Abstract
A high-density genetic map of papaya (Carica papaya L.) was constructed using microsatellite markers derived from BAC end sequences and whole-genome shot gun sequences. Fifty-four F2 plants derived from varieties AU9 and SunUp were used for linkage mapping. A total of 707 markers, including 706 microsatellite loci and the morphological marker fruit flesh color, were mapped into nine major and three minor linkage groups. The resulting map spanned 1069.9 cM with an average distance of 1.5 cM between adjacent markers. This sequence-based microsatellite map resolved the very large linkage group 2 (LG 2) of the previous high-density map using amplified fragment length polymorphism markers. The nine major LGs of our map represent papaya's haploid nine chromosomes with LG 1 of the sex chromosome being the largest. This map validates the suppression of recombination at the male-specific region of the Y chromosome (MSY) mapped on LG 1 and at potential centromeric regions of other LGs. Segregation distortion was detected in a large region on LG 1 surrounding the MSY region due to the abortion of the YY genotype and in a region of LG6 due to an unknown cause. This high-density sequence-tagged genetic map is being used to integrate genetic and physical maps and to assign genome sequence scaffolds to papaya chromosomes. It provides a framework for comparative structural and evolutional genomic research in the order Brassicales.
PAPAYA (Carica papaya L.) is a major fruit crop in tropical and subtropical regions worldwide. It is diploid (2n = 18) with a small genome size of 372 Mbp (Arumuganathan and Earle 1991), which is advantageous in genomic analyses. Papaya is a member of the family Caricaceae in the order Brassicales, sharing a common ancestor with Arabidopsis ∼72 million years ago (Wikström et al. 2001). The phylogenetic positioning of papaya near the major model plant Arabidopsis and within the agronomically significant Brassicales could make it central for comparative structural and evolutional genomic research in this group of dicots.
Papaya is a perennial plant species that flowers as early as 3 months and produces fruit in 9 months; it is trioecious with an intriguing sex determination system with three basic sex types: male, female, and hermaphrodite. A high-density genetic map of papaya, constructed using 1498 AFLP and three morphological markers revealed severe suppression of recombination at the sex determination locus with 225 cosegregating markers accounting for 66% of the markers on linkage group 1 (LG 1) and 15% of the markers genomewide (Ma et al. 2004). This AFLP map provided a critical piece of evidence for defining the primitive Y chromosome in papaya (Liu et al. 2004). Sequence comparison of Y-specific fragments from males and hermaphrodites confirmed that there are two slightly different Y chromosomes in papaya (Liu et al. 2004). To distinguish these two Y chromosomes, the one controlling male was designated as Y, whereas the other controlling hermaphrodites were designated as Yh (Ming et al. 2007). The sex chromosome genotypes of the three sexes are XY for males, XYh for hermaphrodites, and XX for females. The combinations of YY, YYh, or YhYh are embryonic lethal, indicating the loss of essential genes for embryo development during the Y chromosome degeneration process. This notion was reinforced by the gene paucity in the male-specific region of the Y chromosome (MSY) (Yu et al. 2007a). The lethal effect of the YY, YYh, or YhYh genotypes resulted in a distorted 2:1 hermaphrodite and female ratio in F2 populations derived from crosses between females and hermaphrodites. The DNA markers at the MSY and its neighboring regions exhibit the 2:1 segregation ratio, matching the observed phenotype (Ma et al. 2004).
Although the AFLP map was high density, it was constructed with anonymous AFLP markers (Ma et al. 2004). The anonymous dominant markers are not suitable for anchoring bacterial artificial chromosomes (BACs) and whole-genome shotgun sequences. Our attempts to anchor AFLP markers on a papaya physical map using plate, row, column, and diagonal pools of BAC DNA produced an unacceptably high rate of false positives (Q. Yu, P. H. Moore and R. Ming, unpublished results). Thus, developing sequence-tagged DNA markers became a high priority for integrating genetic and physical maps and for aligning papaya genome sequence to individual chromosomes.
Simple sequence repeats (SSRs), or microsatellites, with tandem repeats of di- to tetranucleotide sequence motifs flanked by unique sequences are ubiquitous, abundant, and well distributed in eukaryotic genomes (Tautz 1989; Wang et al. 1994; Cardle et al. 2000; Morgante et al. 2002). Although SSRs were first studied in humans (Weber et al. 1989), they have now been found and widely used in nearly all eukaryotes, including many plant species (Morgante et al. 1993). In recent years, SSRs have become one of the more popular molecular markers with applications in many fields as massive amounts of genomic sequences became available. In contrast to the earlier genetic markers of restriction fragment length polymorphisms (RFLPs), random amplified polymorphic DNAs (RAPDs), and amplified fragment length polymorphisms (AFLPs), SSR markers possess the combined advantages of these three types of DNA markers: they are codominant, highly polymorphic, abundant, distributed throughout the genome, almost always single locus (even in complex genomes), unambiguously and specifically mapped to genomes, and based on efficient PCR-based technology. Microsatellite-based linkage maps have been constructed for a wide variety of species, including mammals and several crop plants (Dib et al. 1996; Dietrich et al. 1996; McCouch et al. 2002; Sharopova et al. 2002; Ihara et al. 2004; Somers et al. 2004; Song et al. 2004). Although development of a high-density SSR map in plants has lagged behind that of mammals, this situation is being improved significantly as more genome sequences become available in selected plant species.
The agricultural significance of papaya in tropical regions, its small genome size, and the unique biology of primitive sex chromosomes were justifications for launching a papaya genome-sequencing project by the Hawaii Papaya Genome Consortium. DNA from female plants of the cultivar SunUp was selected for whole-genome shotgun (WGS) sequencing. In addition, BAC ends of the entire 13× SunUp hermaphrodite BAC library were sequenced. A large collection of nonredundant SSR markers was developed from BAC end sequences (BES) and WGS. Here we report a high-density genetic map of papaya based on sequence-tagged microsatellites. The objectives of this project were to (1) provide anchor markers for integration of genetic and physical maps and for aligning WGS sequences to chromosomes; (2) generate a comprehensive set of low-cost codominant DNA markers for gene tagging and marker-assisted selection; (3) facilitate physical mapping of the MSY region and the corresponding region of the X chromosome; and (4) foster comparative and evolutionary genomic research in Brassicales.
MATERIALS AND METHODS
Plant materials:
An F2 mapping population was derived from a cross between a female tree of the dioecious variety AU9 and a hermaphrodite tree (pollen donor) of the gynodioecious variety SunUp. This population was grown at the Kunia substation on Oahu, Hawaii, along with their parents AU9 and SunUp, and the F1. Young leaves for DNA isolation were collected from 36 hermaphrodite plants, 18 female F2 plants, the two parents, and an F1 plant.
Sequence sources of SSR markers:
A total of 11,976 SSR markers were developed from the following DNA sequence data: (1) 9955 SSRs from papaya WGS of SunUp female, including 9216 from nonredundant individual sequence reads (designated with prefixes P3K, P6K, and P8K to reflect the insert sizes of the WGS libraries) and 739 from assembled contig sequences (designated with the prefix ctg) and (2) 2021 SSRs from 1979 BES and 42 selected subclones of papaya BACs (designated with the prefix CPM).
SSR markers were searched from the whole-genome shotgun sequences using SSR Finder downloaded from http://www.maizemap.org. The same program was used to remove redundant SSR and design primers. The programs used for mining SSR markers from BAC end sequences and designing primers were described previously (Eustice et al. 2008). Primers were synthesized by Invitrogen (Carlsbad, CA).
SSR polymorphism survey and mapping:
All 11,976 SSR markers were screened for polymorphism using the two parents and two pools of bulk segregants. Each pool contained 10 F2 plants of hermaphrodites or females, respectively. The markers exhibiting polymorphism between the parents and confirmed in two pools of bulk segregants were used for mapping.
SSR markers were amplified in a 10-μl PCR mix containing 5 ng of template DNA, 0.15 mm of each dNTP, 1× PCR buffer, 2.0 mm MgCl2, 0.15 μm each of reverse and forward primers, and 0.5 units of Taq polymerase. The PCR reactions were performed using a PTC-225 thermocycler (MJ Research, Watertown, MA), in which the reaction mixture was incubated at 94° for 5 min, then for 35 cycles of 45 sec of denaturing at 94°, 30 sec of annealing at 55°, and 45 sec of extension at 72°, with a final extension at 72° for 7 min. PCR products were separated on 4% super fine resolution (SFR) agarose (Amresco, Solon, OH) gels and visualized by ethidium-bromide staining.
Some SSR markers showed a subtle difference in agarose gels and were separated using fluorescent-tagged SSR primers in a Li-Cor (Lincoln, NE) 4300 DNA analyzer with a 6.5% gel matrix. The PCR reactions were conducted in a 10-μl PCR mix containing 20 ng of template DNA, 0.2 μm each of reverse and M13-tailed forward primers, 0.4 μm fluorescence-labeled M13 primer, 0.2 mm of each dNTP, 1× PCR buffer, 2.0 mm MgCl2, and 0.5 units of Taq polymerase. All marker data were scored by visual inspection and proofread to correct errors.
Map construction:
Chi-square analysis was performed on each segregating marker to test the goodness of fit to the expected segregation ratios of codominant (1:2:1), sex-linked codominant (2:1), and dominant (3:1) markers in the F2 population. A genetic linkage map was constructed using the JoinMap (version 3.0) program (van Ooijen and Voorrips 2001). The linkage map was constructed with a minimum LOD score of 4.0 and a maximum recombination rate (θ) of 0.40 using the Kosambi mapping function. During map construction, markers that appeared in the “suspected linkage” panel of the JoinMap program were manually checked and markers deemed problematic were removed to assure the accuracy of the genetic map.
RESULTS
SSR markers polymorphism:
Among the 11,976 SSR markers surveyed, 8763 (73.2%) amplified successfully, yielding clear and discernible bands, whereas the other 3213 (26.8%) did not amplify or produced nonspecific amplification. Among the 8763 amplified SSRs, 1167 (13.3%) showed polymorphism between the parents of the mapping population and were used for genetic mapping in the F2 population. A total of 886 SSRs (10.1%) produced segregating genotype data for linkage mapping.
Only high-quality SSR markers were used for the genetic map construction. All 886 SSRs were proofread by a different person. Subsequently, 102 of them were eliminated because the PCR products in agarose gels appeared either too faint or too difficult to distinguish from the parental banding patterns in the F2 population. Another 58 SSRs were removed because 34 of them had more than five missing data points and 24 caused spurious linkage in the initial mapping attempts. At the end, a total of 726 markers plus the morphological marker fruit flesh color were used for linkage mapping.
Linkage map construction:
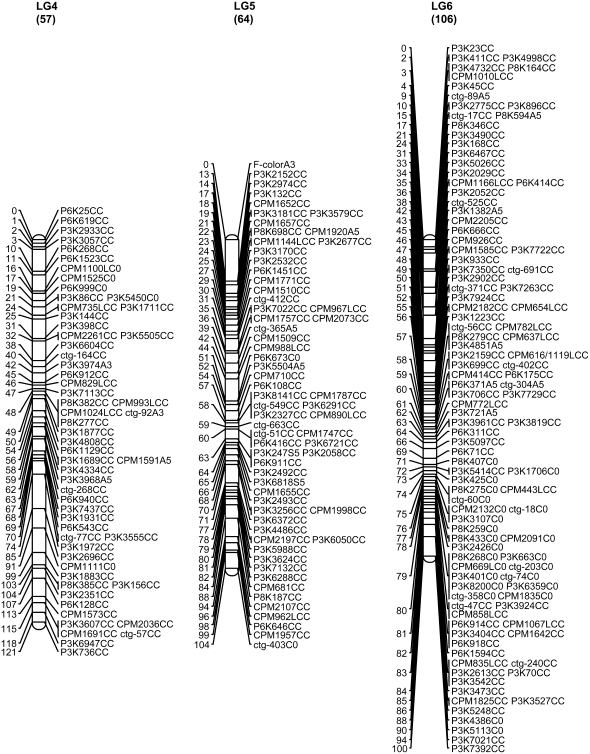
A total of 707 markers, including 706 SSR markers and the morphological marker fruit flesh color, were mapped to 12 LGs, including 9 large and 3 short that collectively span 1068.9 cM with an average distance of 1.51 cM between adjacent markers (Figure 1, Table 1). The 9 major linkage groups, which correspond to 9 chromosomes in the papaya genome, covered a total length of 993.5 cM (92.7%) with 683 mapped loci (96.6%) at an average marker density of 1.45 cM. The three short linkage groups covered a total of 75.4 cM with 24 mapped loci (3.4%) and an average distance of 3.1 cM between adjacent markers (Figure 1). The remaining 20 SSRs (2.8%) were assigned into two-marker linkage groups (4 SSRs) or remained unlinked (16 SSRs).
Figure 1.—
SSR genetic map of papaya linkage groups 1–12. The numerical scale at the left of the map is the cumulative length of the LG in centimorgans. The alphanumeric codes at the right of the map describe the SSR markers. The first three positions of the code represent the source or origin of the SSRs (see materials and methods: P markers from WGS, ctg markers from contig sequences, and CPM markers from BES sequences). The next-to-last element is the letter indicating the parent from which the marker was derived (A, AU9; S, SunUp; C, codominant); the last element of the code is information about the χ2 test (C, codominant fitting a ratio of 1:2:1; 0, not fitting any expected segregation ratio; 2, sex-linked codominant fitting a 2:1 ratio; 3, dominant fitting a 3:1; 5, dominant fitting both 2:1 and 3:1). Eleven SSRs on LG 1 with Y at the third position from the end indicate that they are sex-cosegregating markers. SSRs with L also at the third position from the end indicate that the genotype data were generated by a Li-Cor DNA analyzer. The MSY was mapped on LG 1 and the fruit flesh color was mapped on LG 5.
TABLE 1.
Summary statistics for AU9 × SunUp linkage groups
| Linkage group | Size (cM) | Marker no. | Average distance (cM) | Gaps − Da (cM)
|
Cluster position (cM) (no. of markers) | Codominant markers
|
Dominant markers
|
Total distorted
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 < D < 10 | D > 10 | Distorted | Normal | Total | % | Distorted | Normal | Total | % | No. | % | |||||
| 1 | 145.0 | 77 | 1.88 | 3 | 2 | 47 (11), 120 (5) | 47 | 25 | 72 | 93.5 | 4 | 1 | 5 | 6.5 | 51 | 66.2 |
| 2 | 138.8 | 70 | 1.98 | 5 | 3 | 81 (10), 82 (8) | 1 | 62 | 63 | 90.0 | 0 | 7 | 7 | 10.0 | 1 | 1.4 |
| 3 | 132.4 | 116 | 1.14 | 3 | 0 | 58 (8), 59 (6), 60 (8), 70 (9) | 7 | 103 | 110 | 94.8 | 0 | 6 | 6 | 5.2 | 7 | 6.0 |
| 4 | 120.6 | 57 | 2.12 | 6 | 1 | 48 (5) | 5 | 48 | 53 | 93.0 | 0 | 4 | 4 | 7.0 | 5 | 8.8 |
| 5 | 103.6 | 64 | 1.62 | 3 | 1 | 58 (6), 59 (5) | 2 | 56 | 58 | 90.6 | 0 | 6 | 6 | 9.4 | 2 | 3.1 |
| 6 | 100.2 | 106 | 0.95 | 4 | 0 | 57 (8), 59 (5), 79 (10), 81 (7), 82 (6) | 24 | 75 | 99 | 93.4 | 0 | 7 | 7 | 6.6 | 24 | 22.6 |
| 7 | 96.4 | 59 | 1.63 | 3 | 0 | 56 (6), 80 (5) | 3 | 52 | 55 | 93.2 | 0 | 4 | 4 | 6.8 | 3 | 5.1 |
| 8 | 91.8 | 60 | 1.53 | 4 | 1 | 47 (23) | 0 | 50 | 50 | 83.3 | 5 | 5 | 10 | 16.7 | 5 | 8.3 |
| 9 | 64.5 | 74 | 0.87 | 3 | 0 | 5 (6), 31 (15) | 5 | 68 | 73 | 98.6 | 1 | 0 | 1 | 1.4 | 6 | 8.1 |
| 10 | 27.1 | 12 | 2.26 | 2 | 0 | 26 (5) | 3 | 4 | 7 | 58.3 | 3 | 2 | 5 | 41.7 | 6 | 50.0 |
| 11 | 26.8 | 7 | 3.84 | 1 | 1 | — | 1 | 5 | 6 | 85.7 | 0 | 1 | 1 | 14.3 | 1 | 14.3 |
| 12 | 21.4 | 5 | 4.28 | 2 | 0 | — | 2 | 3 | 5 | 100.0 | 0 | 0 | 0 | 0.0 | 2 | 40.0 |
| Total | 1068.9 | 707 | 1.51 | 39 | 9 | 20 | 100 | 551 | 651 | 92.1 | 13 | 43 | 56 | 7.9 | 113 | 16.0 |
D, gap distance between adjacent markers.
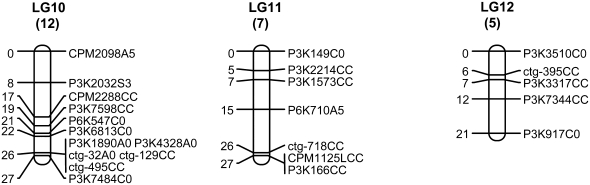
The linkage group of the primitive sex chromosomes, where the MSY is located, was designated LG 1 to be consistent with previous linkage maps (Sondur et al. 1996; Ma et al. 2004). The other linkage groups were designated LG 2–LG 12, in descending order according to the length of each LG generated from our SSR data. Of 78 markers on LG 1, 11 (14%) cosegregated with sex. Cosegregation of the markers with sex was easy to spot as the first 36 lanes of the gel were hermaphrodites and the last 18 lanes were females (Figure 2). These 11 markers were designated with the letter Y at the third position from right in the marker name for denoting their mapping to the MSY region and the LG for the sex chromosomes. Of the 11 Y markers, 10 formed a cluster, while the other marker, P3K2981YC0, with two missing data points appeared to be 1 cM away. LG 1 turned out to be the largest linkage group in our genetic map. Fruit flesh color (F-color) was mapped to one end of LG 5 linked with marker P3K2152 at a distance of 12.7 cM (Figure 1).
Figure 2.—
A gel image generated by SSR marker CPM1815, showing cosegregation with sex phenotype.
Of the 706 SSRs mapped, 651 (92.1%) were codominant and 56 (7.9%) were dominant. See supplemental Table S1 (at http://www.genetics.org/supplemental/) for a complete list of mapped SSRs, including primer sequences, source IDs, and map positions.
Segregation distortion:
Two regions on the genetic map showed significant segregation distortion at the P < 0.05 significant level (chi-square test). One severely distorted region was around the MSY on LG 1 showing 51 (66. 2%) distorted loci of the total loci of 78 (Table 1). The center of this distortion region was at the MSY at the position of 47.0 cM on LG 1, which included 11 sex-cosegregating markers perfectly fitting the 2:1 segregation ratio (Figure 1, Table 1). These SSR markers were tested against 1:2:1 segregation ratio and thus marked as distorted, because they were coded as codominant markers. These distorted markers either completely lack or show severe deficiency of the homozygous pollen-donor SunUp genotype. The second distorted region was on LG 6, containing 24 (22.6%) (Table 1) distorted loci, where the center of distortion was at the position of 79.0 cM near one end of this group (Figure 1). This region showed segregation distortion with a clear deficiency of heterozygote genotype and in favor of homozygous SunUp genotype.
Another notable phenomenon was the high percentages of distorted and dominant markers: 12 (50%) of the 24 markers mapped on the three short groups of LGs 10–12 (Table 1).
SSR markers distribution:
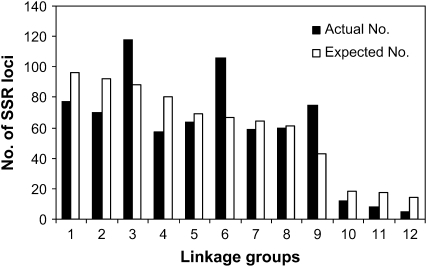
The distribution of the SSR markers varied over the 9 major linkage groups. The larger linkage groups, LGs 1, 2, and 4, had lower marker densities of 1.88, 1.98, and 2.12 cM/marker; whereas the relatively shorter linkage groups, LGs 6 and 9, had higher marker densities of 0.95 and 0.87 cM/marker, respectively. The number of markers per major linkage group ranged from 57 to 116; the length of the major linkage groups ranged from 64.5 to 145.0 cM (Table 1). Chi-square tests of the number of markers mapped to each linkage group indicated a significant deviation from what was anticipated on the basis of the linkage group length (χ2 = 51.14, P ≤ 0.001 in 12 LGs; χ2 = 40.25, P ≤ 0.001 in 9 major LGs). The mapped markers were not evenly distributed (Figure 3). The clustering of SSR loci were observed in each of the 9 major linkage groups, 20 clusters (≥5 loci/cM) were identified on the 9 major linkage groups, including the sex-cosegregating markers and a cluster on LG 10 (Figure 1; Table 1; supplemental Table S1 at http://www.genetics.org/supplemental/). The largest cluster, including 23 loci wthin a 2-cM interval, was located on LG 8. Also, 42 gaps having a distance >5 cM between adjacent markers were spread across the 9 major linkage groups. Among them, 8 gaps with a distance >10 cM between adjacent markers occurred on 5 major linkage groups: one each on LGs 4, 5, and 8; two on LG 1; and three on LG 2 (Figure 1; Table 1; supplemental Table S1).
Figure 3.—
Distribution of expected and actual numbers of SSR loci on 12 LGs.
DISCUSSION
Our high-density genetic map, based on sequence-tagged SSR markers, significantly improves the capacity for structural and functional analyses of the papaya genome. This map includes 706 (97.2%) of the 726 SSR markers used for map construction, indicating that a vast majority of the papaya genome has been represented. The codominant nature of SSR markers significantly increased the resolution and accuracy of previous papaya genetic maps, particularly the map of the papaya sex chromosomes (more details below). Suppression of recombination at the male-specific region of the Y chromosome was validated once again, albeit the suppression of recombination was much less pronounced with SSRs than in the map based on dominant AFLP markers (Ma et al. 2004). In addition, suppression of recombination at the centromeric region was notable in all nine major linkage groups. This DNA sequence-tagged map is being used for integration of genetic and physical maps and for aligning whole-genome shotgun sequences to papaya chromosomes. The BAC end and subclone sequence-derived SSRs immediately connect the genetic and physical maps and serve as anchor markers for assigning supercontigs and scaffolds to linkage groups. The WGS sequence-derived SSRs also anchor the scaffolds to linkage groups through the contigs containing the mapped SSRs. This linkage map provides a framework for gene tagging and map-based cloning. An evenly distributed minimum set of markers can be chosen for mapping quantitative trait loci (QTL) controlling economic and agronomic traits, and linked markers can be used for marker-assisted selection. Sex-cosegregated SSR markers are being used to identify BACs for closing the gaps on the physical maps of the MSY and its corresponding region of the X chromosome.
This sequence-tagged linkage map is being integrated with the papaya physical map and whole-genome shotgun sequence. The papaya physical map was based on a high-information-content fingerprinting method that produces high-resolution fingerprints (Luo et al. 2003) and overgo probes strategically selected from conserved Arabidopsis and Brassica sequences (Q. Yu, P. H. Moore, A. H. Paterson and R. Ming, unpublished data). These anchored Arabidopsis and Brassica sequences linked the papaya genome to other Brassicales genomes. The sequence-based papaya genetic map with anchored genome sequence scaffolds is an effective tool for studying macrosynteny and genome evolution in Brassicales, and papaya is an excellent outgroup for comparison with North American and European Brassicas.
Suppression of recombination in the MSY:
The largest cluster of cosegregating markers on LG 1 is still the MSY that is completely suppressed for recombination (Liu et al. 2004; Ma et al. 2004). However, the 14% sex-cosegregating SSR markers on LG 1 of the map represented a dramatic reduction from the 66% sex-cosegregating AFLP markers previously reported (Ma et al. 2004). SSR markers are known to be located near low-copy or genic sequences (Cardle et al. 2000; Morgante et al. 2002), whereas AFLP markers are distributed randomly in both genic and nongenic regions (Vos et al. 1995). The MSY region of the papaya Y chromosome is known to be extremely gene poor (Yu et al. 2007a), which explains the low abundance of SSR markers. The X and Y sequence divergence is extensive with only 83–87% of the sequences sharing homology (Yu et al. 2007b). This wide divergence provides the molecular basis for the suppression of recombination in the MSY and for the large cluster of polymorphic AFLP markers that were generated by a combination of a 6-base and a 4-base restriction enzyme digestion producing polymorphic target AFLP fragments ranging from 25 to 500 bp (Vos et al. 1995).
A notable feature of the AFLP linkage map of LG 1, the sex chromosome, was the complete presence of dominant markers from the pollen-donor parent SunUp and the absence of markers from the female parent Kapoho (Ma et al. 2004). This bias is due to the unique situation caused by the abortion of the YhYh genotype (homozygous-dominant markers of SunUp), resulting in 2:1 dominant:recessive pollen-donor parent SunUp markers and 3:0 dominant:recessive female parent Kapoho markers (Ma et al. 2004). In this map of mostly codominant SSR markers, five dominant markers derived from the female parent AU9 and one dominant marker from the pollen-donor parent SunUp were mapped. Examination of the genotypes of the five AU9 dominant markers revealed four, five, five, five, and seven recombinants, respectively, among 54 F2 plants. The four markers with either five or seven recombinants each had one recombinant in a single female plant, which was a rare conversion of a homozygous-dominant genotype to a homozygous-recessive genotype; the markers showing four recombinants had no recombination in the homozygous-dominant class (female plants). The rest of the recombinants of these AU9 dominant markers occurred in the heterozygous class (hermaphrodite plants) that changed to the homozygous-recessive class. This type of recombinant, which represents the majority of the recombinants of AU9 dominant markers linked to LG 1, can be distinguished by mapping with codominant markers, but not by mapping with dominant SunUp markers that mixed both homozygous-dominant SunUp (homozygous-recessive AU9 or Kapoho) and heterozygous markers.
SSR markers are usually codominant. Dominant SSR markers were scored mostly from bands in addition to the allelic target bands of the two parents. In a few cases, only one of the two allelic bands was robust and easy to score and in these cases were scored as dominant markers. The dominant SSR markers from multiple bands likely resulted from the residual heterozygosity within each parent. More AU9 dominant markers were mapped on LG 1 because of the greater residual heterozygosity in the improved but not released dioecious AU9 than in the gynodioecious cultivar SunUp that has undergone at least 25 generations of self-pollination (Storey 1969). Among the genomewide 56 dominant markers mapped, 49 (87.5%) were derived from AU9.
The complete suppression of recombination in the MSY region coupled with the loss of the homozygous YhYh genotypes skewed the segregation ratio and linked markers immediately surrounding the MSY region. In addition to the 11 sex-cosegregating SSRs showing a 2:1 segregation ratio, 39 SSRs showed segregation ratios strongly distorted from the expected 1:2:1 segregation ratio: 30 on one side and 9 on the other. Markers farther away from the MSY—57 cM at map position 104 cM of one side and 21 cM at map position 26 cM on the other side—recovered from segregation distortion to fit the expected 1:2:1 ratio. Theoretically, if a marker is 50 cM away from another marker on the same linkage group, it segregates independently, as if it were on a different chromosome. This was the case for markers on one side 57 cM away from the MSY. However, markers on the other side of the MSY recovered from segregation distortion at a distance of 21 cM, much less than the theoretical distance of 50 cM. It is possible that chromotin structure and features on the one side inhibited recombination so that the 21 cM on one side might represent a compatible physical distance of 57 cM on the other side. This possibility is supported by fluorescence in situ hybridization (FISH) mapping of MSY BACs that appear near the middle of the Y chromosome (Yu et al. 2007a), which is in contrast to the unequal genetic distance calculated on the two sides of MSY on LG 1.
Suppression of recombination in centromeric regions:
Another notable feature in this linkage map is the clustering of SSR markers on each of nine major linkage groups in the regions postulated to be centromeric. Each cluster contains more than five cosegregating markers (Table 1). An inhibition of meiotic recombination by centromeres was suggested first by Dobzhansky (1930), and the direct effect of centromeres on suppressing recombination has been demonstrated in yeast where a cloned centromere from the third chromosome (CEN3) has been shown to decrease recombination when it was artificially integrated into new sites in the genome (Lambie and Roeder 1986). Assuming a random distribution of markers, low levels of meiotic recombination would cause markers that are physically well separated to cluster on a linkage map. The clustering of markers in centromeric regions was recognized and physically verified in the genetic maps of tomato and potato (Tanksley et al. 1992), rice (Harushima et al. 1998; Chen et al. 2002), and barley (Ramsay et al. 2000). In Drosophila, up to a 40-fold suppression of recombination has been reported near the centromeres (Roberts 1965). In rice, the physical distance per centimorgan varies with position along the chromosome; the distance averaged 244 kb for the entire rice genome, but in the centromeric regions it was >1 Mb/cM (Chen et al. 2002).
The largest cluster of markers on LG 1 was at the MSY. FISH of MSY BACs mapped the MSY near the centromere; sequence analysis of selected MSY BACs hinted that the MSY might be on only one side of the centromere and not include it (Yu et al. 2007a).
SSR linkage map:
Because SSRs are abundant, codominant, and cost effective for large-scale genetic and QTL mapping projects, there has been considerable effort toward developing microsatellite maps in a variety of plant species. Such maps are already available in major crop plants that have been the subjects in recent years of significant investments to generate genomic resources for crop improvement, including rice (McCouch et al. 2002), maize (Sharopova et al. 2002), wheat (Somers et al. 2004; Song et al. 2005), barley (Ramsay et al. 2000), soybean (Song et al. 2004), and sorghum (Menz et al. 2002). The papaya SSR genetic map was constructed for the papaya genome-sequencing project, which was justified by its agricultural importance in the tropics and its unique reproductive biology. Benefiting from the enormous amount of BAC end and whole-genome shotgun sequences, we constructed a high-density linkage map with an average distance of 1.5 cM between adjacent markers, which is 50% denser than the previous papaya AFLP high-density map that had an average interval of 2.2 cM (Ma et al. 2004). Another major improvement is that this SSR map was able to break the large LG 2 of the AFLP map that likely represented more than one chromosome. The high-quality codominant SSR markers enhanced the accuracy of the linkage map as shown by nine major LGs for the nine pairs of chromosomes.
The papaya SSR map of 1068.9 cM with 707 markers is much more compact than the 3294.2 cM AFLP map with 1501 markers. The threefold reduction of accumulated genetic distance is attributed mainly to the high resolution of codominant SSR markers that separate the three classes of genotypes in the F2 population, which contrasts with the dominant AFLP markers that mix the homozygous-dominant and heterozygous classes to calculate inflated map distances. The AFLP map was constructed from an F2 population of closely related parents, Kapoho and SunUp, which would assure a high recombination rate and thus increase genetic distance (Kim et al. 2002; Ma et al. 2004). The parents of the F2 population used for our SSR map are more distantly related (Kim et al. 2002) and expected to have a lower recombination rate. Another major reason for the inflated genetic distance is missing data as can be seen by the 1-cM genetic distance caused by two missing data points in the genotype of otherwise perfectly sex-cosegregating marker P3K2981YC0 on LG 1. Any markers with more than five missing data points were eliminated in the SSR data set; this limited any artificial inflation of map distances. Finally, the function of “suspected linkage” in JoinMap 3.0 helped eliminate problematic markers that tend to expand genetic distances; a “suspected linkage” function was not available in MAPMAKER 3.0 (Lander et al. 1987) that was used for constructing the AFLP map.
Segregation distortion:
A segregation distorter (SD) gene produces a bias in normal segregation to favor itself, so that the genotype frequency of this gene is increased in a segregating population. The SD system was found first in Drosophila and has been studied extensively (Lyttle 1991). It has also been found in many plant species, including rice (Xu et al. 1997), wheat (Farisa et al. 1998), maize (Lu et al. 2002), barley (Kleinhofs et al. 1993), and coffee (Ky et al. 2000). In our high-density SSR map, only two regions on the nine major LGs showed significant segregation distortion. The first was the MSY region on LG 1, caused by postzygotic selection through the abortion of the YY embryo at 25–50 days after pollination (discussed above). The other distorted segregation region was on LG 6 containing 24 distorted loci (22.6%) spanning 8 cM. The center of this LG 6 distorted region was at 79.0 cM with 10 cosegregating distorted markers. Examining the genotypes of these distorted markers revealed a clear deficiency of heterozygote classes and a high frequency of homozygous SunUp genotypes. It is unknown which genes possess a selective advantage as a homozygous SunUp genotype and whether these genes are associated with abortion of the YY genotype.
In addition to these two distorted regions, another 38 SSRs (5.4%) showed segregation distortion sporadically distributed across the other 10 LGs. These distorted markers could be random events, which is a common feature in plant and animal chromosomes (Taylor and Ingvarsson 2003).
A high percentage of distorted markers was observed on the three small LGs 10–12, even though these three LGs contained only 24 (3.4%) markers on them (Table 1). The high rate of distorted markers on these three LGs might be the consequence of genetic or physical properties in these regions of certain chromosomes that prevented the linkage between these small LGs to the nine major LGs corresponding to individual chromosomes.
SSR distribution in the papaya genome:
The distribution of papaya SSR markers over the nine major linkage groups varied significantly. The marker density ranged from 0.87 to 2.12 cM/interval and chi-square tests of the number of markers mapped to each linkage group indicated a significant deviation from what was expected on the basis of linkage group length. Forty-two gaps with a distance ≥5 cM between adjacent markers were distributed across the nine major linkage groups and, among them, eight gaps ≥10 cM resided on LGs 1, 2, 4, 5, and 8. On the other hand, 19 SSR loci clusters (≥5 loci/cM) were observed in each of the nine major linkage groups, including the cosegregation markers. This phenomenon also occurs in mammals (Dib et al. 1996; Dietrich et al. 1996; Ihara et al. 2004) and many crop plant species (Ramsay et al. 2000; Temnykh et al. 2001; Menz et al. 2002; Sharopova et al. 2002; Song et al. 2004; 2005).
SSRs are not uniformly distributed cross the eukaryotic genomes due to a nonrandom physical distribution of SSRs across the chromosomes (Ramsay et al. 2000; Song et al. 2005). It has been shown that microsatellites are preferentially associated with nonrepetitive DNA and significantly associated with the low-copy fraction of plant genomes on the basis of the estimation of microsatellite density in Arabidopsis thaliana, rice, soybean, maize, and wheat (Cardle et al. 2000; Morgante et al. 2002). Among these species, the overall frequency of microsatellites was negatively correlated with genome size and with the proportion of repetitive DNA. In papaya and other species, the normally recombining regions might represent euchromatic (i.e., gene-rich) regions, while the regions suppressed for recombination represent heterochromatic regions with abundant repetitive sequences.
SSR polymorphism rate:
A previous SSR polymorphism survey resulted in 23.4% polymorphic markers between the parental varieties AU9 and SunUp (Eustice et al. 2008). However, a significant portion of these polymorphism markers detected a subtle difference between 1 and 3 bp. Initially, fluorescent-tagged SSR primers were designed and ordered to map those markers in sequencing gels using a Li-Cor 4300 DNA analyzer, but this practice is slow and costly. This type of marker would be less useful for papaya researchers and breeders. This practice was stopped and only polymorphic markers that can be separated by agarose gels were scored and selected for mapping. Among the 13.3% polymorphic markers selected for genotyping, a fraction of the markers yielded faint bands, abnormal banding patterns, or no polymorphism among progenies. At the end, 10.1% polymorphic markers were scored for genetic mapping. Despite the reduction of polymorphism rate from the initial estimate, SSR markers showed a reasonably high polymorphism rate for a self-pollinated species.
Acknowledgments
We thank Gail L. Uruu, Lydia Fang, Jan Murray, Jianping Wang, Jong-Kuk Na, Andrea Gschwend, and Andrew Wood for technical assistance. This project was supported by a U. S. Department of Agriculture (USDA) T-STAR grant through the University of Hawaii, a USDA-Agricultural Research Service Cooperative Agreement (CA 58-3020-8-134) with the Hawaii Agriculture Research Center, and startup funds from the University of Illinois at Urbana-Champaign.
References
- Arumuganathan, K., and E. D. Earle, 1991. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 93: 208–219. [Google Scholar]
- Cardle, L., L. Ramsay, D. Milbourne, M. Macaulay, D. Marshall et al., 2000. Computational and experimental characterization of physically clustered simple sequence repeats in plants. Genetics 156: 847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., G. Presting, W. B. Barbazuk, J. L. Goicoechea, B. Blackmon et al., 2002. An integrated physical and genetic map of the rice genome. Plant Cell 14: 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib, C., S. Fauré, C. Fizames, D. Samson, N. Drouot et al., 1996. A comprehensive genetic map of the human genome based on 5264 microsatellites. Nature 380: 152–154. [DOI] [PubMed] [Google Scholar]
- Dietrich, W. F., J. C. Miller, R. G. Steen, M. Merchant, D. Damron-Boles et al., 1996. A comprehensive genetic map of the mouse genome. Nature 380: 149–152. [DOI] [PubMed] [Google Scholar]
- Dobzhansky, T., 1930. Translocations involving the third and fourth chromosomes of Drosophila melanogaster. Genetics 15: 347–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustice, M., Q. Yu, C. W. Lai, S. Hou, J. Thimmapuram et al., 2008. Development and application of microsatellite markers for genomic analysis of papaya. Tree Genet. Genomics (in press).
- Farisa, J. D., B. Laddomada and B. S. Gilla, 1998. Molecular mapping of segregation distortion loci in Aegilops tauschii. Genetics 149: 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima, Y., M. Yano, A. Shomura, M. Sato, T. Shimano et al., 1998. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148: 479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara, N., A. Takasuga, K. Mizoshita, H. Takeda, M. Sugimoto et al., 2004. A comprehensive genetic map of the cattle genome based on 3802 microsatellites. Genome Res. 14: 1987–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. S., P. H. Moore, F. Zee, M. M. Fitch, D. L. Steiger et al., 2002. Genetic diversity of Carica papaya as revealed by AFLP markers. Genome 45: 503–512. [DOI] [PubMed] [Google Scholar]
- Kleinhofs, A., A. Kilian, M. A. Saghai, R. M. Biyashev, P. Hayes et al., 1993. A molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theor. Appl. Genet. 86: 705–712. [DOI] [PubMed] [Google Scholar]
- Ky, C.-L., P. Barre, M. Lorieux, P. Trouslot, S. Akaffou et al., 2000. Interspecific genetic linkage map, segregation distortion and genetic conversion in coffee (Coffea sp.). Theor. Appl. Genet. 101: 669–676. [Google Scholar]
- Lambie, E. J., and G. S. Roeder, 1986. Repression of meiotic crossing over by a centromere (Cen3) in Saccharomyces cerevisiae. Genetics 114: 769–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., P. Green, J. Abrahamson, A. Barlow, M. J. Daly et al., 1987. Mapmaker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Liu, Z., P. H. Moore, H. Ma, C. M. Ackerman, R. Makandar et al., 2004. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 427: 348–352. [DOI] [PubMed] [Google Scholar]
- Lu, H., J. Romero-Severson and R. Bernardo, 2002. Chromosomal regions associated with segregation distortion in maize. Theor. Appl. Genet. 105: 622–628. [DOI] [PubMed] [Google Scholar]
- Luo, M. C., C. Thomas, F. M. You, J. Hsiao, S. Ouyang et al., 2003. High-throughput fingerprinting of bacterial artificial chromosomes using the SNaPshot labeling kit and sizing of restriction fragments by capillary electrophoresis. Genomics 82: 378–389. [DOI] [PubMed] [Google Scholar]
- Lyttle, T. W., 1991. Segregation distorters. Annu. Rev. Genet. 25: 511–557. [DOI] [PubMed] [Google Scholar]
- Ma, H., P. H. Moore, Z. Liu, M. S. Kim, Q. Yu et al., 2004. High-density linkage mapping revealed suppression of recombination at the sex determination locus in papaya. Genetics 166: 419–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCouch, S. R., L. Teytelman, Y. Xu, K. B. Lobos, K. Clare et al., 2002. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9: 199–207. [DOI] [PubMed] [Google Scholar]
- Menz, M. A., R. R. Klein, J. E. Mullet, J. A. Obert, N. C. Unruh et al., 2002. A high-density genetic map of Sorghum bicolor (L.) Moench based on 2926 AFLP, RFLP and SSR markers. Plant Mol. Biol. 48: 483–499. [DOI] [PubMed] [Google Scholar]
- Ming, R., Q. Yu and P. H. Moore, 2007. Sex determination in papaya. Semin. Cell Dev. Biol. 18: 401–408. [DOI] [PubMed] [Google Scholar]
- Morgante, M., and A. M. Olivieri, 1993. PCR-amplified microsatellites as markers in plant genetics. Plant J. 1: 175–182. [PubMed] [Google Scholar]
- Morgante, M., M. Hanafey and W. Powell, 2002. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 30: 194–200. [DOI] [PubMed] [Google Scholar]
- Ramsay, L., M. Macaulay, S. Ivanissevich, K. MacLean, L. Cardle et al., 2000. A simple sequence repeat-based linkage map of barley. Genetics 156: 1997–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, P. A., 1965. Difference in the behavior of eu- and hetero-chromatin: crossing over. Nature 205: 725–726. [DOI] [PubMed] [Google Scholar]
- Sharopova, N., M. D. McMullen, L. Schultz, S. Schroeder, H. Sanchez-Villeda et al., 2002. Development and mapping of SSR markers for maize. Plant Mol. Biol. 48: 463–481. [DOI] [PubMed] [Google Scholar]
- Somers, D. J., P. Isaac and K. Edwards, 2004. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 109: 1105–1114. [DOI] [PubMed] [Google Scholar]
- Sondur, S. N., R. M. Manshardt and J. I. Stiles, 1996. A genetic linkage map of papaya based on randomly amplified polymorphic DNA markers. Theor. Appl. Genet. 93: 547–553. [DOI] [PubMed] [Google Scholar]
- Song, Q. J., L. F. Marek, R. C. Shoemaker, K. G. Lark, V. C. Concibido et al., 2004. A new integrated genetic linkage map of the soybean. Theor. Appl. Genet. 109: 122–128. [DOI] [PubMed] [Google Scholar]
- Song, Q. J., J. R. Shi, S. Singh, E. W. Fickus, J. M. Costa et al., 2005. Development and mapping of microsatellite (SSR) markers in wheat. Theor. Appl. Genet. 110: 550–560. [DOI] [PubMed] [Google Scholar]
- Storey, W. B., 1969. Papaya, pp. 389–408 in Outlines of Perennial Crop Breeding in the Tropics, edited by F. P. Ferwerda and F. H. Wit. Veenman & Zonen N. V., Wageningen, The Netherlands.
- Tanksley, S. D., M. W. Ganal, J. P. Prince, M. C. de Vicente, M. W. Bonierbale et al., 1992. High density molecular linkage maps of the tomato and potato genomes. Genetics 132: 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz, D., 1989. Hypervariability of simple sequences as general source for polymorphic DNA markers. Nucleic Acids Res. 17: 6463–6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, D. R., and P. K. Ingvarsson, 2003. Common features of segregation distortion in plants and animals. Genetica 117: 27–35. [DOI] [PubMed] [Google Scholar]
- Temnykh, S., G. DeClerck, A. Lukashova, L. Lipovich, S. Cartinhour et al., 2001. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 11: 1441–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen, J. W., and R. E. Voorrips, 2001. JoinMap 3.0: software for the calculation of genetic linkage maps. Plant Research International, Wageningen, The Netherlands.
- Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee et al., 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., J. L. Weber, G. Zhong and S. D. Tanksley, 1994. Survey of plant short tandem DNA repeats. Theor. Appl. Genet. 88: 1–6. [DOI] [PubMed] [Google Scholar]
- Weber, J. L., and P. E. May, 1989. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am. J. Hum. Genet. 44: 388–396. [PMC free article] [PubMed] [Google Scholar]
- Wikström, N., V. Savolainen and M. W. Chase, 2001. Evolution of the angiosperm: calibrating the family tree. Proc. Biol. Sci. 268: 2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., L. Zhu, J. Xiao, N. Huang and S. R. McCouch, 1997. Chromosomal regions associated with segregation distortion of molecular markers in F2, backcross, doubled haploid, and recombinant inbred populations in rice (Oryza sativa L.). Mol. Gen. Genet. 253: 535–545. [DOI] [PubMed] [Google Scholar]
- Yu, Q., S. Hou, R. Hobza, F. A. Feltus, X. Wang et al., 2007. a Chromosomal location and gene paucity of the male specific region on papaya Y chromosome. Mol. Genet. Genomics 278: 177–185. [DOI] [PubMed] [Google Scholar]
- Yu, Q., S. Hou, F. A. Feltus, M. R. Jones, J. Murray et al., 2007. b Low X/Y divergence in four pairs of papaya sex-liked genes. Plant J. (in press). [DOI] [PubMed]