Abstract
In Saccharomyces cerevisiae, genes placed near telomeres are transcriptionally repressed (telomere position effect, TPE). Although telomeric DNA sequence is the same at all chromosome ends, the subtelomeric elements (STEs) and level of TPE vary from telomere to telomere. We tested whether STEs determine TPE levels. STEs contributed to TPE, as deleting the X element from the VI-R telomere modestly decreased silencing at this telomere. However, STEs were not the major determinant of TPE levels, as inserting the VI-R X element at the truncated VII-L telomere did not increase TPE. These data suggest that the TPE levels of individual telomeres are dependent on some aspect of chromosome context.
IN Saccharomyces cerevisiae and other organisms, including humans, genes placed near telomeres are transcriptionally repressed, a phenomenon termed telomere position effect (TPE; reviewed in Mondoux and Zakian 2005). There are at least six telomere-associated proteins that are essential for TPE in yeast: Rap1p, a sequence-specific telomeric DNA binding protein, Sir2p, Sir3p, Sir4p, and the Ku heterodimeric complex. All of these proteins except Ku are also required for silencing at the two silent mating-type (HM) loci.
In most organisms, including S. cerevisiae, middle repetitive subtelomeric elements (STEs) are found immediately proximal to the simple sequence TG1–3 telomere repeats. In yeast, there are two types of STEs, X and Y′ (reviewed in Louis 1995). X is heterogeneous, ranging in size from ∼300 bp to 3 kb, but each X element contains a “core-X” repeat that is found at essentially all telomeres. The core X consists of an autonomously replicating sequence (ARS) consensus sequence (ACS), which can bind the multisubunit origin recognition complex (ORC), and the transcription factor Abf1p. ORC and Abf1p are both important for silencing at the HM loci (Diffley and Stillman 1989; Kurtz and Shore 1991; reviewed in Haber 1998). Most X elements also contain antisilencing X-combinatorial repeats (XCRs) that have recognition sites for Reb1p, a transcription factor (Morrow et al. 1989; Wang et al. 1990) and Tbf1p, an essential protein of unknown function (Brigati et al. 1993). Sites bound by Tbf1p act as heterochromatin boundaries that keep silencing from spreading to more internal parts of the chromosome (Fourel et al. 1999, 2001). The combination, number, and arrangement of XCRs vary from telomere to telomere. In contrast to the ubiquitous X element, the Y′ element is found at only one-half to two-thirds of yeast telomeres. The Y′ element also has binding sites for Reb1p and Tbf1p. When present, Y′ is distal to X and is found in up to four tandem copies (Chan and Tye 1983). Although there are only two classes of STEs, the X and Y′ elements are sufficiently diverse that each of the 32 yeast telomeres can be thought of as having a distinct identity.
TPE was discovered by placing reporter genes immediately adjacent to the telomeric TG1–3 tract, which generates telomeres that lack both the X and Y′ elements (Gottschling et al. 1990). Two methods are used to study TPE at native yeast telomeres: (1) inserting a reporter gene into the X-ACS, keeping the Y′ and X elements largely intact (Fourel et al. 1999; Pryde and Louis 1999; hereafter referred to as “native” telomeres) and (2) observing RNA levels of subtelomeric genes that are naturally near chromosome ends (Vega-Palas et al. 1997, 2000; Wyrick et al. 1999, 2001; Barton and Kaback 2006). A subset of native telomeres and all truncated telomeres that have been tested exhibit TPE (for summary of silencing status of individual telomeres, see http://www.molbio1.princeton.edu/labs/zakian/assets/2007-08-mondoux-phenotypes.pdf). However, in the same strain background, the level of TPE can vary substantially from telomere to telomere: at some telomeres, telomere-adjacent genes are repressed in a small percentage and at others in ∼100% of cells. Thus, in addition to having different subtelomeric structures, telomeres exhibit different levels of TPE.
In addition to varied TPE levels, requirements for silencing proteins are different at different telomeres. Sir1p acts in the establishment of silencing at the HM loci (Pillus and Rine 1989) via binding to ORC (Foss et al. 1993; Triolo and Sternglanz 1996). Sir1p is not necessary for TPE at truncated telomeres (Aparicio et al. 1991). In contrast, TPE is reduced at the native XI-L telomere in a sir1Δ strain (Fourel et al. 1999; Pryde and Louis 1999). In addition, if Sir1p is tethered to the truncated VII-L telomere, silencing increases (Chien et al. 1993). Sir1p could promote silencing at native telomeres by associating with ORC bound to the X-ACS (Wyrick et al. 2001; Xu et al. 2006). Since Sir1p interacts with Sir4p (Triolo and Sternglanz 1996), its binding can recruit other silencing proteins to the telomere. Mutation of the X-ACS or its Abf1p binding site reduces TPE at native telomere XI-L (Pryde and Louis 1999). In addition, the core X element can improve silencing at a weakened HM locus and can counteract the antisilencing effects of XCRs when both are integrated at the truncated VII-L telomere (Lebrun et al. 2001). Given that the subtelomeric DNA can recruit different trans-acting factors to telomeres, the sequence composition of the subtelomere might determine the silencing profile of individual telomeres. This model is not unique to yeast, because subtelomeric regions of Drosophila and human chromosomes are also diverse, and there are differences in TPE phenotypes between Drosophila telomeres with different subtelomeric sequences (Shanower et al. 2005).
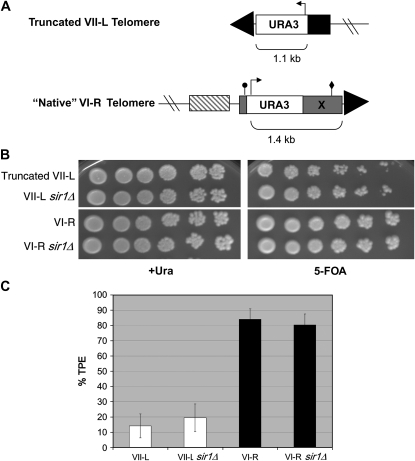
To determine the effects of STEs on TPE, we compared silencing at the truncated VII-L telomere (Gottschling et al. 1990) to silencing at the VI-R telomere, which has the minimal subtelomeric core X element, but no XCRs or Y′ elements. TPE is biologically relevant at the VI-R telomere, as YFR057w, the uncharacterized ORF that is the closest unique sequence to the VI-R telomere, is transcriptionally silenced in wild-type strains, and is expressed upon deletion of the Sir proteins (Vega-Palas et al. 2000). At VI-R, URA3 was integrated into the subtelomeric X element in a manner that largely maintains its structure (as in Pryde and Louis 1999; Figure 1A). We then determined the TPE phenotypes of these marked truncated VII-L and native VI-R strains.
Figure 1.—
Telomere structure and TPE analysis at the truncated VII-L and VI-R telomeres. (A) The VI-R subtelomere contains a 380-bp “core X” element (shaded), containing an ARS consensus sequence (ACS; circle) and Abf1p binding site (diamond). The URA3 TPE reporter was introduced at the VI-R telomere X-ACS in a manner analogous to the creation of native TPE reporters described in Pryde and Louis (1999). PCR primers whose 5′ ends corresponded to VI-R X-element sequence surrounding the X-ACS were used to amplify URA3 from ADH4UCAIV, the same plasmid used to create the truncated VII-L telomere reporter (Gottschling et al. 1990). Unlike the system that truncated the VII-L telomere at ADH4 (solid), the “native” VI-R TPE reporter preserves the subtelomeric structure. The closest upstream gene is YFR057w (striped box). Both strains also contain upstream lac operator arrays for visualization studies (Mondoux et al. 2007). All strains used in this study were constructed in the YPH background (ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1; Sikorski and Hieter 1989) and grown at 30° in yeast complete (YC) synthetic medium and plates (Zakian and Scott 1982). All TPE reporter strains were verified for correct integration by Southern blotting and pulsed-field gel electrophoresis. SIR1 was deleted in each strain background using a PCR-mediated knockout that eliminated the complete open reading frame, replacing it with a hygromycin resistance cassette (Goldstein and McCusker 1999). (B) TPE assays. Tenfold serial dilutions of the VI-R, truncated VII-L, and sir1Δ versions of these strains were plated onto +Ura or 5-FOA plates to assay silencing and photographed after 3 days' growth. TPE is higher at VI-R compared to truncated VII-L and is not dependent on Sir1p at either telomere. (C) Quantitation of TPE. +Ura-grown cells were plated onto +Ura and 5-FOA plates and colonies counted after 3 days of growth. The percentage of total cells (+Ura) that grew on 5-FOA plates is represented as % TPE. Error bars represent standard deviations. TPE at VI-R is significantly higher than TPE at VII-L by Student's t-test (P < 7 × 10−5). There was no significant difference in TPE at either the VI-R or VII-L telomere in the absence of Sir1p.
In our strain, the URA3 gene was silenced at the truncated VII-L telomere in ∼15% of yeast complete (YC)-grown cells (14.2 ± 7.9%; Figure 1C). Although this average TPE level was lower than the average reported for truncated VII-L in some studies (e.g., Gottschling et al. 1990; Hediger et al. 2002), these analyses were in different strain backgrounds and/or under different growth conditions. Moreover, our average value is within the range of TPE values for VII-L in earlier studies (e.g., 3–78% TPE for YC-grown cells; Hediger et al. 2002), and the TPE value reported here was very similar to the TPE level for haploid YC-grown PPR1 cells in the same strain background (Tham et al. 2001).
In contrast, in the same strain background, the URA3 gene at the native VI-R telomere was silenced in ∼85% of the cells (84.0 ± 6.8%; Figure 1C). Thus, the VI-R telomere had a higher level of TPE than the VII-L telomere, despite the fact that the URA3 transcription start site was 328 bp farther away from the telomere at VI-R compared to VII-L (Figure 1A), and TPE levels are known to decrease exponentially with distance at truncated telomeres (Gottschling et al. 1990; Renauld et al. 1993). The level of TPE at the native VI-R telomere is among the highest reported for a yeast telomere.
Virtually all X elements have an Abf1p binding site and an ACS that can bind ORC and thereby recruit Sir1p to the telomere. Although Abf1p binding at telomeres has not been tested, the VI-R X-ACS is bound by ORC (Xu et al. 2006). We therefore predicted that the VI-R X element is responsible for the high TPE phenotype of this telomere. We also predicted that the VI-R X element would be able to confer a strong TPE phenotype on another chromosome end.
As expected (Aparicio et al. 1991), we observed no significant difference in TPE at the truncated VII-L telomere in the absence of SIR1 (19.5 ± 9.1%; Figure 1C). However, there was also no significant difference in TPE levels at the native VI-R telomere in the absence of SIR1 (80.2 ± 7.2%). This Sir1p independence is in contrast to what is seen at the XI-L telomere (Pryde and Louis 1999), but consistent with expression of YFR057w, the telomere proximal ORF on VI-R, which shows wild-type TPE in a sir1Δ strain by Northern analysis (Vega-Palas et al. 2000). Thus, the very high level of silencing seen at the VI-R telomere is not due to a Sir1p-dependent mechanism that is absent at the truncated VII-L telomere.
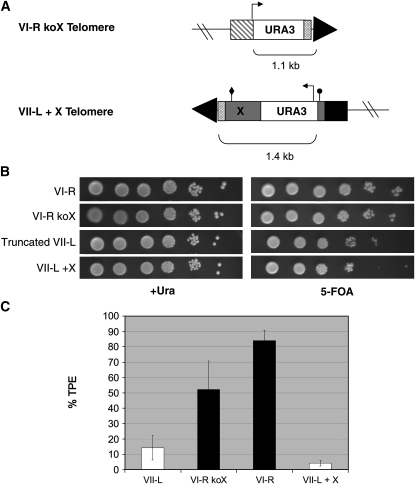
The X element plays a role in TPE at telomere XI-L that is distinct from the recruitment of Sir1p, as mutating the X-ACS or the Abf1p binding site results in a larger (∼10–100-fold) decrease in TPE (Pryde and Louis 1999). Since deleting SIR1 did not reduce TPE at the native VI-R telomere, we decided to eliminate the X element. We designed primers that eliminated X from the VI-R telomere and replaced it with URA3, leaving behind the last 56 bp of the X element but removing all known binding and regulatory sites, including the X-ACS and the Abf1p binding site (strain VI-R koX; Figure 2A). URA3 then serves as the TPE reporter, with the transcription start site at approximately the same distance from the telomere as it is at the truncated VII-L telomere. Surprisingly, deleting the X element did not have a large effect on TPE at VI-R (Figure 2B), as silencing was decreased less than twofold in its absence (52.2 ± 18.7%; Figure 2C). Therefore, Abf1p binding did not appear to make a major contribution to TPE, nor did ORC binding via the X-ACS site. Either the X element is largely dispensable for TPE at VI-R or there is some unidentified activity in the final 56 bp of the X element left at the VI-R telomere that contributes positively to TPE.
Figure 2.—
The X element requires chromosomal context to function. (A) Construction of VI-R koX and VII-L + X telomere reporter strains. The VI-R koX TPE reporter was created via integration of the URA3 gene flanked by an upstream segment of unique DNA (YFR057w, striped box) and the final 56 bp of the X element (crosshatched box), knocking out most of the X element, including the X-ACS and Abf1p binding site. The VII-L + X TPE reporter was created via truncation of the VII-L telomere seed at an upstream segment of unique DNA (ADH4 gene, solid). The VI-R X element was cloned using an upstream primer with the same sequence as the primer used to integrate URA3 at the X-ACS and a downstream primer with the same sequence used to knockout the VI-R X element. This PCR product replaced the URA3 fragment in the VII-L truncation plasmid, pADH4UCAIV (Gottschling et al. 1990). The transcription start site (arrow), X-ACS (circle), and Abf1p binding site (diamond) are positioned identically to their locations in the VI-R strain (see Figure 1A). (B) TPE assays. Tenfold serial dilutions were plated as in Figure 1B. TPE appeared unchanged in the absence of the X element at VI-R and was not enhanced by the presence of the X element on truncated VII-L. (C) Quantitation of TPE. TPE at VI-R was reduced (P < 0.02) in the absence of the X element, but was still significantly higher than TPE at VII-L (P < 0.002), which also lacks the X element. The addition of the X element to the truncated VII-L telomere did not increase the level of TPE at that telomere.
We next did the reciprocal experiment, asking whether the X element could confer a high level of TPE at the truncated VII-L telomere. We wanted to clone the VI-R X element specifically, so as to directly compare its function at the VI-R telomere with its potential function at the truncated VII-L telomere. Previous studies used X elements cloned from different telomeres (II-R and XI-L) and inserted fragments of these X elements between the URA3 reporter and the truncated VII-L telomere (Fourel et al. 1999). Both of these elements contain XCRs, which the VI-R X element lacks. The VI-R X element and URA3 TPE reporter were cloned via genomic PCR from the native VI-R telomere strain and integrated at the truncated VII-L telomere (truncated VII-L + X; Figure 2A). We sequenced the cloned VI-R X element and found that it was virtually identical to the sequence of the VI-R X in the Saccharomyces Genome Database (SGD), differing at only 3 bp, none of which was in the ACS or Abf1p binding sites (data not shown). Thus, the transcription start site and sequence and spacing of regulatory sites were identical between the telomeres in the native VI-R and truncated VII-L + X strains.
The insertion of the VI-R X element at the truncated VII-L telomere did not increase TPE at this telomere. Rather, the TPE level was actually decreased approximately threefold relative to the truncated VII-L telomere (4.3 ± 1.9%; Figure 2C). This decrease was probably due to the increased distance of the URA3 transcription start site from the telomere end. Since this construct included the 56-bp fragment of the VI-R X element that was left behind in the X-element knockout at VI-R (Figure 2A), neither this fragment nor the entire X element from telomere VI-R was sufficient to confer a higher level of TPE on the VII-L telomere.
We propose that the different responses by the VI-R and XI-L telomeres to genetic perturbations are explained by differences in STE content. Although the X elements at both telomeres appear to bind ORC to similar extents (Xu et al. 2006), antisilencing XCR elements are present at the XI-L telomere but not at the VI-R telomere. Perhaps the core X contributes substantively to TPE only at telomeres that contain XCR elements because it acts by countering their negative effects.
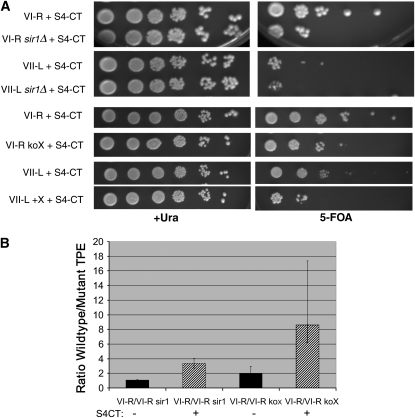
Since Sir1p had no effect on silencing at the VI-R telomere in a wild-type strain, despite the telomere's ability to bind ORC, we next asked whether Sir1p could contribute to TPE at the native VI-R telomere if TPE were compromised. Overexpressing the C terminus of Sir4p (S4-CT) greatly reduces TPE at truncated telomere VII-L as well as HM silencing (Marshall et al. 1987). S4-CT interacts with itself (Chien et al. 1991), Sir2p (Strahl-Bolsinger et al. 1997), Sir3p (Moazed et al. 1997), Rap1p (Moretti et al. 1994), and yKu70p (Tsukamoto et al. 1997). Sir4p recruits Sir2p and Sir3p to the telomere (Bourns et al. 1998; Luo et al. 2002), so the reduction in TPE observed with S4-CT overexpression presumably occurs via the titration of other silencing factors away from telomeres. TPE at native telomere VI-R was reduced ∼20-fold in the presence of S4-CT (Figure 3A). In the absence of SIR1, TPE was reduced an additional 3.5-fold in the S4-CT strain (Figure 3B). Thus, Sir1p does affect TPE at the X-bearing VI-R telomere, but this contribution is only seen when silencing proteins are limiting.
Figure 3.—
Sir1p and the X element contribute to TPE at VI-R when silencing is compromised. (A) TPE assays. Tenfold serial dilutions were plated to assay silencing. Strains were grown in +Ura −His medium to maintain the S4-CT plasmid. TPE is compromised by the overexpression of the Sir4p C terminus, and this phenotype was exacerbated in the absence of Sir1p or the VI-R X element. (B) Quantitation of TPE. Error bars represent standard deviations. When possible, standard deviations were calculated separately above and below the mean. Values are presented as ratios of TPE level at the wild-type VI-R telomere to TPE level at VI-R in the absence of Sir1p or the X element. When TPE was compromised by S4-CT (striped bars), there was a statistically significant increase in the TPE ratio at VI-R in the absence of Sir1p, in comparison to sir1Δ alone (P < 0.008). When TPE was compromised by SIR4-CT, there was a statistically significant increase in the TPE ratio at VI-R koX, in comparison to the X-element knockout alone (P < 0.04).
Although deletion of the core-X element slightly decreased TPE levels at the VI-R telomere, TPE was nonetheless higher at VI-R koX than at either truncated VII-L (Figure 2C) or XI-L with a mutant X element (Pryde and Louis 1999). Therefore, we next examined whether, like Sir1p, the X element would make a larger contribution to the TPE status of the VI-R telomere in the presence of excess S4-CT. Deletion of the VI-R X element resulted in an 8.5-fold reduction in TPE when silencing was compromised (VI-R/VI-R koX + S4-CT; Figure 3B). In contrast, the X element did not enhance TPE at the VII-L telomere even in the presence of S4-CT (Figure 3A).
Thus, our data suggest that the role of core X in silencing increases when TPE is compromised by the limitation of Sir proteins, just as it plays a role in the presence of XCRs. Both Sir1p and the VI-R X element can contribute to silencing at the VI-R telomere, but their effects are minimal (koX) or undetectable (sir1Δ) unless silencing is compromised by overexpression of the carboxyl terminus of Sir4p. If the X element at the VI-R telomere recruits Sir1p and other silencing proteins, this recruitment pathway must be redundant in a wild-type cell, becoming important only when silencing proteins are limiting. However, at the truncated VII-L telomere, which has a much lower inherent level of silencing than the VI-R telomere, neither the VI-R X element nor Sir1p improved its TPE phenotype, even when silencing was compromised by S4-CT expression (Figure 3A). In addition, in the same strain two different truncated telomeres, neither of which has STEs, can have quite different levels of TPE (∼3% vs. ∼30%; Gottschling et al. 1990). We conclude that there must be an aspect (or aspects) of an individual chromosome end, other than the identity of its STEs, that is a major determinant of its TPE level.
What aspects of chromosome identity could contribute to TPE? One possibility is that control of TPE might act in trans, through higher-order chromatin structure, chromosome dynamics, or nuclear localization. Elsewhere we demonstrate that the truncated VII-L and native VI-R telomeres localize similarly to both the nuclear periphery and to pools of silencing proteins (Mondoux et al. 2007, accompanying article, this issue). Thus, differential localization of the two telomeres to these structures does not explain their different TPE phenotypes. In addition, there is no difference in the level of telomere binding of proteins that promote (Rap1p, yKu80p) or antagonize (Rif1p, Rif2p) TPE between the truncated VII-L and native VI-R telomeres (Sabourin et al. 2007). We also do not observe any difference in replication timing between the two telomeres (Sabourin et al. 2007).
Another model for the different TPE levels at different chromosome ends is the presence of proximal cis-acting sequences that either promote or repress TPE. Heterochromatin formation is thought to originate at the telomere and spread inward, with distance of spread determined by the concentration of available Sir3p (Renauld et al. 1993) and the activity of protosilencers and antisilencers in the subtelomeric region (Fourel et al. 1999, 2001; Pryde and Louis 1999). Although there are no known differences in the distal chromatin structure of these two telomeres that explain their different TPE phenotypes, it is possible that there are differences in the proximal chromatin structure that influence TPE. Asymmetric nucleosome spacing on either side of the HML-I and HMR-E silencers correlates with the preferential association of the Sir proteins to one side (Zou et al. 2006). The establishment of asymmetry precedes the formation of heterochromatin and is dependent on ORC and Abf1p (Zou et al. 2006). It is also possible that the distance of heterochromatin spread, and therefore level of TPE, depends on other proximal elements, which could include the identity and transcriptional activity of nearby genes. For example, the level of silencing at the VI-R X-ACS may be high in part because of the repression of the proximal gene, YFR057w (Wyrick et al. 1999; Vega-Palas et al. 2000).
Acknowledgments
We thank David Shore and Rolf Sternglanz for strains and plasmids, and Ed Louis, Jane Phillips, and Michelle Sabourin for helpful discussions and advice. We also thank James Broach, Paul Schedl, and Eugenia Xu for providing suggestions on the manuscript. This work was supported by grants from the National Institutes of Health to V.A.Z. and a National Science Foundation predoctoral fellowship to M.A.M.
References
- Aparicio, O. M., B. L. Billington and D. E. Gottschling, 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66: 1279–1287. [DOI] [PubMed] [Google Scholar]
- Barton, A. B., and D. B. Kaback, 2006. Telomeric silencing of an open reading frame in Saccharomyces cerevisiae. Genetics 173: 1169–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourns, B. D., M. K. Alexander, A. M. Smith and V. A. Zakian, 1998. Sir proteins, Rif proteins and Cdc13p bind Saccharomyces telomeres in vivo. Mol. Cell. Biol. 18: 5600–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigati, C., S. Kurtz, D. Balderes, G. Vidali and D. Shore, 1993. An essential yeast gene encoding a TTAGGG repeat-binding protein. Mol. Cell. Biol. 13: 1306–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, C. S. M., and B.-K. Tye, 1983. Organization of DNA sequences and replication origins at yeast telomeres. Cell 33: 563–573. [DOI] [PubMed] [Google Scholar]
- Chien, C. T., P. L. Bartel, R. Sternglanz and S. Fields, 1991. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. USA 88: 9578–9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien, C. T., S. Buck, R. Sternglanz and D. Shore, 1993. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell 75: 531–541. [DOI] [PubMed] [Google Scholar]
- Diffley, J. F., and B. Stillman, 1989. Similarity between the transcriptional silencer binding proteins ABF1 and RAP1. Science 246: 1034–1038. [DOI] [PubMed] [Google Scholar]
- Foss, M., F. J. McNally, P. Laurenson and J. Rine, 1993. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cereivisae. Science 262: 1838–1844. [DOI] [PubMed] [Google Scholar]
- Fourel, G., C. Boscheron, E. Revardel, E. Lebrun, Y. F. Hu et al., 2001. An activation-independent role of transcription factors in insulator function. EMBO Rep. 2: 124–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel, G., E. Revardel, C. E. Koering and E. Gilson, 1999. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 18: 2522–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Gottschling, D. E., O. M. Aparicio, B. L. Billington and V. A. Zakian, 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762. [DOI] [PubMed] [Google Scholar]
- Haber, J. E., 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32: 561–599. [DOI] [PubMed] [Google Scholar]
- Hediger, F., F. R. Neumann, G. Van Houwe, K. Dubrana and S. M. Gasser, 2002. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 12: 2076–2089. [DOI] [PubMed] [Google Scholar]
- Kurtz, S., and D. Shore, 1991. RAP1 protein activates and silences transcription of mating-type genes in yeast. Genes Dev. 5: 616–628. [DOI] [PubMed] [Google Scholar]
- Lebrun, E., E. Revardel, C. Boscheron, R. Li, E. Gilson et al., 2001. Protosilencers in Saccharomyces cerevisiae subtelomeric regions. Genetics 158: 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis, E. J., 1995. The chromosome ends of Saccharomyces cerevisiae. Yeast 11: 1553–1574. [DOI] [PubMed] [Google Scholar]
- Luo, K., M. A. Vega-Palas and M. Grunstein, 2002. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 16: 1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, M. D., D. Mahoney, A. Rose, J. B. Hicks and J. R. Broach, 1987. Functional domains of SIR4, a gene required for position effect regulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 4441–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed, D., A. Kistler, A. Axelrod, J. Rine and A. D. Johnson, 1997. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl. Acad. Sci. USA 94: 2186–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondoux, M. A., and V. A. Zakian, 2005. Telomere position effect: silencing near the end, pp. 261–316 in Telomeres, Ed. 2, edited by T. de Lange, V. Lundblad and E. H. Blackburn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Mondoux, M. A., J. G. Scaife and V. A. Zakian, 2007. Differential nuclear localization does not determine the silencing status of Saccharomyces cerevisiae telomeres. Genetics 177: 2019–2029. [DOI] [PMC free article] [PubMed]
- Moretti, P., K. Freeman, L. Coodly and D. Shore, 1994. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 8: 2257–2269. [DOI] [PubMed] [Google Scholar]
- Morrow, B. E., S. P. Johnson and J. R. Warner, 1989. Proteins that bind to the yeast rDNA enhancer. J. Biol. Chem. 264: 9061–9068. [PubMed] [Google Scholar]
- Pillus, L., and J. Rine, 1989. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell 59: 637–647. [DOI] [PubMed] [Google Scholar]
- Pryde, F. E., and E. J. Louis, 1999. Limitations of silencing at native yeast telomeres. EMBO J. 18: 2538–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renauld, H., O. M. Aparicio, P. D. Zierath, B. L. Billington, S. K. Chhablani et al., 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7: 1133–1145. [DOI] [PubMed] [Google Scholar]
- Sabourin, M., C. T. Tuzon and V. A. Zakian, 2007. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol. Cell 27: 550–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanower, G. A., M. Muller, J. L. Blanton, V. Honti, H. Gyurkovics et al., 2005. Characterization of the grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics 169: 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger, S., A. Hecht, K. Luo and M. Grunstein, 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11: 83–93. [DOI] [PubMed] [Google Scholar]
- Tham, W. H., J. S. Wyithe, P. K. Ferrigno, P. A. Silver and V. A. Zakian, 2001. Localization of yeast telomeres to the nuclear periphery is separable from transcriptional repression and telomere stability functions. Mol. Cell 8: 189–199. [DOI] [PubMed] [Google Scholar]
- Triolo, T., and R. Sternglanz, 1996. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature 381: 251–253. [DOI] [PubMed] [Google Scholar]
- Tsukamoto, Y., J. Kato and H. Ikeda, 1997. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature 388: 900–903. [DOI] [PubMed] [Google Scholar]
- Vega-Palas, M. A., E. Martin-Figueroa and F. J. Florencio, 2000. Telomeric silencing of a natural subtelomeric gene. Mol. Gen. Genet. 263: 287–291. [DOI] [PubMed] [Google Scholar]
- Vega-Palas, M. A., S. Venditti and E. Di Mauro, 1997. Telomeric transcriptional silencing in a natural context. Nat. Genet. 15: 232–233. [DOI] [PubMed] [Google Scholar]
- Wang, H., P. R. Nicholson and D. J. Stillman, 1990. Identification of a Saccharomyces cerevisiae DNA-binding protein involved in transcriptional regulation. Mol. Cell. Biol. 10: 1743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick, J. J., J. G. Aparicio, T. Chen, J. D. Barnett, E. G. Jennings et al., 2001. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294: 2357–2360. [DOI] [PubMed] [Google Scholar]
- Wyrick, J. J., F. C. Holstege, E. G. Jennings, H. C. Causton, D. Shore et al., 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402: 418–421. [DOI] [PubMed] [Google Scholar]
- Xu, W., J. G. Aparicio, O. M. Aparicio and S. Tavare, 2006. Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics 7: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakian, V. A., and J. F. Scott, 1982. Construction, replication and chromatin structure of TRP1 RI circle, a multiple-copy synthetic plasmid derived from Saccharomyces cerevisiae chromosomal DNA. Mol. Cell. Biol. 2: 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, Y., Q. Yu and X. Bi, 2006. Asymmetric positioning of nucleosomes and directional establishment of transcriptionally silent chromatin by Saccharomyces cerevisiae silencers. Mol. Cell. Biol. 26: 7806–7819. [DOI] [PMC free article] [PubMed] [Google Scholar]