Abstract
In plants, many major regulatory genes that control plant growth and development have been identified and characterized. Despite a detailed knowledge of the function of these genes little is known about how they contribute to the natural variation for complex traits. To determine whether major regulatory genes of maize contribute to standing variation in Balsas teosinte we conducted association mapping in 584 Balsas teosinte individuals. We tested 48 markers from nine candidate regulatory genes against 13 traits for plant and inflorescence architecture. We identified significant associations using a mixed linear model that controls for multiple levels of relatedness. Ten associations involving five candidate genes were significant after correction for multiple testing, and two survive the conservative Bonferroni correction. zfl2, the maize homolog of FLORICAULA of Antirrhinum, was associated with plant height. zap1, the maize homolog of APETALA1 of Arabidopsis, was associated with inflorescence branching. Five SNPs in the maize domestication gene, teosinte branched1, were significantly associated with either plant or inflorescence architecture. Our data suggest that major regulatory genes in maize do play a role in the natural variation for complex traits in teosinte and that some of the minor variants we identified may have been targets of selection during domestication.
PLANT developmental geneticists have identified and characterized a large number of major regulatory genes that control diverse aspects of plant growth and form. These genes are known via major mutants that can dramatically transform a plant—for example, tall to dwarf, branched to unbranched, or bisexual to unisexual. What is unknown is the extent to which these major regulatory genes contribute to the small phenotypic differences between plants in natural populations. Previous studies in animals have provided evidence that major regulatory genes contribute to the natural variation for complex traits. For example, in Drosophilia melanogaster, scabrous, a signaling protein involved in lateral inhibition of the developing nervous system, also has a large effect on sternoplural and abdominal bristle number (Lai et al. 1994). In humans, genetic variation within T(Brachyury), a nortochord gene involved in posterior axis development, associates with susceptibility to neural tube defects such as spina bifida and anencephaly (Morrison et al. 1996, 1998). By conducting association mapping in Balsas teosinte, we hope to determine if major regulatory genes of maize contribute to natural variation in its wild ancestor. This study will also allow us to investigate if teosinte branched1 (tb1), a major regulatory gene involved in the domestication of maize, contributes to natural variation in Balsas teosinte.
Association mapping, also known as linkage disequilibrium (LD) mapping, is a powerful tool for the genetic dissection of complex traits (Long and Langley 1999; Gupta et al. 2005). Association mapping involves the identification of statistical associations between genotypic and phenotypic variation. Unlike conventional quantitative trait locus (QTL) mapping, association mapping utilizes historical recombination events within a population, allowing much higher mapping resolution. Recently, association mapping has been applied to a wide range of organisms and phenotypes, including traits related to human health (Wellcome Trust Case Control Consortium 2007), agricultural performance in maize and other crops (Wilson et al. 2004), classical quantitative traits in Drosophila (Lai et al. 1994), and adaptive traits in natural populations of mice (Nachman et al. 2003).
Despite these successes, a major concern for association mapping is the occurrence of false-positive associations due to population structure or unrecognized familial relatedness (coancestry) among individuals. If genetic variants that do not influence phenotype are distributed among populations or individuals in a manner similar to that of the causal genetic variants, then these noncausal variants can be falsely associated with the trait. There are multiple analytical methods to reduce the frequency of false-positive associations such as using genomic control to adjust the raw test statistic (Devlin and Roeder 1999), incorporation of population structure into a likelihood-ratio test comparing the likelihood of the null hypothesis of genotype–phenotype independence to the likelihood of dependence (Pritchard et al. 2000), and incorporation of both population structure and familial relatedness into a mixed linear model (Yu et al. 2006; Zhao et al. 2007).
Maize (Zea mays ssp. mays) and its wild ancestor, Balsas teosinte (Z. mays ssp. parviglumis), are an attractive system for study of the inheritance of complex traits as well as domestication for several reasons. First, teosinte grows in large populations over a broad region of southwestern Mexico and harbors high levels of molecular genetic variation (Fukunaga et al. 2005; Wright et al. 2005). Second, LD in teosinte generally degrades to near background levels within a single gene (A. Weber and J. Doebley, unpublished results), and thus markers that associate with traits are apt to be within or immediately nearby causal genes. Third, there is strong interest in understanding the genetic control of phenotypic variation in teosinte since it is this pool of variation from which maize was selected. Fourth, genes controlling domestication traits may be fixed for a single functional allele in maize, making it impossible to perform association genetics with these genes in maize itself. Fifth, association mapping in teosinte can be used to identify favorable alleles that were lost during domestication (Tanksley and McCouch 1997).
In this study, we performed tests of associations between markers in nine candidate regulatory genes and 13 complex traits in teosinte. For these analyses, we used a mixed linear model that incorporates both population structure and familial relatedness as inferred with a large set of randomly selected SNPs. This model was effective in eliminating false positives due to population structure or familial relatedness and had enough power to detect 10 significant marker–trait associations after a correction for multiple testing. We observed significant associations between markers in five of the nine candidate genes with various morphological traits that were likely under selection during domestication. Notably, five markers and two different genic regions in the regulatory gene teosinte branched1 (tb1), known to play a major role in domestication, were significantly associated with variation in either plant or inflorescence architecture. By identifying genetic variation associated with phenotypic variation in teosinte, we hope to better understand the role of major regulatory genes in the genetic architecture of complex traits. Finally, we describe the necessary resources for others to perform association mapping in Balsas teosinte.
MATERIALS AND METHODS
Teosinte sample:
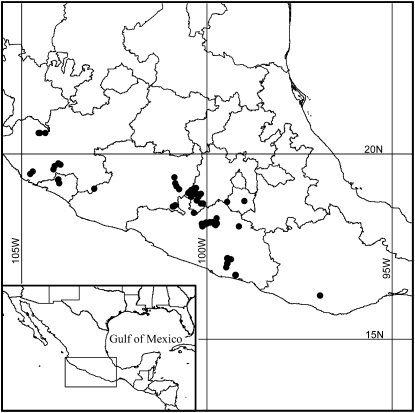
A sample of 584 plants of Z. mays ssp. parviglumis (Race Balsas) were grown in 2002–2003 on the island of Molokai, Hawaii. The seed for these plants came from 74 local populations that represent the entire geographic range of Balsas teosinte in Mexico (Figure 1; supplemental Table 1 at http://www.genetics.org/supplemental/). Our goal was to sample 592 plants in total (8 from each local population); however, 8 plants with substantial missing data were dropped from the analysis, giving a total of 584. The field was divided into two blocks each with 74 randomized plots containing 4 plants each. The plots within each block were arranged in a grid that was 18 plots (columns) wide and 10 plots long (rows).
Figure 1.—
Map showing region of Mexico where the local populations of Balsas teosinte were sampled. Each dot represents the location of one of the 74 local populations from which the seed for the 584 plants were collected.
Phenotypes:
Thirteen phenotypes were measured that can be grouped into three categories: flowering time (days to pollen, POLL; and days to silk, SILK), plant architecture (branch length, BRLN; mean lateral branch internode length, LBIL; plant height, PLHT; and tiller number, TILL), and inflorescence architecture (fruitcase weight, FCWT; proportion of female ear length, FELN; female ear length, FERL; lateral inflorescence branch number, LIBN; number of female cupules, NFCP; proportion of female cupules, PRFCP; and staminate score, STAM) (Table 1). All of the primary lateral branch and primary lateral inflorescence traits were measured on the uppermost well-formed lateral branch of the plant.
TABLE 1.
List of traits analyzed
| Trait | Descriptiona | Units |
|---|---|---|
| Branch length (BRLN) | Length of the lateral branch not including the length of the inflorescence and peduncle at the tip of that branch | Centimeter |
| Female ear length (FERL) | Length of the female portion of the basal-most ear on the lateral branch | Millimeter |
| Fruitcase weight (FCWT) | Average fruitcase weight based on 50 mature fruitcases | Gram |
| Mean lateral branch internode length (LBIL) | Length of the lateral branch divided by the number of internodes in that branch | Centimeter |
| Lateral inflorescence branch number (LIBN) | Number of branches in the tassel or inflorescence that terminates the lateral branch | Count |
| Number of female cupules (NFCP) | Number of female cupules in the basal-most ear on the lateral branch | Count |
| Proportion of female cupules (PRFCP) | Ratio of the number of female cupules in the basal-most ear on the lateral branch divided by the total number of cupules (internodes) including any male portions | Proportion |
| Proportion of female ear length (FELN) | Length of the female portion of the basal-most ear on the lateral branch divided by the total length of the ear including any male spikelets | Proportion |
| Plant height (PLHT) | Length of the primary stalk from the ground to the tip of the primary tassel | Centimeter |
| Days to pollen (POLL) | Days from planting to first visible anthers on a single plant | Days |
| Days to silk (SILK) | Days from planting to first visible silks on a single plant | Days |
| Staminate score (STAM) | Fraction of the male spikelets in the inflorescence that terminates the lateral branch | Percentage |
| Tiller number (TILL) | Number of tillers at time of pollen shed | Count |
All lateral branch traits were measured on the uppermost well-formed lateral branch.
Genotyping:
Two classes of genes were assayed: random and candidate. A set of 495 random genes was picked from ∼10,000 low-copy-number maize ESTs without consideration as to gene function or gene type (Gardiner et al. 2004). These genes were sequenced using a discovery panel that consisted of 14 maize inbred lines and 16 teosinte partial inbreds (Wright et al. 2005). A set of 706 SNPs was selected from sequence alignments for the random genes (supplemental Table 2 at http://www.genetics.org/supplemental/) and was used to control for population structure in the association analyses. Criteria for selecting the 706 random SNPs included (1) a minimum of 20 bp on one side of the target SNP that was devoid of polymorphism to allow for primer design, (2) base-call quality scores suggesting that the SNP of interest was not an artifact, and (3) a minor allele that was present in at least two individuals within the discovery panel. A set of 9 candidate regulatory genes was selected because they have possible effects on the phenotypes under study given their known mutant phenotype in maize or other plants (Table 2). We used a set of previously published sequence alignments for these genes (http://www.panzea.org). Because the candidate gene alignments come from diverse sources, the discovery panels were variable. Similar criteria to those used for random SNP selection were used in the selection of candidate gene SNPs (or other markers). A total of 48 markers including SNPs, indels, and cleaved-amplified polymorphic sequence (CAPS) were developed in the 9 candidate genes (supplemental Table 3 at http://www.genetics.org/supplemental/). DNA extractions and scoring of CAPS and indel markers on agarose gels were accomplished using standard procedures (Briggs et al. 2007). SNP genotyping was performed at Genaissance Pharmaceuticals using the Sequenome MassARRAY system (Jurinke et al. 2002). Sequence alignments and marker context sequences are available at http://www.panzea.org.
TABLE 2.
List of candidate genes analyzed
| Gene | Gene symbol | Description |
|---|---|---|
| dwarf plant8 | d8 | A transcription factor known to affect plant height and flowering time in maize (Peng et al. 1999) |
| indeterminate growth1 | id1 | A transcription factor that affects inflorescence architecture and flowering time in maize (Colasanti et al. 1998) |
| teosinte branched1 | tb1 | A transcription factor that affects branching and inflorescence architecture in maize (Doebley et al. 1997) |
| terminal ear1 | te1 | A RNA-binding gene known to affect inflorescence sex and plant architecture in maize (Veit et al. 1998) |
| tassel seed2 | ts2 | A short-chain alcohol dehydrogenase that affects inflorescence architecture in maize (Delong et al. 1993) |
| zea agamous-like1 | zagl1 | A MADS-box transcription factor homologous to SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 that affects flowering time in Arabidopsis (Samach et al. 2000) |
| zea apetala homolog1 | zap1 | A transcription factor homologous to the floral homeotic gene APETALA1 that affects inflorescence architecture in Arabidopsis (Mandel et al. 1992) |
| zea centroradialis1 | zen1 | A maize homolog of CENTRORADIALIS that affects inflorescence architecture and flowering time in Antirrhinum (Bradley et al. 1996) |
| zea floricaula leafy2 | zfl2 | A transcription factor that affects floral development and flowering time in Arabidopsis (Weigel et al. 1992) |
Population structure:
To determine the presence and extent of population structure for our sample of 584 plants, we evaluated several indicators of population structure using PowerMarker (Liu and Muse 2005). First, deviations from Hardy–Weinberg expectations for each of the 706 random SNPs were assessed using Fisher's exact test. Second, FST was used to measure the extent of differentiation among local populations. Confidence intervals for FST were generated using 10,000 bootstrap resamplings over loci. Third, FIS was calculated as a measure of recent coancestry among individuals within local populations. Again, 10,000 bootstrap resamplings over loci were used to generate confidence intervals. Fourth, we assessed the degree of correlation between geographic and genetic distance since population structure resulting from isolation-by-distance would produce such a correlation. Great circle distances between individuals using latitude and longitude were calculated using an R module (Nychka 2007). The correlation coefficient between the geographic and the genetic (negative log of the proportion of shared alleles) distances was computed and its significance evaluated with the Mantel test (10,000 permutations).
Principal component analysis and a kinship matrix were computed to control for population structure and recent coancestry, respectively (Zhao et al. 2007). Principal component analysis was conducted with the random markers using the program EIGENSTRAT (Price et al. 2006). We eliminated 51 of the random 706 SNPs because they were in high LD (r2 > 0.5) with another random SNP. The r2 values were calculated using PowerMarker. The remaining set of 655 SNPs was used for principal component analysis. The statistical significance of each principal component vector generated by EIGENSTRAT was assessed by comparing the corresponding transformed eigenvalue with the Tracy–Widom distribution (Patterson et al. 2006). This analysis identified 29 significant principal components that we are using to describe population structure. To correct for recent coancestry or familial relatedness, a kinship matrix composed of the proportion of shared alleles for all pairwise combinations of the 584 plants was generated. These values were calculated with PowerMarker using the full set of 706 random SNPs.
Testing of marker–trait associations:
A mixed linear model was used to test marker–trait associations,
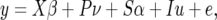 |
where y is a vector of phenotypic values, β is a vector of fixed effects concerning the position of plants within the field, ν is a vector of fixed effects regarding population structure, α is the fixed effect for the candidate marker, u is a vector of the random effects pertaining to recent coancestry, and e is a vector of residuals. X describes the position of plants within the field and includes a row, a column, and a row–column interaction term (see above). P is a matrix of the 29 significant principal component vectors. S is the vector of genotypes at the candidate marker, and I is an identity matrix. The variances of the random effects are assumed to be Var(u) = 2KVg and Var(e) = IVR, where K is the kinship matrix consisting of the proportion of shared allele values, I is an identity matrix, Vg is the genetic variance, and VR is the residual variance. For markers that were significantly associated with a trait, a general linear model with all of the fixed-effect terms described above was used to estimate the amount of phenotypic variation explained by each of the candidate markers, as measured by R2. Significant associations were also tested with a reduced mixed linear model that included only 5 of the 29 principal components to verify that they were not artifacts of overfitting the model. If two markers associated with the same trait, the above model was expanded to test for epistasis by including two Sα terms for the two markers, as well as an interaction term to test for epistasis between the two markers.
This mixed linear model was used to test 523 marker–trait pairs of the 624 possible pairs (13 traits × 48 markers). Not all markers were tested against all traits. Rather, knowledge of the known functions or mutant phenotypes of the candidate genes was used to determine which marker–trait pairs were appropriate to test (supplemental Table 4 at http://www.genetics.org/supplemental/). For each marker–trait association, the model was run in SAS using PROC MIXED (SAS Institute 1999). The F test, with the denominator degrees of freedom determined by the Satterthwaite method, was used to assess the significance of the marker effect of each marker–trait pair. No pattern was found in any of the resulting residual plots, verifying that no transformation of the phenotypic data was necessary (data not shown). The false discovery rate was used to correct for multiple testing (Storey 2002; Storey and Tibshirani 2003). LD, as described by r2 values among candidate markers in the same gene, was assessed using PowerMarker.
RESULTS
Population structure:
Several measures of population structure using the 706 random SNPs indicated that our sample of 584 Balsas teosinte plants does not represent a single unstructured Hardy–Weinberg population. First, the genotypic frequencies at 665 of these 706 SNPs show significant deviations from Hardy–Weinberg expectations (P < 0.05; data not shown). Second, population differentiation (FST) among the 74 local populations is high (FST = 0.1690 ± 0.005). Third, FIS is also relatively high (FIS = 0.0999 ± 0.011), suggesting more frequent matings among relatives than expected by chance. Fourth, we observed a strong correlation (r = 0.4605, P < 0.001) between geographical and genetic distance, indicating that genetic variation is geographically structured in Balsas teosinte.
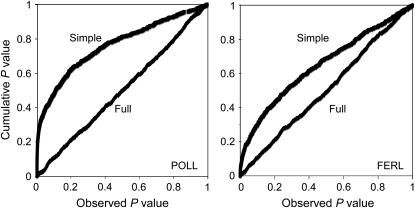
Since our sample of 584 Balsas teosinte plants shows indications of a considerable degree of population structure, we assessed the ability of a mixed linear model that incorporates both measures of population structure and recent coancestry among individuals to reduce the number of false-positive associations. A plot of the ranked raw P-values by the cumulative P-values for associations between our 706 random SNPs and POLL and FERL using the simple model that does not incorporate population structure shows an excess of small P-values as indicated by the curved line in the P–P plot (Figure 2; supplemental Table 4 at http://www.genetics.org/supplemental/). However, when the associations are tested using the full model that includes terms for population structure (the 29 principal components) and recent coancestry (the K matrix), the P-values fall along a diagonal line, indicating that our random SNPs follow the expected distribution under the null hypothesis of independence between the SNP and the phenotype. We conclude, therefore, that the full model is effective at reducing the number of false-positive associations.
Figure 2.—
Plots of the cumulative distributions of P-values for the associations between our 706 random SNPs and days to pollen shed (POLL) and female ear length (FERL). The curved line within each plot represents the cumulative distribution of P-values under a simple model that does not incorporate population structure or recent coancestry. The diagonal line represents the cumulative distribution of P-values under the full model that includes 29 principal components that account for population structure and a kinship matrix that accounts for recent coancestry. The P-value distribution for the full model follows the expected distribution under the null hypothesis of independence between the SNPs and the traits.
Genetic associations:
We used the full mixed linear model incorporating the principal components and kinship matrix to test for associations between 48 markers in nine candidate regulatory genes and 13 traits. The traits measured included those related to plant and inflorescence architecture, as well as to flowering time (Table 1; supplemental Table 4 at http://www.genetics.org/supplemental/). The nine candidate genes were selected on the basis of known function or mutant phenotype that, in maize and/or other species, suggests they might control traits measured in this study (Table 2). Only 523 marker–trait pairs of the 624 possible pairs were tested (supplemental Table 5 at http://www.genetics.org/supplemental/). The selection of which marker–trait pairs to test was based upon a priori knowledge of the known function of a candidate gene or its mutant phenotype.
Thirty-seven of the 523 marker–trait pairs demonstrated detectable associations (P ≤ 0.05, supplemental Table 6 at http://www.genetics.org/supplemental/). Ten of these 37 associations are significant after correction for multiple testing (Q ≤ 0.10) using the false discovery rate (Table 3). To verify that these significant associations are not artifacts of overfitting the model with 29 principal components, we retested these associations with a reduced model that included only 5 principal components and the kinship matrix. All 10 remained significant and survived the correction for multiple testing with the reduced model. Two of these associations, an association between zfl2.3 and plant height and an association between zen1.1 and branch length, are significant after the conservative Bonferroni correction. The 10 significant associations include 10 markers from five of the nine candidate genes and 4 of the 13 traits. Below we discuss these 4 traits.
TABLE 3.
List of significant marker–trait pairs after correction for multiple testing
| Trait | Gene | Marker | Na | F | R2 | P | FDR Q-valueb |
|---|---|---|---|---|---|---|---|
| PLHT | zfl2 | zfl2.3 | 501 | 7.53 | 0.028 | 0.0006 | 0.0090 |
| BRLN | zen1 | zen1.1 | 449 | 7.43 | 0.028 | 0.0007 | 0.0202 |
| BRLN | tb1 | tb1.19 | 440 | 6.11 | 0.026 | 0.0024 | 0.0347 |
| BRLN | tb1 | tb1.15 | 449 | 5.70 | 0.021 | 0.0036 | 0.0347 |
| BRLN | tb1 | tb1.20 | 442 | 4.63 | 0.020 | 0.0103 | 0.0743 |
| FERL | te1 | te1.3 | 335 | 7.02 | 0.031 | 0.0011 | 0.0257 |
| FERL | tb1 | tb1.5 | 332 | 5.32 | 0.023 | 0.0052 | 0.0607 |
| FERL | te1 | te1.4 | 337 | 4.73 | 0.023 | 0.0095 | 0.0624 |
| FERL | tb1 | tb1.8 | 320 | 4.61 | 0.022 | 0.0107 | 0.0624 |
| LIBN | zap1 | zap1.6 | 436 | 6.40 | 0.027 | 0.0018 | 0.0648 |
Number of individuals with both trait and marker data.
Only marker–trait associations with FDR Q-values <0.10 are shown.
PLHT:
Twenty-five markers were tested for association with plant height. Significant associations (P < 0.05) with two of these markers, zfl2.3 and te1.4, were detected (supplemental Table 6 at http://www.genetics.org/supplemental/). Only the association with zfl2.3 survives correction for multiple testing (Table 3). Moreover, this association survives the conservative Bonferroni correction. No other markers in the candidate gene zfl2 were included in our study. At marker zfl2.3, the mean plant height for the heterozygous class is greater (164 ± 2 cm for AG) than the mean plant height for either homozygous class (157 ± 1 cm for AA and 158 ± 2 cm for AG), which suggests overdominant gene action. This marker explains 2.8% of the phenotypic variance for our sample.
BRLN:
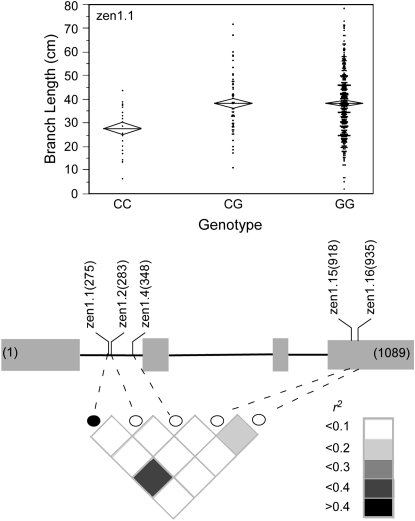
Forty-four markers were tested for association with branch length, resulting in seven significant associations (supplemental Table 6 at http://www.genetics.org/supplemental/). Four markers in two candidate genes, tb1 and zen1, are significantly associated with branch length after correction for multiple testing (Table 3). zen1.1, in candidate gene zen1, has the strongest association with branch length and this association survives a Bonferroni correction. The 3 additional markers tested within zen1 resulted in no detectable associations. Marker zen1.1 is in moderate LD (r2 = 0.3461) with one of these 3 markers, zen1.15. At marker zen1.1, the mean branch lengths of the genotypic classes (27.6 ± 2.3 cm for CC, 38.3 ± 1.8 cm for CG, and 38.1 ± 0.7 cm for GG) suggest that the G allele is dominant to the C allele (Figure 3). Of the 4 markers that were significantly associated with branch length, zen1.1 accounted for the largest fraction (2.8%) of the phenotypic variance.
Figure 3.—
Schematic of zen1 showing the location in base pairs of the markers assayed and the degree of linkage disequilibrium between markers, as measured by r2. The marker that associates with branch length (BRLN) is represented by a solid circle. Shading indicates the magnitude of the linkage disequilibrium. The scatterplot shows the trait distribution for each genotypic class at marker zen1.1. The mean for each genotypic class is represented by a horizontal line. The diamonds depict the standard errors of the means.
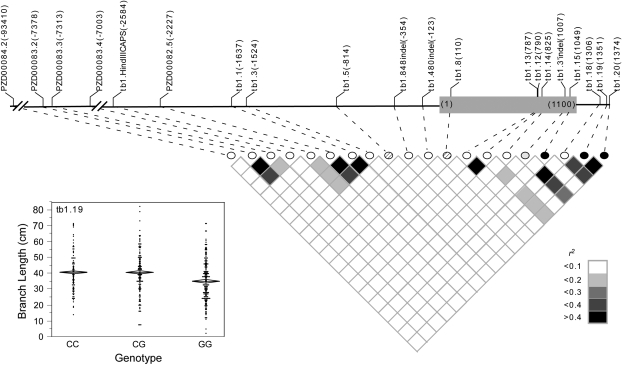
Three of the 4 markers that significantly associate with branch length after correction for multiple testing are located in the maize domestication gene, tb1. Of the 17 additional markers tested in tb1, only 1 other marker, tb1.480indel, has a raw P-value <0.05 for association with branch length; however, this association does not survive correction for multiple testing (supplemental Table 6 at http://www.genetics.org/supplemental/). Two of the 3 markers within tb1 (tb1.19 and tb1.20), which significantly associate with branch length (Table 3), are in LD with each other as well as with 3 other markers, tb1.12, tb1.3′indel, and tb1.18 (Figure 4). At the marker within tb1 that has the strongest association with branch length, tb1.19, the C allele is dominant to the G allele (40.6 ± 1.4 cm for CC, 39.9 ± 1.1 cm for CG, and 35.0 ± 0.9 cm for GG, Figure 4). A similar trend is observed at marker tb1.15 where the G allele is dominant to the A allele (32.6 ± 1.7 cm for AA, 38.5 ± 0.9 cm for AG, and 38.8 ± 1.0 cm for GG) as well as at marker tb1.20 where the C allele is dominant to the G allele (39.6 ± 1.4 cm for CC, 40.5 ± 1.2 cm for CG, and 35.6 ± 0.9 cm for GG). Marker tb1.19 accounted for a larger fraction of the phenotypic variance (2.6%) than either tb1.15 (2.1%) or tb1.20 (2.0%). Tests for epistasis between all possible pairs of the 4 markers that significantly associated with branch length failed to detect any significant interaction terms, suggesting no epistasis.
Figure 4.—
Schematic of tb1 showing the location in base pairs of the markers assayed and the degree of linkage disequilibrium between markers, as measured by r2. The markers that associate with branch length (BRLN) are represented by solid circles and the markers that associate with female ear length (FERL) are represented by hatched circles. Shading indicates the magnitude of the linkage disequilibrium. The scatterplot shows the trait distribution for each genotypic class at marker tb1.19. The mean for each genotypic class is represented by a horizontal line. The diamonds depict the standard errors of the means.
FERL:
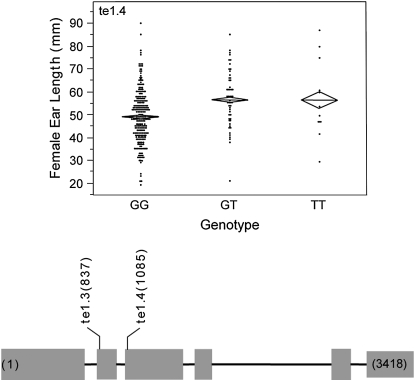
Forty-seven markers were tested for association with female ear length. Of these markers, 5 have significant associations with the trait including 3 markers in tb1 and 2 in te1 (Table 3; supplemental Table 6 at http://www.genetics.org/supplemental/). Four of these associations survive the correction for multiple testing: tb1.5, tb1.8, te1.3, and te1.4. At marker te1.3, which has the strongest association with female ear length, the mean female ear lengths of the genotypic classes (50 ± 1.7 mm for AA, 50 ± 1.1 mm for AG, and 52 ± 0.94 mm for GG) indicate that the A allele is dominant to the G allele. The difference between the genotypic classes is more striking at marker te1.4 (57 ± 3.3 mm for TT, 57 ± 1.7 mm for GT, and 49 ± 0.69 mm for GG) where the T allele is dominant to the G allele (Figure 5). Marker te1.3 accounted for 3.1% of the phenotypic variance and marker te1.4 accounted for 2.3%.
Figure 5.—
Schematic of te1 showing the location in base pairs of the markers assayed. The scatterplot shows the trait distribution for each genotypic class at marker te1.4. The mean for each genotypic class is represented by a horizontal line. The diamonds depict the standard errors of the means.
As stated above, 2 markers in maize domestication gene tb1 are significantly associated with female ear length after correction for multiple testing. These 2 markers, tb1.5 and tb1.8 are not in linkage disequilibrium with each other or with any of the other 18 markers tested in the gene. At tb1.5, which has the strongest association with female ear length, the C allele is dominant to the T allele (49 ± 0.97 mm for TT, 54 ± 1.2 mm for CT, and 52 ± 1.3 mm for CC). The markers in tb1 accounted for similar fractions of the phenotypic variance: tb1.5 accounted for 2.3% and tb1.8 accounted for 2.2%. All possible pairs of the 4 significant markers (2 in tb1 and 2 in te1) were tested for evidence of epistasis. None of the pairs showed any evidence for epistasis.
LIBN:
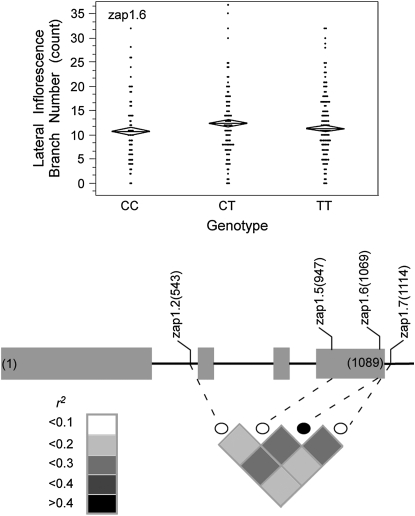
Thirty-six markers were tested for association with lateral inflorescence branch number. Only one significant association, with marker zap1.6, was detected. This association remains significant after correction for multiple testing. The 3 other markers tested in candidate gene zap1 were not significantly associated with the trait, despite the fact that zap1.6 is in moderate LD with each of these 3 other markers (Figure 6). The mean lateral inflorescence branch numbers of the genotypic classes at zap1.6 indicate that the T allele is dominant to the C allele (11 ± 0.65 for CC, 12 ± 0.52 for CT, 12 ± 0.47 for TT). This marker accounted for 2.7% of the phenotypic variance.
Figure 6.—
Schematic of zap1 showing the location in base pairs of the markers assayed and the degree of linkage disequilibrium between markers, as measured by r2. The marker that associates with lateral inflorescence branch number (LIBN) is represented by a solid circle. Shading indicates the magnitude of the linkage disequilibrium. The scatterplot shows the trait distribution for each genotypic class at marker zap1.6. The mean for each genotypic class is represented by a horizontal line. The diamonds depict the standard errors of the means.
Allelic effects and frequencies:
The majority of assayed traits measure aspects of morphology that changed during domestication and were likely under human selection. For all markers that were significantly associated with these traits, we calculated allele frequencies and the average additive effects of the maize-like allele in our teosinte sample (Table 4). We also obtained the frequencies of these alleles in maize landraces from the Maize Diversity Database (http://www.panzea.org). This database includes a sample of >1200 plants representing noncommercial varieties collected throughout the pre-Columbian range of maize in the Americas. The average additive effects are relatively large, suggesting that teosinte harbors considerable genetic variation in domestication traits upon which selection could act. For example, tb1.15 has an additive effect of reducing branch length by 1.5 cm, and zen1.1 reduces it by 0.75 cm. Interestingly, in all cases, the allele associated with the maize-like phenotype in teosinte exhibits a higher frequency in maize than in teosinte, suggesting that these alleles may have been driven to a higher frequency by selection. For zen1.1, a relatively rare allele in teosinte has become the predominant allele in maize.
TABLE 4.
Frequency of and additive effects associated with the maize-like alleles
| Frequencya
|
|||||
|---|---|---|---|---|---|
| Marker | Allele | Trait | Teosinte | Maize | αb |
| tb1.15 | A | BRLN (cm) | 0.33 | NA | −1.4974 |
| tb1.19 | G | BRLN | 0.64 | 0.90 | −0.9792 |
| tb1.20 | G | BRLN | 0.64 | NA | −0.5280 |
| zen1.1 | C | BRLN | 0.13 | 0.74 | −0.7511 |
| tb1.5 | C | FERL (mm) | 0.39 | 0.83 | 2.0009 |
| tb1.8 | A | FERL | 0.17 | NA | −0.3499 |
| te1.3 | G | FERL | 0.88 | 0.93 | 0.5718 |
| te1.4 | T | FERL | 0.10 | 0.53 | 6.0987 |
| zap1.6 | C | LIBN (count) | 0.41 | 0.59 | −0.0882 |
NA, not available.
The additive effect was calculated as 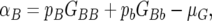 where pi is the allele frequency of allele i, Gij is the phenotypic mean of the genotypic class ij, and μG is the overall phenotypic mean.
where pi is the allele frequency of allele i, Gij is the phenotypic mean of the genotypic class ij, and μG is the overall phenotypic mean.
DISCUSSION
In this study, we tested for associations between nine major regulatory genes and 13 plant and inflorescence architecture traits hypothesized to have been under selection during domestication. We detected a total of 10 associations including markers in five of the nine regulatory genes and 4 of the traits assayed. These results suggest that major regulatory genes of maize contribute to small-scale phenotypic differences in natural populations of its ancestor, Balsas teosinte. Our results also included 5 associations between 2 traits and the maize domestication gene, tb1. This is evidence that a major regulatory gene controlling dramatic morphological change during domestication also contributes to the natural variation of complex traits in teosinte. We caution that these results are only a first step toward determining how major regulatory genes contribute to the natural variation of complex traits. Further work will be needed not only to confirm these associations but to also determine the genetic architecture of complex traits and the specific role played by major regulatory genes.
Association mapping is an emerging methodology that is being used with an increasing number of organisms, traits, and genes (Gupta et al. 2005; Wellcome Trust Case Control Consortium 2007). The application of association mapping to any new organism presents unique challenges because of differences among organisms in factors such as population structure, mating systems, and extent of LD. Our analyses indicate that candidate-gene-based association mapping is a feasible tool for the dissection of complex traits in teosinte despite a considerable degree of population structure. Using a mixed model that incorporates terms for both population structure and recent coancestry (Yu et al. 2006; Zhao et al. 2007), the distribution of P-values for trait–marker pairs was found to approximate that expected under the null hypothesis of genotype–phenotype independence at random loci. This result indicates that the model sufficiently controls for false positives or type I error (rejecting the null hypothesis of independence of the trait and marker when it is true). At the same time, the full model retains sufficient power to detect associations that withstand a correction for multiple testing. Moreover, we were able to detect significant associations with a relatively moderate sample size of 584 plants, indicating that association mapping can be successfully applied by research groups with relatively modest resources.
In addition to type I error, we also considered the extent of false negatives or type II error (failing to reject the null hypothesis when it is false) for our analyses. We suspect that false negatives are a significant concern. This suspicion is especially true for traits related to flowering time and other traits related to local adaptation in teosinte. We observed a strong correlation between genetic and geographic distance and high FST values in teosinte for random markers. For this reason, variation in genes controlling traits such as flowering time is apt to be correlated with variation in any randomly selected gene and filtered out by the principal components in the full model. This correlation between flowering time and genetic variation in randomly selected genes is evident by the inflated number of small P-values observed when flowering-time traits were tested with the simple model (supplemental Table 4 at http://www.genetics.org/supplemental/). The loss of power due to the confounding of these traits and population structure could explain why we observed no associations for POLL and SILK despite testing candidate genes such as d8, id1, and zfl2 that are known to influence flowering time in maize (Colasanti et al. 1998; Thornsberry et al. 2001; Bomblies and Doebley 2006).
Below we discuss the five regulatory genes containing markers that significantly associate with one of the traits. In doing so, we present our hypotheses for why variation in these regulatory genes might be expected to associate with these traits.
teosinte branched1 (tb1):
We chose to assay tb1 because of its known effects on plant and inflorescence architecture and because of its role as a gene of large effect involved in maize domestication (Doebley et al. 1997; Doebley 2004). tb1 is a member of the TCP transcriptional regulator family, and it is hypothesized to function as a negative regulator of organ growth (Cubas et al. 1999). Plants homozygous for tb1 loss-of-function alleles have long lateral branches tipped with tassels, many tillers, and tassel-like inflorescence structures as opposed to ears. We observed associations between multiple markers in tb1 with BRLN and FERL, suggesting that this gene contributes to natural phenotypic variation in teosinte.
The pattern of the associations for tb1 suggests that different genic regions are associated with different traits. The three markers associated with BRLN are all near the 3′ end of the ORF: a synonymous-site change in the 3′ end of the ORF and two markers just 3′ of the stop codon. The two markers that associate with FERL are near the 5′ end of the ORF: one is in the putative promoter and the other is a non-synonymous-site change in the 5′ end of the ORF. The observed associations with BRLN and FERL are not surprising given that alterations in lateral branch length and inflorescence structure are key features of the tb1 mutant (Doebley et al. 1997) and are also the key differences conferred by the maize vs. teosinte alleles of this gene (Clark et al. 2006).
terminal ear1 (te1):
Two markers resulting in synonymous-site changes within te1 associate with FERL, suggesting that this gene contributes to natural phenotypic variation in teosinte. te1 is a putative RNA-binding protein that affects plant architecture and inflorescence sex (Veit et al. 1998). The recessive mutant in maize has an increase in node number in the main stalk as well as shortened internodes. In mutant plants, the base of the tassel that terminates the main stalk can also be converted into an ear. Our observed association of te1 with FERL seems plausible given the effect of te1 on internode number and elongation since variation in teosinte ear length is a function of the number and lengths of the internodes that compose the ear.
zea floricaula leafy2 (zfl2):
An association between a synonymous-site change in zfl2 and PLHT survives the Bonferroni correction, providing evidence that this gene contributes to natural phenotypic variation in teosinte. zfl2 is an ortholog of FLORICAULA in Antirrhinum and LEAFY in Arabidopsis (Bomblies and Doebley 2006). FLORICAULA/LEAFY were first identified for their role in the regulation of ABC floral organ identity genes (Coen et al. 1990; Weigel et al. 1992); however, subsequent studies have shown that they influence a variety of traits from leaf morphology to flowering time in different species (Blazquez et al. 1997; Hofer et al. 1997; Bomblies and Doebley 2006). In maize, these genes influence flowering time, leaf number, and inflorescence structure (Bomblies et al. 2003; Bomblies and Doebley 2006). We may not have detected an association between zfl2 and flowering time because flowering time is correlated with population structure in teosinte as discussed above. However, our observed association of zfl2 with plant height is consistent with the known effect of zfl2 on flowering time and leaf number in maize since these are traits that are typically correlated with plant height.
zea centroradialis1 (zen1):
We observed an association between a marker in zen1 and the length of the lateral branches (BRLN) in teosinte. zen1 is a maize gene with DNA sequence homology to CENTRORADIALIS in Antirrhinum, which is involved in the programming of the apical meristem by acting antagonistically to the floral meristem identity gene FLORICAULA (Bradley et al. 1996). In plants that are mutant for CENTRORADIALIS, a determinate flower replaces the normally indeterminate inflorescence. CENTRORADIALIS homologs have been studied in the grasses, rice, and ryegrass (Zhang et al. 2005). In these grasses, CENTRORADIALIS-like genes have complex expression patterns, including expression in branch meristems. They also control the elongation of shoot internodes. Thus, our observed association with lateral branch length is consistent with what is known of the function or expression pattern of CENTRORADIALIS-like genes in other grasses.
zea apetala homolog1 (zap1):
A synonymous-site change in zap1 is associated inflorescence branching (LIBN). zap1 is a maize homolog of the MADS-box transcription factor APETALA1 in Arabidopsis (Mena et al. 1995). The principal function of APETALA1 in Arabidopsis is its role in controlling floral organ identity; however, it also influences inflorescence architecture (Bowman et al. 1993). No zap1 mutants are available in maize; nevertheless, RNA blot analysis has found that zap1 expression is found in the husk leaves and both male and female inflorescences (Mena et al. 1995). Similar to APETALA1 in Arabidopsis, zap1 RNA in maize is excluded from the stamens and pistils, but is expressed in the first (lemma and palea) and second (lodicules) whorl floral organs. The association found in our study suggests that zap1 may control aspects of inflorescence structure in teosinte. This functionality seems plausible given the expression pattern of zap1 in maize and the known effect of APETALA1 on inflorescence architecture in Arabidopsis.
Our results provide some suggestive information about genetic variation in teosinte and the domestication of maize. First, the fact that we detected associations for five of the nine regulatory genes tested and 4 of the 13 traits studied suggests that major maize regulatory genes contribute to natural variation in teosinte. These results also indicate that teosinte possesses considerable standing genetic variation upon which selection could have acted during domestication. Although no marker explained >4% of the phenotypic variation for any of the traits, the additive effects associated with the markers are reasonably large (Table 4). The additive effects on branch length range from 0.5 to 1.5 cm, which represents a considerable portion of the ∼40-cm reduction in branch length during domestication (Doebley et al. 1995). Second, our results also suggest that tb1, a gene that controlled a large step in the domestication of maize, also contributes to the relatively minor standing variation within the wild species. Finally, our observation that the allele associated with the maize-like phenotype at all six markers for which we have data has a higher frequency in maize than in teosinte (Table 4) hints that these minor variants may have been under selection during domestication even if they never reached fixation in maize.
While our results are encouraging in regard to the application of association mapping in teosinte, we caution that there are several concerns and caveats to consider. We consider the significant marker–trait associations as putative and in need of verification by independent experiments. Ideally, one would identify the causal polymorphism and directly demonstrate that the alternate alleles confer distinct phenotypes by genetic fine mapping. Otherwise, additional correlative evidence could be obtained that strengthens but does not prove that the marker–trait association is real. For example, one might show that the different haplotypes confer different mRNA levels or that different amino acids in the gene alter protein function. Similarly, one might show the same marker–trait association in an independent sample of plants, perhaps from another type of teosinte or maize.
To facilitate further association mapping in Balsas teosinte, we have deposited seed from 85 local teosinte populations with the U.S. Department of Agriculture North Central Regional Plant Introduction Station in Ames, Iowa. These seed are available as kits of 25 seed from each of these 85 local populations. Association mapping with this resource should allow others to test additional traits and genes, identify useful alleles for maize breeding, or test whether genes that show associations with traits in maize show the same associations in teosinte. One important use of these materials would be to test trait–gene associations with genes for which maize possesses only a single haplotype due to either selection or drift during domestication. Through these analyses a better understanding of the genetic architecture for traits important to maize domestication and breeding can be obtained.
Acknowledgments
We thank Jeff Glaubitz, Qiong Zhao, and Bill Briggs for comments and discussion. We also thank Jaime Stoltenberg for her work on Figure 1. This work was funded by National Science Foundation grant DBI-0321467 and National Institutes of Health grant GM-58816. Jianming Yu's contribution to this research was supported in part by a grant from the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service–National Research Initiative Plant Genome Program.
References
- Blazquez, M. A., L. N. Soowal, I. Lee and D. Weigel, 1997. LEAFY expression and flower initiation in Arabidopsis. Development 124: 3835–3844. [DOI] [PubMed] [Google Scholar]
- Bomblies, K., and J. F. Doebley, 2006. Pleiotropic effects of the duplicate maize FLORICAULA/LEAFY genes zfl1 and zfl2 on traits under selection during maize domestication. Genetics 172: 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies, K., R.-L. Wang, B. A. Ambrose, R. J. Schmidt, R. B. Meeley et al., 2003. Duplicate FLORICAULA/LEAFY homologs zfl1 and zfl2 control inflorescence architecture and flower patterning in maize. Development 130: 2385–2395. [DOI] [PubMed] [Google Scholar]
- Bowman, J. L., J. Alvarez, D. Weigel, E. M. Meyerowitz and D. R. Smyth, 1993. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743. [Google Scholar]
- Bradley, D., R. Carpenter, L. Copsey, C. Vincent, S. Rothstein et al., 1996. Control of inflorescence architecture in Antirrhinum. Nature 379: 791–797. [DOI] [PubMed] [Google Scholar]
- Briggs, W. H., M. D. McMullen, B. S. Gaut and J. Doebley, 2007. Linkage mapping of domestication loci in a large maize–teosinte backcross resource. Genetics 177: 1915–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, R. M., T. N. Wagler, P. Quijada and J. Doebley, 2006. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat. Genet. 38: 594. [DOI] [PubMed] [Google Scholar]
- Coen, E. S., J. Romero, S. Doyle, R. Elliott, G. Murphy et al., 1990. floricaula: a homeotic gene required for flower development in Antirrhinum majus. Cell 63: 1311. [DOI] [PubMed] [Google Scholar]
- Colasanti, J., Z. Yuan and V. Sundaresan, 1998. The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 93: 593–603. [DOI] [PubMed] [Google Scholar]
- Cubas, P., N. Lauter, J. Doebley and E. Coen, 1999. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18: 215–222. [DOI] [PubMed] [Google Scholar]
- DeLong, A., A. Calderon-Urrea and S. L. Dellaporta, 1993. Sex determination gene TASSELSEED2 of maize encodes a short-chain dehydrogenase required for stage-specific organ abortion. Cell 74: 757–768. [DOI] [PubMed] [Google Scholar]
- Devlin, B., and K. Roeder, 1999. Genomic control for association studies. Biometrics 55: 997–1004. [DOI] [PubMed] [Google Scholar]
- Doebley, J., 2004. The genetics of maize evolution. Annu. Rev. Genet. 38: 37–59. [DOI] [PubMed] [Google Scholar]
- Doebley, J., A. Stec and C. Gustus, 1995. teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141: 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., A. Stec and L. Hubbard, 1997. The evolution of apical dominance in maize. Nature 386: 485–488. [DOI] [PubMed] [Google Scholar]
- Fukunaga, K., J. Hill, Y. Vigouroux, Y. Matsuoka, J. Sanchez et al., 2005. Genetic diversity and population structure of teosinte. Genetics 169: 2241–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, J., S. Schroeder, M. L. Polacco, H. Sanchez-Villeda, Z. Fang et al., 2004. Anchoring 9,371 maize expressed sequence tagged unigenes to the bacterial artificial chromosome contig map by two-dimensional overgo hybridization. Plant Physiol. 134: 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, P., S. Rustgi and P. Kulwal, 2005. Linkage disequilibrium and association studies in higher plants: present status and future prospects. Plant Mol. Biol. 57: 461–485. [DOI] [PubMed] [Google Scholar]
- Hofer, J., L. Turner, R. Hellens, M. Ambrose, P. Matthews et al., 1997. UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr. Biol. 7: 581–587. [DOI] [PubMed] [Google Scholar]
- Jurinke, C., D. van den Boom, C. R. Cantor and H. Koster, 2002. The use of MassARRAY technology for high throughput genotyping. Adv. Biochem. Eng./Biotech. 77: 58–74. [DOI] [PubMed] [Google Scholar]
- Lai, C., R. F. Lyman, A. D. Long, C. H. Langley and T. F. C. Mackay, 1994. Naturally occurring variation in bristle number and DNA polymorphism at the scabrous locus of Drosophila melanogaster. Science 266: 1697–1702. [DOI] [PubMed] [Google Scholar]
- Liu, K., and S. V. Muse, 2005. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21: 2128–2129. [DOI] [PubMed] [Google Scholar]
- Long, A. D., and C. H. Langley, 1999. The power of association studies to detect the contribution of candidate genetic loci to variation in complex traits. Genome Res. 9: 720–731. [PMC free article] [PubMed] [Google Scholar]
- Mandel, M. A., C. Gustafson-Brown, B. Savidge and M. F. Yanofsky, 1992. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360: 273–277. [DOI] [PubMed] [Google Scholar]
- Mena, M., M. A. Mandel, D. R. Lerner, M. F. Yanofsky and R. J. Schmidt, 1995. A characterization of the MADS-box gene family in maize. Plant J. 8: 845–854. [DOI] [PubMed] [Google Scholar]
- Morrison, K., C. Papapetrou, J. Attwood, F. Hol, S. A. Lynch et al., 1996. Genetic mapping of the human homologue (T) of mouse T(Brachyury) and a search for allele association between human T and spina bifida. Hum. Mol. Genet. 5: 669–674. [DOI] [PubMed] [Google Scholar]
- Morrison, K., C. Papapetrou, F. A. Hol, E. C. M. Mariman, S. A. Lynch et al., 1998. Susceptibility to spina bifida; an association study of five candidate genes. Ann. Hum. Genet. 62: 379–396. [DOI] [PubMed] [Google Scholar]
- Nachman, M. W., H. E. Hoekstra and S. L. D'Agostino, 2003. The genetic basis of adaptive melanism in pocket mice. Proc. Natl. Acad. Sci. USA 100: 5268–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nychka, D., 2007. Fields: tools for spatial data, R package version 3.5. (http://www.image.ucar.edu/GSP/Software/Fields).
- Patterson, N., A. L. Price and D. Reich, 2006. Population structure and eigenanalysis. PLoS Genet. 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., D. E. Richards, N. M. Hartley, G. P. Murphy, K. M. Devos et al., 1999. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261. [DOI] [PubMed] [Google Scholar]
- Price, A. L., N. J. Patterson, R. M. Plenge, M. E. Weinblatt, N. A. Shadick et al., 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909. [DOI] [PubMed] [Google Scholar]
- Pritchard, J. K., M. Stephens, N. A. Rosenberg and P. Donnelly, 2000. Association mapping in structured populations. Am. J. Hum. Genet. 67: 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., H. Onouchi, S. E. Gold, G. S. Ditta, Z. Schwarz-Sommer et al., 2000. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616. [DOI] [PubMed] [Google Scholar]
- SAS Institute, 1999. SAS/STAT User's Guide, Version 8. SAS Institute, Cary, NC.
- Storey, J. D., 2002. A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B 64: 479–498. [Google Scholar]
- Storey, J. D., and R. Tibshirani, 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley, S. D., and S. R. McCouch, 1997. Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277: 1063–1066. [DOI] [PubMed] [Google Scholar]
- Thornsberry, J. M., M. M. Goodman, J. Doebley, S. Kresovich, D. Nielson et al., 2001. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28: 286–289. [DOI] [PubMed] [Google Scholar]
- Veit, B., S. P. Briggs, R. J. Schmidt, M. F. Yanofsky and S. Hake, 1998. Regulation of leaf initiation by the terminal ear 1 gene of maize. Nature 393: 166–168. [DOI] [PubMed] [Google Scholar]
- Weigel, D., J. Alvarez, D. R. Smyth, M. F. Yanofsky and E. M. Meyerowitz, 1992. LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium, 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, L. M., S. R. Whitt, A. M. Ibanez, T. R. Rocheford, M. M. Goodman et al., 2004. Dissection of maize kernel composition and starch production by candidate gene association. Plant Cell 16: 2719–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S. I., I. V. Bi, S. G. Schroeder, M. Yamasaki, J. F. Doebley et al., 2005. The effects of artificial selection on the maize genome. Science 308: 1310–1314. [DOI] [PubMed] [Google Scholar]
- Yu, J., G. Pressoir, W. H. Briggs, I. Vroh Bi, M. Yamasaki et al., 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208. [DOI] [PubMed] [Google Scholar]
- Zhang, S., W. Hu, L. Wang, C. Lin, B. Cong et al., 2005. TFL1/CEN-like genes control intercalary meristem activity and phase transition in rice. Plant Sci. 168: 1393–1408. [Google Scholar]
- Zhao, K., M. J. Aranzana, S. Kim, C. Lister, C. Shindo et al., 2007. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 3: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]