome sequence of Nipponbare, which is a temperate japonica variety (International Rice Genome Sequencing Project 2005). The O. sativa accessions were partitioned into 21 indica, 18 tropical japonica, and 22 temperate japonica strains. These accessions are mainly landraces, but 5 are modern cultivars. We analyzed each O. sativa group (indica, tropical japonica, and temperate japonica) and O. rufipogon separately.
Fragments sequenced:
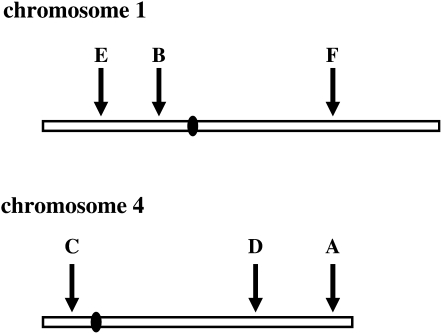
Variation was assessed in six genomic regions of ∼500 kb from chromosomes 1 and 4 (Figure 1), which were the first two rice chromosomes to be completely sequenced (Feng et al. 2002; Sasaki et al. 2002). These regions are designated A–F (Table 1). In each genomic region, a focal gene in the middle of the region was completely sequenced (including ∼1.5 kb of upstream sequence and 1.0 kb of downstream sequence). Twelve ∼500- to 600-bp gene fragments on both sides of these focal genes and spaced ∼40 kb apart were sequenced to provide coverage across the 500-kb genomic region (Figure 2; supplemental Table S2 at http://www.genetics.org/supplemental/). Our analysis of genomewide background LD used 111 sequence-tagged site (STS) gene fragments randomly distributed throughout the genome as reported in a previous study (Caicedo et al. 2007).
Figure 1.—
Relative position of targeted genomic regions in chromosomes 1 and 4.
TABLE 1.
Genomic regions used in the study
| Genomic region | Chromosome | Chromosomal position | Focal gene |
|---|---|---|---|
| A | 4 | 33,623,004–34,103,236 | Os04g57210: actin-6, putative, expressed |
| B | 1 | 10,262,474–10,746,999 | Os01g18640: isopenicillin N epimerase, putative, expressed |
| C | 4 | 652,837–1,138,768 | Os04g02510: nucleic-acid-binding protein, putative, expressed |
| D | 4 | 24,924,845–25,412,071 | Os04g42900: expressed protein |
| E | 1 | 4,500,001–4,980,708 | Os01g09320: NADP-dependent malic enzyme, chloroplast, precursor, putative, expressed |
| F | 1 | 35,031,158–35,503,032 | Os01g60410: ubiquitin-conjugating enzyme E2-17 kDa, putative, expressed |
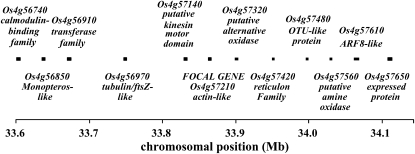
Figure 2.—
Map of targeted genomic region A on chromosome 4. Relative positions of the sequenced genes/gene fragments in the region are indicated. The sizes of the genes are not to scale.
Primers for amplification and sequencing across the focal gene and in the flanking gene fragments were designed using Primer 3 (http://frodo.wi.mit.edu/) (Rozen and Skaletsky 2000) from the Nipponbare sequence at Gramene (http://www.gramene.org); whenever possible, primers for flanking gene fragments were positioned in exons to span an intron. All PCR primers were searched against the rice genome sequence in Gramene using BLAST analysis to ensure that multiple regions would not be amplified and genes annotated as transposable elements were not selected (however, see discussion of the C region in the results and discussion). Amplification and sequencing reactions were conducted by Cogenics (Morrisville, NC) as previously described (Olsen et al. 2006; Caicedo et al. 2007).
Analysis of nucleotide variation:
The population mutation parameter θW (Watterson 1975) was calculated for silent sites within each gene or gene fragment. Each O. sativa accession and a Malaysian O. rufipogon accession (RA2747) were considered to contribute a single haplotype because heterozygotes are rare in O. sativa and both the O. sativa and the Malaysian O. rufipogon accessions had been selfed for several generations. The remaining O. rufipogon accessions contributed two haplotypes to the θW estimate.
LD analysis:
For the analysis of LD, only biallelic SNPs of at least 10% frequency and present in the data set at least three times for each group under analysis were considered, as rare alleles can have large variances in LD estimates. SNPs typed in <75% of the individuals in a group were excluded from analysis. Insertion/deletion polymorphisms were treated as missing data. We ran the LD analyses excluding the five elite cultivars and results were the same (data not shown).
We calculated the LD as the correlation coefficient r2 between each SNP pair (Hartl and Clark 1997). Heterozygous SNPs were rare in O. sativa individuals (0.53% of SNP genotypes), but ∼6.7% of O. rufipogon SNP genotypes were heterozygotes. When only one SNP in a pair is heterozygous, it is possible to infer the haplotypes and calculate a r2 value. We excluded individuals from analysis if both SNPs in a pair were heterozygous so that only unambiguous haplotypes were used in the analysis. In the three O. sativa groups, very few SNP pairs contained doubly heterozygous individuals (2.59% in indica, 0.47% in tropical japonica, and 4.36% in temperate japonica) and were thus excluded. In the vast majority of these cases, only a single individual was heterozygous at both sites. In O. rufipogon, 24.5% of SNP pairs contained individuals with double heterozygotes, but the majority of these pairs (73.78%) had only a single doubly heterozygous individual to exclude. We also used the previously described STS data set (Caicedo et al. 2007) to calculate the genomewide background LD, which is the level of disequilibrium between unlinked SNPs.
Due to the large amount of variance in the estimates of LD for any particular SNP pair, we combined SNP pairs into distance intervals to reduce the influence of outliers and to obtain a better visual description of the LD decay with distance. For the estimate of genomewide LD using the STS data set, the distance classes are <1 kb, 0.001–0.5 Mb, 0.5–2.0 Mb, and 2 Mb distance windows for SNP pairs >2 Mb in distance. For the study of the targeted genomic regions, a distance window of 40 kb was used. Due to our sampling scheme, the majority of SNP pairs in the genomic regions are close to the middle of the interval, so we plotted the median r2 for each distance window. We consider a particular intermarker distance interval to have LD elevated above the background level if it contains >10 SNP pairs and the interval median r2 exceeds the 75th percentile of the unlinked pairs. We chose the 75th percentile as a compromise between criteria that were too stringent and too relaxed. Clearly, the median of unlinked pairs is too low: by this criterion we would infer elevated LD for half of all intervals with LD at background levels. Given the great variance in LD values for any particular distance interval, there are bound to be pairs with low LD values, so requiring half of the pairs in a given interval to exceed the 95th percentile of unlinked pairs, for example, will only find evidence for LD that greatly exceeds genomewide background levels. We felt this was too stringent for our purposes.
Recombination analysis:
A composite-likelihood method (Hudson 2001) as implemented in the LDhat software (McVean et al. 2002) was used to estimate the population recombination parameter ρ = 4Ner for each target region. A likelihood permutation test was performed for each ρ estimate and the corresponding maximum-likelihood test for significant evidence of recombination. The minimum number of recombination events (Hudson and Kaplan 1985) was estimated across each target region using LDhat.
RESULTS AND DISCUSSION
Levels and patterns of nucleotide variation among six genomic regions:
We sequenced a total of six entire genes and 70 gene fragments in six genomic regions (Figure 1) for a total of 72,341 bp of aligned sequence data; these genomic regions ranged in size from ∼472 to ∼487 kb in length. In O. rufipogon, 1364 SNPs were identified, while 522 where found in the O. sativa variety group indica, 250 in tropical japonica, and 219 in temperate japonica.
Levels of nucleotide variation (θw) are given in Table 2. As expected, variation is greatest in O. rufipogon (average silent site θW = 0.0053) and lowest in temperate japonica (silent θw = 0.0010). The mean values of silent θW in indica and tropical japonica are 0.0023 and 0.0012, respectively. These are similar to the genomewide estimates of diversity reported in two previous studies (Caicedo et al. 2007; Rakshit et al. 2007). The levels of nucleotide variation also differed widely among the six genomic regions; in indica, for example, silent variation ranged from a mean of θw = 0.0007–0.0046. In O. sativa, generally the A region had the lowest variation and the C region had the highest.
TABLE 2.
Nucleotide variation (θw) of targeted genomic regions
| Region | O. rufipogon | O. sativa (indica) | O. sativa (tropical japonica) | O. sativa (temperate japonica) |
|---|---|---|---|---|
| A | 0.0025 (0.0016) | 0.0007 (0.0018) | 0.0001 (0.0004) | 0.0002 (0.0004) |
| B | 0.0105 (0.0062) | 0.0035 (0.0039) | 0.0005 (0.0012) | 0.00005 (0.0002) |
| C | 0.0070 (0.0048) | 0.0046 (0.0056) | 0.0036 (0.0044) | 0.0037 (0.0027) |
| D | 0.0016 (0.0013) | 0.0009 (0.0005) | 0.0003 (0.0004) | 0.0001 (0.0004) |
| E | 0.0051 (0.0042) | 0.0020 (0.0028) | 0.0008 (0.0011) | 0.0006 (0.0009) |
| F | 0.0050 (0.0050) | 0.0022 (0.0041) | 0.0023 (0.0042) | 0.0016 (0.0028) |
| Mean | 0.0052 (0.0051) | 0.0023 (0.0036) | 0.0012 (0.0028) | 0.0010 (0.0021) |
Standard deviations are in parentheses.
Unlinked SNPs indicate low background linkage disequilibrium in O. sativa and O. rufipogon:
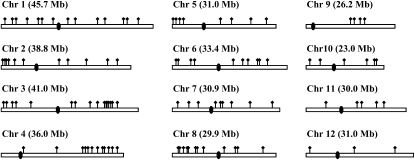
In addition to physical linkage, a number of other mainly demographic factors can influence the level of linkage disequilibrium between a pair of SNPs in the genome. These demographic influences are on a genomewide scale and so their impact on LD can be estimated by measuring the background LD in the genome. This information can then be used to determine whether specific values of LD among SNPs are elevated relative to background levels. We used a previously described data set for 111 gene fragments across the genome (Caicedo et al. 2007) (Figure 3) to calculate the background LD, which we define as the distribution of LD between unlinked loci (such as SNPs on different chromosomes). In O. rufipogon, there were 394 biallelic SNPs with a frequency of at least 10% present in at least three copies in the 111 STS fragments, with 63,187 SNP pairs on different chromosomes and therefore unlinked. Fewer biallelic SNPs met our criteria in O. sativa (134 in indica, 137 in tropical japonica, and 39 in temperate japonica) and consequently fewer unlinked SNP pairs: 6855 in indica, 3964 in tropical japonica, and 422 in temperate japonica.
Figure 3.—
The relative positions of the STS genes sequenced in a genome study of variation (Caicedo et al. 2007). This data set was used in the study of genomewide LD between unlinked SNPs, as well as SNPs in the same chromosome.
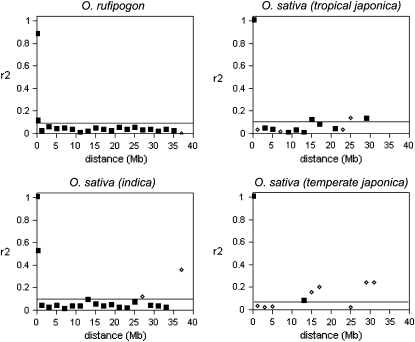
For these unlinked SNPs, r2 ranges between 0 and 1 for temperate japonica, tropical japonica, and O. rufipogon and between 0 and 0.583 for indica. Figure 4 shows the distribution of r2 values for the unlinked SNP pairs for each subgroup. Distributions are shifted toward low r2 values, which are expected as these SNPs are unlinked and the r2 values reflect disequilibrium due to either chance or evolutionary forces that affect variation across the entire genome. Median r2 values are similar in all groups, ∼0.04, while the 75th percentile for background LD (which we are using to define elevated LD) is ∼0.07–0.10 in all the studied genomes.
Figure 4.—
Distribution of median r2 values between unlinked SNPs from the STS data set. The distributions for O. rufipogon and each major O. sativa variety group are shown. The r2's corresponding to the 75th percentiles are indicated.
LD among SNPs on the same chromosome:
To get an initial estimate of LD among linked SNPs, we used the same STS data set (Figure 3) (Caicedo et al. 2007) and estimated LD between SNP pairs located on the same chromosome. These included 820 pairs in indica, 1191 pairs in tropical japonica, 138 pairs temperate japonica, and 8587 pairs in O. rufipogon. The distance between STS SNPs range from 1 bp to 37.3 Mb, with a median distance of 8.0 Mb. Figure 5 demonstrates that LD is quite elevated in all groups (median r2 = 1 in the three O. sativa groups and median r2 = 0.88 in O. rufipogon) at the shortest distance scale (<0.001 Mb). In indica, tropical japonica, and O. rufipogon, the median LD for intervals with at least 10 SNP pairs declines to below the 75th percentile of background LD level between the 0.001- to 0.5-Mb and 0.5- to 2.0-Mb intervals (with two exceptions in tropical japonica with median r2 at the 78th and 80th percentiles). LD decreases most rapidly in O. rufipogon, with the lowest median for the 0.001- to 0.5-Mb distance interval both in actual number (r2 = 0.11) and as compared to the distribution of unlinked SNP pairs (80th percentile). In contrast, the median r2 for this interval is 0.53 in indica and 1 in tropical japonica, which are both at the 99th percentile of LD values for unlinked SNPs in these groups.
Figure 5.—
The relationship of median r2 vs. distance for genomewide SNPs using the STS data set. Symbols represent binned data in 2-Mb intervals, with the first three bins being <1 kb, 1–500 kb, and 0.5–2.0 Mb. Solid squares have at least 10 SNP pairs and open diamonds have <10 SNP pairs in the distance interval. A horizontal line indicates the 75th percentile from the distribution of unlinked SNPs from the STS data set (Figure 4).
The smaller number of SNP pairs in the temperate japonica varieties obscures the underlying LD pattern, but suggests that disequilibrium among SNP pairs remains high at greater distances in this variety group. The only two intervals with >10 SNP pairs in temperate japonica are the <0.001 Mb and the 12- to 14-Mb distance intervals. The median r2 for the 12- to 14-Mb interval is elevated slightly above the 75th percentile for unlinked pairs in this variety group. Without any other distance intervals containing sizable numbers of data points, it is difficult to draw strong conclusions, but the data are consistent with LD extending over much greater distances in temperate japonica than in the other O. sativa groups or in O. rufipogon.
LD decay in targeted genomic regions:
Data from the genomewide panel of 111 STS loci (Caicedo et al. 2007) indicate that LD decays at <2 Mb for the O. sativa variety groups indica and tropical japonica and <500 kb for O. rufipogon. However, the STS data are fairly coarse grained, as the gene fragments are generally spaced apart at a megabase scale. We conducted a finer scale analysis of the extent of LD using five ∼500-kb regions (Figure 1), with gene fragment markers spaced ∼40 kb apart (Figure 2). One region (A) had very low variation in O. sativa (only two haplotypes in each variety group using SNPs that were frequent enough to be used for LD analysis) and thus was not considered in this analysis. Excluding SNPs in the A region, 423 in O. rufipogon, 262 in indica, 136 in tropical japonica, and 99 in temperate japonica had frequencies of at least 10%, were present in at least three copies, and were typed in at least 75% of the individuals in a group, making them suitable for assaying linkage disequilibrium.
The relationships of median r2 values with distance for each of these genomic regions are shown in supplemental Figure S1 at http://www.genetics.org/supplemental/. Variation in O. sativa is low (Table 2), and in many instances there are not a sufficient number of data points within each distance interval to draw strong conclusions for particular regions in particular groups. For example, the D region in temperate japonica and the B region in tropical japonica and temperate japonica each contained only a single biallelic SNP with a minor allele frequency >10%, so LD could not be assessed for these regions in these groups.
We combined the data from four genomic regions (regions B, D, E, and F) to estimate the average extent of LD within these different O. sativa variety groups and O. rufipogon (Figure 6). This does not weight each region equally, and regions with more data (for example, the F region) contribute more to the overall pattern. We did not include the data from genomic region C, since the pattern for this region appears atypical; LD in region C is only weakly correlated with intermarker distance and is exceptionally high in O. sativa: the median r2 values in the three O. sativa variety groups exceeds the 75th percentile for unlinked markers in all but one tropical japonica distance interval and four indica distance intervals. This region appears to have a large number of transposable elements. Although at the outset, attempts were made to exclude transposable elements, annotation updates of the rice genome during the course of this investigation meant that several flanking gene fragments in the C region were redone when annotation changed or sequencing demonstrated that more than one region was being amplified. New annotation since the conclusion of sequence indicates that three flanking gene fragments (5_01, 5_04, and 3_05) in the C region are putative transposable elements (see supplemental Table S2 at http://www.genetics.org/supplemental/). The atypical LD pattern (as well as high variation) in O. sativa may be explained by a high number of transposable elements in this region.
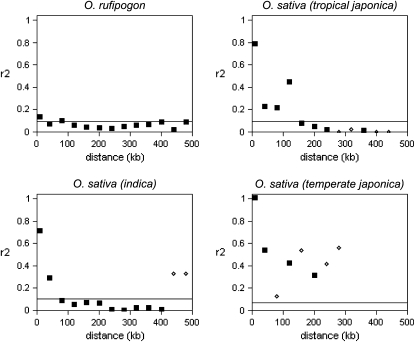
Figure 6.—
The relationship of median r2 vs. distance for SNPs in the targeted genomic regions. The data are for four genomic regions. Symbols represent binned data in 40-kb intervals, with the first bin being <7.5 kb. Solid squares have at least 10 SNP pairs and open diamonds have <10 SNP pairs in the distance interval. A horizontal line indicates the 75th percentile from the distribution of unlinked SNPs from the STS data set (Figure 4).
As expected, the combined data from the four genomic regions show a decrease in LD with increasing intermarker distance. In O. rufipogon, the median r2 exceeds only the 75th percentile of unlinked SNPs for the smallest interval (<7.5 kb), but this r2 value is low and close to background levels (Figure 6). The O. sativa groups display a greater extent of LD across the genome. Median r2 remains below the 75th percentile of unlinked SNPs in indica rice for all distance intervals >40 kb, the exception being the two intervals that contain <10 SNP pairs at distances >400 kb. In tropical japonica, LD is elevated above background levels for the first 120 kb, and the median r2 never exceeds the 63rd percentile of unlinked SNP pairs for intervals >120 kb. These patterns indicate that LD decay occurs at the shortest distance scales in the ancestral wild rice species, but in indica and tropical japonica extends to between ∼75 and 150 kb.
As with the genomewide sampling, there is low variation in temperate japonica, but the overall pattern in this group shows that LD extends to a much greater distance than in the other groups. For all distance intervals with >10 SNP pairs (out to the 200-kb distance interval), median r2 remains elevated above the 90th percentile of unlinked temperate japonica SNP pairs. It is likely that LD extends much farther, probably even to >500 kb in this O. sativa variety group.
Recombination and the decay of LD:
The lower bound on the number of recombination events (RM) and composite-likelihood estimates of ρ are given in Table 3. Recombination rates are quite low in both domesticated and wild rice. Across regions of ∼500 kb, RM varies between 0 and 22 and ρ estimates are <30 (0.06 × 10−3/bp) in all but two cases. Estimates of ρ in other plants are higher: 7–8 × 10−3/bp in wild barley (Morrell et al. 2006), ∼16–19 × 10−3/bp in maize (Tenaillon et al. 2002), and 0.2–0.8 × 10−3/bp in Arabidopsis thaliana (Nordborg et al. 2005; Kim et al. 2007). Effective population size, outcrossing rate, domestication, and demographic history all play a role in shaping ρ. It is therefore difficult to explain the exact species differences that give rise to different population recombination estimates, but some patterns are apparent. Maize utilizes an outcrossing mating system, consistent with higher estimates of the population recombination parameter. Although wild barley self-fertilizes at a very high rate (probably ∼98%), which decreases apparent recombination, the high ρ could be attributed perhaps to a recent transition to selfing (Morrell et al. 2006). The estimate of ρ in A. thaliana is higher than in rice, but less dramatically higher than the other plant species considered here. A. thaliana shows signs of a population expansion (Innan et al. 1997), increasing the opportunities for crossing over, which may elevate the recombination estimates despite an inbreeding mating system. Low rates of outcrossing in cultivated and wild rice and bottlenecks associated with domestication in cultivated rice likely explain the very low effective recombination rates detected. Although recombination is very low, for most regions in O. rufipogon and indica, rates are significantly different from zero.
TABLE 3.
Population recombination (ρ) and minimum recombination event (RM) estimates of targeted genomic regions
| Region | O. rufipogon | O. sativa (indica) | O. sativa (tropical japonica) | O. sativa (temperate japonica) |
|---|---|---|---|---|
| B | 19.494**** (16) | 2.442**** (3) | 1.421 (0) | NAa (0) |
| C | >100**** (18) | 21.570**** (17) | 8.150 (9) | 7.489*** (8) |
| D | 2.073 (3) | 7.473**** (2) | 6.129 (1) | NAa (0) |
| E | 27.131**** (22) | 47.571**** (4) | 2.409**** (0) | 8.869**** (3) |
| F | 11.447**** (12) | 85.697**** (5) | 1.602**** (5) | 0.000 (3) |
RM are in parentheses. ***Significantly different from zero at the 0.005 level; ****significantly different from zero at the 0.001 level.
Three or fewer SNPs.
Within cultivated rice varieties, recombination in indica in all regions is significantly different from zero. Recombination in the other two cultivated rice variety groups, tropical japonica and temperate japonica, is very much lower and is significantly different from zero in only two regions in each group (regions C and E in temperate japonica and regions E and F in tropical japonica). The minimum number of recombination events, RM, is higher in indica than in the other two O. sativa varieties, consistent with higher ρ estimates.
In all five regions, O. rufipogon appears to have more recombination than the O. sativa groups, consistent with greater outcrossing rates (Oka 1988; Gao et al. 2007) and a larger effective population size (Caicedo et al. 2007). O. rufipogon RM values are higher than O. sativa RM estimates, often to a great extent (as in region E). O. rufipogon ρ estimates are higher than either tropical or temperate japonica (or as in the case in region D, none of the three are significantly different from zero) and vary from 0 to >100. When comparing O. rufipogon and indica ρ estimates, O. rufipogon rates are not consistently greater, but due to the low overall variation, ρ cannot be precisely estimated from these data, and these estimates are not likely to be significantly different from each other.
Recombination breaks down LD, so the higher recombination rates in O. rufipogon explain the lower LD levels in this species compared to cultivated rice and, within O. sativa, lower recombination rates in tropical and temperate japonica as compared to the indica variety explain the greater extent of LD in the two japonica groups. As well as variation in recombination rates among varieties, rate heterogeneity is present among regions, likely because of recombination hotspots, as have been observed in humans (McVean et al. 2004).
The extent of linkage disequilibrium in rice:
Both linkage disequilibrium and the high densities of SNPs have combined to facilitate a new genomics strategy for identifying and mapping genes responsible for quantitative trait variation (Terwilliger and Weiss 1998; Kruglyak 1999; Jorde 2000; Remington et al. 2001; Kim et al. 2007). This mapping strategy, referred to as LD or association mapping, has been applied to humans (Martin et al. 2000; Puca et al. 2001) Drosophila (Long et al. 1998, 2000; Geiger-Thornsberry and Mackay 2002), and several plant species (Thornsberry et al. 2001; Palaisa et al. 2003; Olsen et al. 2004; Wilson et al. 2004). There has been considerable interest in using LD mapping or candidate gene association studies in identifying genes underlying variation in agronomically important phenotypes (Garris et al. 2003; Rostoks et al. 2006; Yu and Buckler 2006). The ability to detect significant associations between molecular polymorphism(s) and particular phenotypes, as well as the resolving power of LD mapping techniques, depends on knowledge of the extent of linkage disequilibrium in species genomes and the rate of decay of LD with physical distance (Long et al. 1998; Terwilliger and Weiss 1998; Kruglyak 1999; Jorde 2000; Pritchard et al. 2000; Remington et al. 2001).
We examined the levels of LD in cultivated rice in part to determine the level of resolution of LD mapping and candidate gene association studies in this important crop species. We found that the extent of linkage disequilibrium differs in significant ways between domesticated Asian rice and its wild ancestor O. rufipogon. Both in genomewide LD and in targeted genomic regions, there is substantially less LD in O. rufipogon compared to the O. sativa variety groups. The fact that O. rufipogon outcrosses at a higher rate than O. sativa, which we see for the most part reflected in higher recombination rates, and that both population bottlenecks and selection are associated with the domestication of the latter (Caicedo et al. 2007) probably accounts for the difference in disequilibrium between the two species. In particular, the transition to selfing in O. sativa leads to a decrease in the effective recombination rate, while bottlenecks and selection will reduce the number of haplotypes in domesticated rice, all of which would inflate LD in the species. A similar pattern of greater LD in domesticated crops compared to their wild ancestors has been observed in several other crop species (e.g., barley) (Remington et al. 2001; Tenaillon et al. 2001; Caldwell et al. 2006; Hyten et al. 2007), indicating that the process of domestication is a key force in the rise of LD in crop species.
Among the three variety groups, there is more LD in temperate japonica rice than in indica or tropical japonica. Temperate japonica is closely related to tropical japonica (Khush 1997; Garris et al. 2005), and the bottleneck associated with adaptation to the temperate environment resulted in a smaller effective population size compared to the other two rice groups (S. Williamson, unpublished results). Interestingly, the extent of LD in indica is less than that of tropical japonica, which may also arise from differences in the severity of the population bottleneck or the intensity of selection accompanying the domestication of these two groups. Modeling studies based on the nucleotide polymorphism site-frequency spectrum, for example, suggest a less severe bottleneck and less intense selection in indica compared to tropical japonica (S. Williamson and A. L. Caicedo, unpublished results), which would also account for the greater extent of LD in the latter group.
Most studies to date in humans (Huttley et al. 1999; Gabriel et al. 2002) and maize (Remington et al. 2001; Tenaillon et al. 2001) have documented variation in levels and patterns of LD across the genome, and our work in rice supports these observations in other species. Differences in recombination may account for this pattern, and for the most part we find that higher recombination rates are associated with regions and groups with lower linkage disequilibrium. The low number of targeted genomic regions that we used makes it difficult to draw strong inferences, but from our sample there appears to be a correlation between estimated recombination rate and extent of LD.
There appears to be a correspondence between levels of selfing and extent of LD in crop species. LD in maize, which is outcrossing, decays within 2 kb (Remington et al. 2001; Tenaillon et al. 2001), while LD in sorghum, which has an outcrossing rate of 10–20%, can extend to 20 kb (Hamblin et al. 2005). Barley and soybean, which are both selfers, can have high LD levels that extend to several hundred kilobases (Caldwell et al. 2006; Hyten et al. 2007), a pattern similar to that for O. sativa. However, wild barley, which is also a selfer, has LD decay scales of only a few kilobases (Morrell et al. 2005; Caldwell et al. 2006), suggesting that the process of domestication may amplify the effects of self-fertilization, resulting in increasing LD levels across larger genomic regions in crop species.
These results provide insights into the possible resolution of LD mapping in domesticated rice. In maize, the short extent of LD allows association studies to localize SNPs that are significantly correlated to trait phenotypes to specific candidate genes, including those in flowering time (Thornsberry et al. 2001), starch content (Wilson et al. 2004), and kernel coloration (Palaisa et al. 2003). Rice has a greater extent of LD and thus candidate gene association studies may not be as successful. If we take 75 kb as the average resolution scale in indica and 150 kb in tropical japonica, this corresponds to genomic regions encompassing ∼9 to 17 genes, respectively. We should note, however, that these represent mean estimates of genomewide LD, and different genomic regions and groups do have different LD decay patterns (supplemental Figure S1 at http://www.genetics.org/supplemental/). The resolving power of LD mapping in rice thus will differ across genomic regions and population samples.
Nevertheless, our study suggests that a modest number of SNPs across the genome may be sufficient for undertaking genomewide LD mapping studies in rice. On the basis of the LD decay range in the O. sativa variety groups, LD mapping would be possible in indica and tropical japonica; in temperate japonica, there is too little polymorphism and the extent of LD is too large for this mapping approach to be readily feasible. It appears that placement of SNP markers every 75 kb for indica for a total of ∼5200 markers stands a reasonable chance of genomewide coverage in this variety group. For tropical japonica, markers can be placed ∼150 kb apart, and a total of 2600 markers can result in good coverage. Other approaches, such as the use of tag SNPs (Carlson 2004) will require larger numbers of markers to achieve genomewide coverage, but more extensive resequencing is necessary to determine the tag-SNP densities required. It does appear, however, that rice association studies can be achieved with reasonable SNP marker densities and, given genotyping technologies, at relatively low cost.
Acknowledgments
We are grateful to Chris Smith, Adi Fledel-Alon, and Xianfa Xie for developing programs to process data and conduct some analyses. This project was funded in part by a grant from the National Science Foundation Plant Genome Research Program to M.D.P., S.M., Carlos Bustamante, and Rasmus Nielsen.
References
- Caicedo, A., S. Williamson, R. D. Hernandez, A. Boyko, A. Fledel-Alon et al., 2007. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 3: e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell, K. S., J. Russell, P. Langridge and W. Powell, 2006. Extreme population-dependent linkage disequilibrium detected in an inbreeding plant species, Hordeum vulgare. Genetics 172: 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, C. S., M. A. Eberle, M. J. Rieder, Q. Yi, L. Kruglyak et al., 2004. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 74: 106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, G. W., and S. Chen, 1998. The origins of rice agriculture: recent progress in East Asia. Antiquity 72: 858–866. [Google Scholar]
- Feng, Q., Y. Zhang, P. Hao, S. Wang, G. Fu et al., 2002. Sequence and analysis of rice chromosome 4. Nature 420: 316–320. [DOI] [PubMed] [Google Scholar]
- Gabriel, S. B., S. F. Schaffner, H. Nguyen, J. M. Moore, J. Roy et al., 2002. The structure of haplotype blocks in the human genome. Science 296: 2225–2229. [DOI] [PubMed] [Google Scholar]
- Gao, H., S. Williamson and C. D. Bustamante, 2007. A Markov chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics 176: 1635–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris, A. J., S. R. McCouch and S. Kresovich, 2003. Population structure and its effect on haplotype diversity and linkage disequilibrium surrounding the xa5 locus of rice (Oryza sativa L.). Genetics 165: 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris, A. J., T. H. Tai, J. Coburn, S. Kresovich and S. McCouch, 2005. Genetic structure and diversity in Oryza sativa L. Genetics 169: 1631–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger-Thornsberry, G. L., and T. F. Mackay, 2002. Association of single-nucleotide polymorphisms at the Delta locus with genotype by environment interaction for sensory bristle number in Drosophila melanogaster. Genet. Res. 79: 211–218. [DOI] [PubMed] [Google Scholar]
- Hamblin, M. T., M. G. Salas Fernandez, A. M. Casa, S. E. Mitchell, A. H. Paterson et al., 2005. Equilibrium processes cannot explain high levels of short- and medium-range linkage disequilibrium in the domesticated grass Sorghum bicolor. Genetics 171: 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl, D. L., and A. G. Clark, 1997. Principles of Population Genetics. Sinauer Associates, Sunderland, MA.
- Hudson, R. R., 2001. Two-locus sampling distributions and their application. Genetics 159: 1805–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., and N. L. Kaplan, 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttley, G. A., M. W. Smith, M. Carrington and S. J. O'Brien, 1999. A scan for linkage disequilibrium across the human genome. Genetics 152: 1711–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten, D. L., I.-Y. Choi, Q. Song, R. C. Shoemaker, R. L. Nelson et al., 2007. Highly variable patterns of linkage disequilibrium in multiple soybean populations. Genetics 175: 1937–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan, H., R. Terauchi and N. T. Miyashita, 1997. Microsatellite polymorphism in natural populations of the wild plant Arabidopsis thaliana. Genetics 146: 1441–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project, 2005. The map-based sequence of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- Jorde, L. B., 2000. Linkage disequilibrium and the search for complex disease genes. Genome Res. 10: 1435–1444. [DOI] [PubMed] [Google Scholar]
- Khush, G. S., 1997. Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 35: 25–34. [PubMed] [Google Scholar]
- Kim, S., V. Plagnol, T. T. Hu, C. Toomajian, R. M. Clark et al., 2007. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 39: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Kruglyak, L., 1999. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat. Genet. 22: 139–144. [DOI] [PubMed] [Google Scholar]
- Long, A. D., R. F. Lyman, C. H. Langley and T. F. Mackay, 1998. Two sites in the Delta gene region contribute to naturally occurring variation in bristle number in Drosophila melanogaster. Genetics 149: 999–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, A. D., R. F. Lyman, A. H. Morgan, C. H. Langley and T. F. Mackay, 2000. Both naturally occurring insertions of transposable elements and intermediate frequency polymorphisms at the achaete-scute complex are associated with variation in bristle number in Drosophila melanogaster. Genetics 154: 1255–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, E. R., J. R. Gilbert, E. H. Lai, J. Riley, A. R. Rogala et al., 2000. Analysis of association at single nucleotide polymorphisms in the APOE region. Genomics 63: 7–12. [DOI] [PubMed] [Google Scholar]
- McVean, G., P. Awadalla and P. Fearnhead, 2002. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 160: 1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean, G. A., S. R. Myers, S. Hunt, P. Deloukas, D. R. Bentley et al., 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304: 581–584. [DOI] [PubMed] [Google Scholar]
- Morrell, P. L., D. M. Toleno, K. E. Lundy and M. T. Clegg, 2005. Low levels of linkage disequilibrium in wild barley (Hordeum vulgare ssp. spontaneum) despite high rates of self-fertilization. Proc. Natl. Acad. Sci. USA 102: 2442–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell, P. L., D. M. Toleno, K. E. Lundy and M. T. Clegg, 2006. Estimating the contribution of mutation, recombination and gene conversion in the generation of haplotypic diversity. Genetics 173: 1705–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg, M., T. T. Hu, Y. Ishino, J. Jhaveri, C. Toomajian et al., 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, H. I., 1988. Origin of Cultivated Rice. Elsevier, Amsterdam.
- Olsen, K. M., S. S. Halldorsdottir, J. R. Stinchcombe, C. Weinig, J. Schmitt et al., 2004. Linkage disequilibrium mapping of Arabidopsis CRY2 flowering time alleles. Genetics 167: 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, K. M., A. L. Caicedo, N. Polato, A. McClung, S. McCouch et al., 2006. Selection under domestication: evidence for a sweep in the rice Waxy genomic region. Genetics 173: 975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaisa, K., M. Morgante, M. Williams and A. Rafalski, 2003. Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytoene synthase loci. Plant Cell 15: 1795–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., M. Stephens and P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puca, A. A., M. J. Daly, S. J. Brewster, T. C. Matise, J. Barrett et al., 2001. A genome-wide scan for linkage to human exceptional longevity identifies a locus on chromosome 4. Proc. Natl. Acad. Sci. USA 98: 10505–10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakshit, S., A. Rakshit, H. Matsumura, Y. Takahashi, Y. Hasegawa et al., 2007. Large-scale DNA polymorphism study of Oryza sativa and O. rufipogon reveals the origin and divergence of Asian rice. Theor. Appl. Genet. 114: 731–743. [DOI] [PubMed] [Google Scholar]
- Remington, D. L., J. M. Thornsberry, Y. Matsuoka, L. M. Wilson, S. R. Whitt et al., 2001. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl. Acad. Sci. USA 98: 11479–11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostoks, N., L. Ramsay, K. MacKenzie, L. Cardle, P. R. Bhat et al., 2006. Recent history of artificial outcrossing facilitates whole-genome association mapping in elite inbred crop varieties. Proc. Natl. Acad. Sci. USA 103: 18656–18661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S., and H. J. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers, pp. 365–386 in Bioinformatics Methods and Protocols: Methods in Molecular Biology, edited by S. Misener and S. Krawetz. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Sasaki, T., T. Matsumoto, K. Yamamoto, K. Sakata, T. Baba et al., 2002. The genome sequence and structure of rice chromosome 1. Nature 420: 312–316. [DOI] [PubMed] [Google Scholar]
- Tenaillon, M. I., M. C. Sawkins, A. D. Long, R. L. Gaut, J. F. Doebley et al., 2001. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc. Natl. Acad. Sci. USA 98: 9161–9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, M. I., M. C. Sawkins, L. K. Anderson, S. M. Stack, J. Doebley et al., 2002. Patterns of diversity and recombination along chromosome 1 of maize (Zea mays ssp. mays L.). Genetics 162: 1401–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, J. D., and K. M. Weiss, 1998. Linkage disequilibrium mapping of complex disease: Fantasy or reality? Curr. Opin. Biotechnol. 9: 578–594. [DOI] [PubMed] [Google Scholar]
- Thornsberry, J. M., M. M. Goodman, J. Doebley, S. Kresovich, D. Nielsen et al., 2001. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28: 286–289. [DOI] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Wilson, L. M., S. R. Whitt, A. M. Ibanez, T. R. Rocheford, M. M. Goodman et al., 2004. Dissection of maize kernel composition and starch production by candidate gene association. Plant Cell 16: 2719–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., and E. S. Buckler, 2006. Genetic association mapping and genome organization of maize. Curr. Opin. Biotechnol. 17: 155–160. [DOI] [PubMed] [Google Scholar]
- Yu, J., S. Hu, J. Wang, G. K. Wong, S. Li et al., 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92. [DOI] [PubMed] [Google Scholar]