Abstract
Understanding the mechanisms controlling the generation and maintenance of biodiversity provides some of the planet's greatest and most pressing challenges. Variation in resource concentration, which varies widely at multiple scales, may cause biodiversity to increase, decrease, or exhibit a unimodal response and underlying mechanisms remain obscure. We established experimental cultures of long-term stationary phase (LTSP) Escherichia coli to test whether per capita heterozygosity varies with resource concentration, and, if so, whether population sizes associated with different resource concentrations contributed to these patterns. Our results provide the clearest example to date of increasing per capita heterozygosity with increasing resource concentration. Further, our experimental manipulations of population size, independent of resource concentration, provide the first unequivocal evidence that population size is one of the underlying factors controlling per capita heterozygosity along such resource gradients. Specifically, we show that cultures with higher maximum population sizes, associated with higher resource concentrations, have higher per capita heterozygosity. These experiments provide the first experimental evidence for an underappreciated factor controlling biodiversity along resource gradients—population size. This direct evidence of population size influencing diversification rates has implications for regional and global scale patterns of biodiversity.
BIODIVERSITY varies widely across the planet, along gradients of temperature and productivity (Mittelbach et al. 2001; Willig et al. 2003; Hillebrand 2004; Pennisi 2005; Suding et al. 2005; Jablonski et al. 2006). The near-infinite complexity and variability of natural ecosystems has, perhaps, slowed our ability to both characterize patterns and elucidate mechanisms underlying the origin and maintenance of this biodiversity. Here we use a simple model system to show unambiguously how diversification rates increase monotonically with both resource availability and population size.
Dozens of hypotheses have been generated to explain variation in diversity along resource gradients, and most of them have focused on how resource concentration, per se, can influence competitive interactions and predator–prey dynamics (Waide et al. 1999). In nature, resource concentration gradients inevitably confound changes in ratios of potentially limiting factors, and this variation probably contributes to the variation in the patterns of diversity we see along resource and productivity gradients. It is critical that we determine experimentally the baseline patterns of diversity along gradients that are under our complete control. Only this will allow us to observe what tends to happen along resource gradients where only concentration, and not ratios, vary.
Resource gradients create not only gradients in biodiversity, but also gradients in population size, and this variation in population size may generate patterns of both genetic and species diversity (hereafter diversity) (Stevens and Carson 1999; Vandermeulen et al. 2001; Suding et al. 2005; Bazin et al. 2006). Neutral theory predicts that larger populations (perhaps associated with high resource levels) will have greater heterozygosity (Crow and Kimura 1970). Further, we know that larger populations have higher rates of adaptation (de Visser and Rozen 2005), and this has the potential to increase the rate of adaptive diversification. Clonal interference in asexual populations (Gerrish and Lenski 1998) and the Hill–Robertson effect in sexual populations (Hill and Robertson 1966), in which more beneficial mutations compete with each other and slow the rate of selective sweeps, will also maintain greater diversity in larger populations. It is also widely accepted, however, that microbial populations can rapidly lose diversity through selective sweeps (Atwood et al. 1951) and hitchhiking (Gillespie 2001)—increased evolutionary rates caused by larger population size would serve only to exacerbate this loss of diversity. Experimental verification of the effect of population size on diversification rate is lacking, yet would have profound implications for global patterns of biodiversity along resource and latitudinal gradients.
Here we describe dynamics of population size and per capita genetic diversity (hereafter simply “heterozygosity”) in highly dynamic long-term stationary phase (LTSP) cultures of Escherichia coli. The LTSP of microbial population dynamics follows the better characterized lag, exponential, stationary, and death phases (Finkel 2006). LTSP is characterized by relatively low population sizes (106–107 cells · ml−1), rapid turnover, and dynamic evolutionary change (Zambrano and Kolter 1996; Yeiser et al. 2002; Finkel 2006). This population turnover and evolutionary dynamism begins to appear almost immediately, even in overnight cultures, and continues indefinitely for days, weeks, and years.
One of the most notable changes is the accumulation of beneficial mutations that confer a competitive advantage to surviving cells (Zambrano et al. 1993; Finkel and Kolter 1999; Finkel et al. 2000; Yeiser et al. 2002). Frequently, when cells are grown under aerobic conditions in Luria–Bertani medium, the first gene to receive a detectable mutation is rpoS, the gene encoding the stationary phase-specific or stress-response sigma factor RpoS (Zambrano et al. 1993; Hengge-Aronis 2000; Farrell and Finkel 2003). For example, in one experiment, individual LTSP cultures contained 6–7 rpoS mutant alleles after only 10 days of incubation (M. J. Farrell, V. Palchevskiy, T. Jessner and S. E. Finkel, unpublished results). Such mutations frequently result in different degrees of attenuated RpoS activity.
We used well-mixed LTSP cultures of E. coli populations to test (i) whether heterozygosity varies with resource concentration in an unstructured environment, and (ii) whether larger populations associated with high resource concentrations could account for the observed pattern of heterozygosity. Within a single culture, different rpoS mutations frequently result in different degrees of attenuated RpoS activity, and we can observe directly the degree of attenuation using a reporter gene. For our experiments, observed variation (based on subsamples of equal cell numbers) in the expression of this reporter served as our measure of the heterozygosity of LTSP cultures of E. coli. In our first experiment, we established an initial resource-concentration gradient and assayed population size and heterozygosity for 10 days. In our second experiment, we manipulated both initial population size and initial resource concentration independently and then assayed the response of both population size and heterozygosity to this manipulation for 10 days.
MATERIALS AND METHODS
Bacterial cultures:
Following standard methods for long-term stationary phase culture of E. coli (Zambrano and Kolter 1996; Finkel et al. 2000; Finkel 2006), all experiments used strains derived from ZK918, an E. coli K-12 (nonmating) strain, that carries a bolA∷lacZ reporter fusion to monitor the appearance of rpoS mutants within the populations (see below; Bohannon et al. 1991). SF2054 and SF2055 are otherwise isogenic versions of ZK918, expressing resistance to either nalidixic acid (NalR) or streptomycin (StrR), respectively (Farrell and Finkel 2003). The parental strain and its derivatives are commonly used in long-term survival and evolution experiments (Zambrano and Kolter 1993, 1996; Finkel and Kolter 1999; Finkel et al. 2000). The NalR and StrR markers (encoded by mutant alleles of gyrA and rpsL, respectively) are effectively neutral in the absence of drug selection, having no effect on competitive advantage under these growth conditions (Finkel and Kolter 1999; Finkel et al. 2000). In addition, we routinely assay for the spontaneous appearance of both drug markers in either strain and have not observed a NalR strain become StrR, or vice versa. All bacterial cultures were grown in Luria–Bertani medium (LB; Difco, Detroit), a standard complex medium for E. coli experiments. Cultures were grown in 7.0 ml of LB broth in 18 × 150 mm glass test tubes, incubated at 37°, mounted at a 45° angle, and shaken at 200 rpm on a New Brunswick shaker platform for constant aeration.
Identification of rpoS mutants under different growth conditions:
Derivatives of the RpoS-reporter strain ZK918 were used to observe phenotypic changes due to mutation at rpoS. These strains carry a bolA∷lacZ reporter fusion (Bohannon et al. 1991). bolA gene expression is under control of the general regulatory protein RpoS (Hengge-Aronis 2000); maximal lacZ expression is dependent on wild-type alleles of rpoS. To determine the RpoS phenotype of evolved strains, individual colonies are plated on MacConkey agar (Difco). Colonies are scored as wild type or a category of attenuated expression on the basis of their color: red for wild-type cells, and dark pink, light pink, or white for different attenuated mutant alleles (Farrell and Finkel 2003). The status of the rpoS gene can be further confirmed by performing the catalase “bubble assay” (expression of the KatE catalase is dependent on RpoS) (Bohannon et al. 1991; Zambrano and Kolter 1993).
Use of the bolA∷lacZ reporter as a phenotypic marker of genetic variation, provides a very robust and reliable indicator of rpoS activity. RpoS activity is a trait that comprises a critical component of a cell's complex fitness response to stressful environments (Hengge-Aronis 2000; Notley-McRobb et al. 2002; Farrell and Finkel 2003). As a measure of phenotypic, rather than genotypic, diversity, it reflects traits that more directly influence interactions and responses to abiotic factors. Our observations of the reporter trait, however, comprise only four categories derived from a continuum of likely types: “red,” “dark pink,” “light pink,” and “white.” This may be akin to describing the diversity of a plant community with four functional types, for instance, annual weeds, perennial weeds, early-successional trees, and late-successional trees. Such gross differences provide a sound starting point for understanding ecologically relevant phenotypic diversity in an assemblage of species or phenotypes within a species. Nonetheless, such a coarse classification may result in a poor resolution of true taxonomic diversity and composition. If we observe patterns of diversity at such a coarse resolution, they are likely to represent very robust patterns.
Assaying population size and diversity of rpoS mutants:
Each culture was sampled, serially diluted, and spread onto MacConkey agar plates to determine the color of individual colonies and population densities. One plate per culture, containing 150–300 colonies, was selected for enumeration. The viable cells were counted and scored by their colors (red for wild type and dark pink, light pink, and white colonies for different levels of attenuated activity).
Nei's heterozygosity (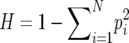 , aka Simpson's diversity) measured per capita culture diversity (Simpson 1949; Nei 1987; Stevens et al. 2003) using the proportions, pi, of sample colonies of each color. Because we have a finite number of observed mutant phenotypes (white, light pink, dark pink, plus the red wild type), maximum heterozygosity (
, aka Simpson's diversity) measured per capita culture diversity (Simpson 1949; Nei 1987; Stevens et al. 2003) using the proportions, pi, of sample colonies of each color. Because we have a finite number of observed mutant phenotypes (white, light pink, dark pink, plus the red wild type), maximum heterozygosity (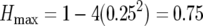 ), as well as the heterozygosity expected under neutrality (Crow and Kimura 1970) are both <1. All assays of replicate cultures were based on approximately equal numbers of cells, and a regression (not shown) of sample size and diversity revealed no pattern.
), as well as the heterozygosity expected under neutrality (Crow and Kimura 1970) are both <1. All assays of replicate cultures were based on approximately equal numbers of cells, and a regression (not shown) of sample size and diversity revealed no pattern.
Because of our use of both a diversity index and similar sample sizes across replicates, our results reflect the relative abundance of phenotypes, i.e., diversity or population-level heterozygosity. The results do not reflect sampling artifacts, wherein larger populations contain more total mutants—rather, they reflect variation in diversity.
Resource gradient experiment:
In the first experiment, LB concentrations for each experiment included the standard concentration (1×) and four twofold serial dilutions (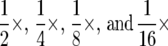 ). Each strain was replicated twice at each resource concentration, for a total of 20 tubes. Sterile distilled water was added to each culture periodically to maintain constant volume. Population sizes and heterozygosity were assayed each day for 10 days, using methods described above. Although we report results for heterozygosity, we obtained directly analogous results when using other measures of diversity and dominance, such as the proportion of the sample composed of the wild type (red colonies). General linear mixed models with repeated measures tested effects of resource concentration, sampling day, and their interaction on the logarithm of total population size and the arcsin square root of diversity. The most parsimonious random-effects structure was determined with likelihood ratio tests (Pinheiro and Bates 2000). The most parsimonious models included culture tubes as independent random variables, and variances estimated independently for each nutrient concentration. All analyses were performed with the R language (R Development Core Team 2007) using the nlme package (Pinheiro and Bates 2000).
). Each strain was replicated twice at each resource concentration, for a total of 20 tubes. Sterile distilled water was added to each culture periodically to maintain constant volume. Population sizes and heterozygosity were assayed each day for 10 days, using methods described above. Although we report results for heterozygosity, we obtained directly analogous results when using other measures of diversity and dominance, such as the proportion of the sample composed of the wild type (red colonies). General linear mixed models with repeated measures tested effects of resource concentration, sampling day, and their interaction on the logarithm of total population size and the arcsin square root of diversity. The most parsimonious random-effects structure was determined with likelihood ratio tests (Pinheiro and Bates 2000). The most parsimonious models included culture tubes as independent random variables, and variances estimated independently for each nutrient concentration. All analyses were performed with the R language (R Development Core Team 2007) using the nlme package (Pinheiro and Bates 2000).
Population size manipulation:
We tested directly the independent effects of population size and resource concentration on diversity by growing E. coli populations at different volumes, at different resource concentrations. The selected volumes and resource concentrations (see below) manipulated population size independently of resource concentration. In particular, they allowed the population size of the low-concentration–high-volume treatment to exceed the population size of the high-concentration–low-volume treatment. We established two replicates of each strain at each unique volume–concentration combination: 7 ml and 70 ml of 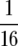 × standard LB concentration, and 0.7 ml and 7 ml of standard LB. Population sizes and Nei's heterozygosity were assayed each day for 10 days, using methods described above. Statistical methods are the same as those described above, and in addition, single degree-of-freedom contrasts used “dummy” (i.e., “treatment”) coding to test differences among unique treatment combinations for total population size on day 1, and heterozygosity on day 10.
× standard LB concentration, and 0.7 ml and 7 ml of standard LB. Population sizes and Nei's heterozygosity were assayed each day for 10 days, using methods described above. Statistical methods are the same as those described above, and in addition, single degree-of-freedom contrasts used “dummy” (i.e., “treatment”) coding to test differences among unique treatment combinations for total population size on day 1, and heterozygosity on day 10.
RESULTS
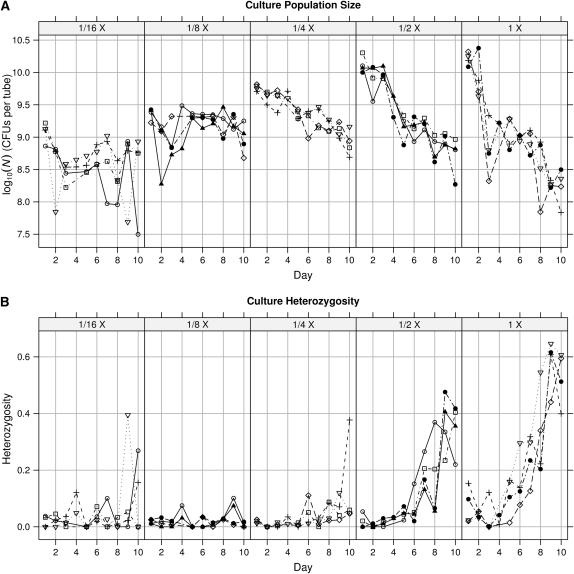
In the resource-concentration experiment, E. coli population size increased with resource concentration, but only in relatively young cultures (Figure 1; day × resource-concentration interaction: F36,125 = 6.53, P < 0.0001). Population size declined markedly through time in higher resource concentrations. E. coli heterozygosity increased over time (Figure 1), and to a much higher degree in higher resource concentrations (day × concentration: F36,125 = 5.73, P < 0.0001). This strong and consistent response is only the second such monotonic diversification–resource-concentration relation of which we are aware (Kassen et al. 2000). Number of different phenotypes also showed this pattern, although it is a less sensitive measure of changing phenotype frequencies (data not shown). Day 10 heterozygosity was strongly correlated with maximum population size (r = 0.74, P = 0.0002).
Figure 1.—
Time series of each replicate culture (line equals replicate) in the resource gradient experiment. Dilution of LB medium is indicated at the top. (A) Maximum (day 1) population size (CFUs per tube) increases significantly with increasing resource concentration. Over time population sizes tend to become similar among resource concentrations. (B) Heterozygosity tends to increase over time and does so more rapidly with higher resource concentrations.
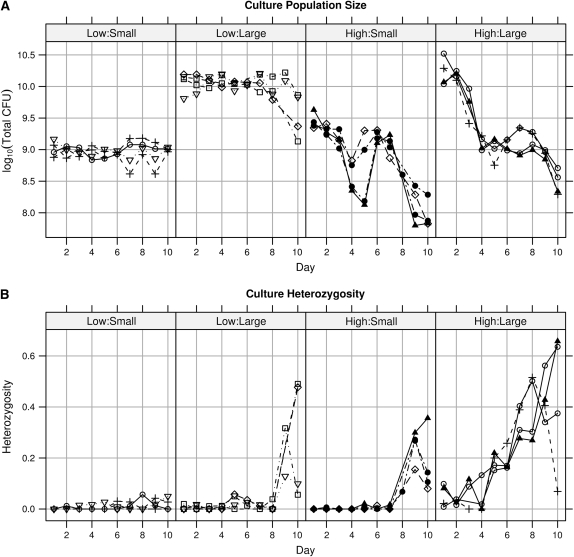
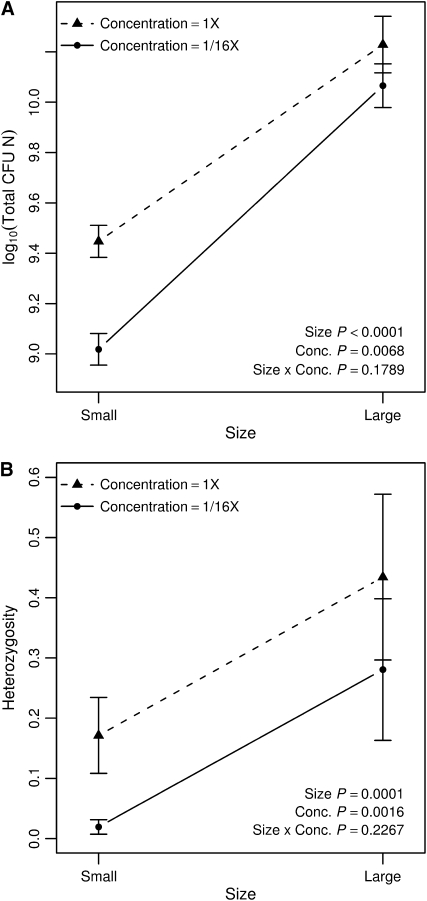
In the population size manipulation experiment, increasing flask size and resource concentration both caused increased population size and increased heterozygosity (Figure 2). Our experimental manipulation of total population size was successful in controlling day 1 (maximum) population size (Figure 3A). Resource concentration and culture flask size had independent and additive positive effects on day 1 total population size (Figure 3A) (general linear model coefficients: concentration, bConc = 0.429, t12 = 3.26, P = 0.0068; size, bSize = 1.047, t12 = 7.96, P < 0.0001; concentration × size interaction, bC×S = −0.266, t12 = 1.43, P = 0.1789). Resource concentration and culture flask size also had independent and additive positive effects on day 10 E. coli heterozygosity (Figure 3B) (model coefficients: concentration, bConc = 0.314, t12 = 4.05, P = 0.0016; size, bSize = 0.426, t12 = 5.50, P = 0.0001; concentration × size, bC×S = −0.140, t12 = 1.27, P = 0.2266).
Figure 2.—
Time series of each replicate culture (line equals replicate) in the population size experiment. LB concentration (low, 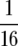 ×; high, 1×) and relative flask size (small, large, see text) are shown at the top. (A) Maximum (day 1) population size (CFUs per tube) increases with both flask size and resource concentration. Relative flask size has a large and persistent effect on population size. (B) Heterozygosity tends to increase over time and does so more rapidly in larger flasks and in higher resource concentrations.
×; high, 1×) and relative flask size (small, large, see text) are shown at the top. (A) Maximum (day 1) population size (CFUs per tube) increases with both flask size and resource concentration. Relative flask size has a large and persistent effect on population size. (B) Heterozygosity tends to increase over time and does so more rapidly in larger flasks and in higher resource concentrations.
Figure 3.—
Both day 1 (maximum) population size (A) and day 10 (final) heterozygosity (B) increase with flask size and with resource concentrations. Error bars are ±1 SE.
DISCUSSION
These experiments provide novel and fundamental contributions to our understanding of biodiversity gradients. Specifically, they reveal simultaneously (i) that diversification rate can increase along a resource gradient in an unstructured environment, and (ii) an explanation for the observed pattern—variation in maximum population size. Our first experiment demonstrates that higher resource concentration can lead to higher diversification in a thoroughly mixed, unstructured environment. This confirms expectations of both neutral theory and adaptive speciation. This is the second such experimental demonstration in a well-mixed enviornment of which we are aware (Figure 4a in Kassen et al. 2000) and provides important unambiguous confirmation of this result in a different model system, over a longer time period. This is important because the role of spatial heterogeneity in generating biodiversity patterns has a long and distinguished history (Hedrick et al. 1976; Tilman 1982; Hedrick 1986), and many hypotheses for natural diversity patterns rely on a significant role of spatial heterogeneity. Demonstrating the existence of diversity patterns in the absence of spatial heterogeneity may provide clues about natural patterns. Most importantly, our second experiment demonstrates that greater total population size contributes to the increased heterozygosity associated with higher resource concentrations. This is, to our knowledge, the first study demonstrating a mechanism (increased population size) underlying such a pattern of diversification along a resource concentration gradient.
Increased population size can enhance heterozygosity in several ways, and yet this flies in the face of conventional wisdom of laboratory microbial populations. Sweeps by mutants with higher fitness are often the expectation in such populations. In contrast, our results are consistent with expectations with several other mechanisms, including neutral mutation. In addition, clonal interference (Gerrish and Lenski 1998), mutator phenotypes (Tenaillon et al. 1999), and divergent selection (Dieckmann and Doebeli 1999) may all contribute to increased heterozygosity of larger populations in unstructured environments. This increased heterozygosity arose in spite of mechanisms that have the potential to reduce heterozygosity in large populations, such as increased hitchhiking (Gillespie 2001) and more rapid adaptive evolution that could result in more rapid sweeps. Future research must partition the relative importance of these mechanisms in unstructured environments. Regardless, however, of the details of these processes, population size does not explain all of the increased heterozygosity at higher resource concentrations.
Higher resource concentration alone, independent of its effect on initial population size, caused higher heterozygosity, and several processes might contribute to this. First, populations in higher resource concentrations may have higher mutation rates for at least two reasons aside from the effect of population size. Higher resource concentrations may cause more rapid turnover of individuals within populations thereby increasing the number of mutants appearing in a culture per unit time. Higher resource concentrations may also, through a variety of mechanisms, result in a higher concentration of free radicals (e.g., 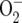 , ·OH) that may increase DNA damage and mutation rates (Finkel 2003; Nemoto and Finkel 2004).
, ·OH) that may increase DNA damage and mutation rates (Finkel 2003; Nemoto and Finkel 2004).
In addition to its effects on mutation rates, high resource concentrations may enhance the maintenance of heterozygosity in at least three ways. First, increased population size increases the absolute abundance of the rarest types, decreasing the probability of stochastic extinction (Abrams 1995). For this to be significant, however, there would need to be a mechanism for these types to increase in frequency enough to be observed in our samples. Second, increased resources may increase the abundance of rare resources or resource combinations used by rare phenotypes and allow these specialists to increase in frequency (Abrams 1995). Third, it has long been suggested that even with no change in the relative abundance or ratios of resources, increased concentration of all resources favors specialization (MacArthur and Pianka 1966; Dieckmann and Doebeli 1999). This specialization would permit a greater variety of mutants to persist on the same variety (but greater concentration) of resources. This adaptive diversification associated with narrowing of niche breadth is most likely to occur if there are not other costs associated with this change (Ackermann and Doebeli 2004). This seems quite plausible in this system with a moderately complex medium (LB broth) and in which we altered resource concentration without changing the initial relative concentrations of any of the substrates.
Recent work on adaptive diversification in both asexual and sexually reproducing populations has demonstrated clearly the plausibility of sympatric diversification (Doebeli et al. 2005). These advances suggest that the work presented here, using a clonally reproducing organism in a highly simplified unstructured laboratory environment has implications for not only bacteria, but for sexually reproducing, charismatic megafauna and flora.
The population size mechanism we describe here may have implications for the latitudinal gradient of taxonomic richness (Willig et al. 2003). If this mechanism applies generally, then regions or habitats with larger numbers of individuals will contain more species (or genera or genotypes) by virtue of having more individuals. Such areas may have more individuals by virtue of greater area alone or by virtue of higher population densities. The tropics, defined by climate, have a larger land surface (Rosenzweig 1992) and therefore may have more individuals than any other similarly uniform terrestrial habitat. Our population size hypothesis would therefore predict, all else being equal, higher diversity. Further, as climate periodically causes bottlenecks at higher latitudes, this would dramatically lower the effective population sizes at high latitudes, reducing the potential for diversification (Jablonski et al. 2006).
The population size mechanism could also have implications for patterns of diversity along other gradients. A unimodal pattern of diversity along productivity gradients might arise, for instance, if such habitats contain more individuals by virtue of greater area (Vandermeulen et al. 2001). It might also contribute to a mid-domain effect (Rangel and Diniz-Filho 2005) in which diversity peaks at the midpoint of any region with physical geographic constraints. Dispersal limitation due to physical boundaries might limit effective population size, so that effective population size would be lower near boundaries. The population size hypothesis would predict lower rates of diversification near these boundaries.
The above list of plausible implications suffers from a paradoxical chicken-and-egg problem: if larger populations exhibit more rapid diversification, this will reduce the size of the derived populations through lineage splitting. The resulting smaller populations would have correspondingly slower rates of diversification. This possible slowing of diversification, however, could well be offset by increased diversification stemming from greater diversity itself (Emerson and Kolm 2005). This might lead us to speculate that life seems destined for diversity: large populations diverge because they are large, and the resulting divergent groups further diversify because of their differences.
This work examines the processes of diversification and its subsequent maintenance over many generations, with implications for extant patterns on the globe. Extant patterns of diversity, however, reflect not only patterns of both diversification and its in situ maintenance, but also community assembly and species sorting following dispersal of allopatrically derived forms. Nonetheless, the implications of our results for community assembly seem clear: we can hypothesize that if particular conditions maintain a greater variety of sympatrically derived phenotypes, as we have shown, then it seems plausible that a greater variety of allopatrically derived phenotypes can be maintained following dispersal-mediated assembly. Whether this is generally the case, however, is untested.
It seems difficult to reconcile the arguably small differences in population size observed in the “real” world with the amazing differences in diversity between some contrasting regions of the planet. Other factors, such as temperature, may be more important than resource availability. Nonetheless, we might consider that speciation is an exponential (albeit slow) process (Emerson and Kolm 2005), because a greater variety of genotypes results in a wider base from which to diverge. Therefore, small differences in diversification rates may result, over time, in large differences in diversity.
Acknowledgments
We are grateful to P.J. Morin and M. Nordborg for comments on an earlier draft, and to M. Fields and G. Janssen for discussion and lab assistance. This work was funded in part by an NSF grant (DBI-0353915). S.F. was supported in part by the W.M. Keck Foundation.
References
- Abrams, P. A., 1995. Monotonic or unimodal diversity-productivity gradients: What does competition theory predict? Ecology 76: 2019–2027. [Google Scholar]
- Ackermann, M., and M. Doebeli, 2004. Evolution of niche width and adaptive diversification. Evolution 58: 2599–2612. [DOI] [PubMed] [Google Scholar]
- Atwood, K. C., L. K. Schneider and F. J. Ryan, 1951. Periodic selection in Escherichia coli. Proc. Natl. Acad. Sci. USA 37: 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin, E., S. Glemin and N. Galtier, 2006. Population size does not influence mitochondrial genetic diversity in animals. Science 312: 570–572. [DOI] [PubMed] [Google Scholar]
- Bohannon, D. E., N. Connell, J. Keener, A. Tormo, M. Espinosa-Urgel et al., 1991. Stationary-phase-inducible “gearbox” promoters: differential effects of katF mutations and role of sigma 70. J. Bacteriol. 173: 4482–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow, J. F., and M. Kimura, 1970. An Introduction to Population Genetics Theory. Harper & Row, New York.
- de Visser, J. A. G. M., and D. E. Rozen, 2005. Limits to adapation in asexual populations. J. Evol. Biol. 18: 779–788. [DOI] [PubMed] [Google Scholar]
- Dieckmann, U., and M. Doebeli, 1999. On the origin of species by sympatric speciation. Nature 400: 354–357. [DOI] [PubMed] [Google Scholar]
- Doebeli, M., U. Dieckmann, J. A. J. Metz and D. Tautz, 2005. What we have also learned: adaptive speciation is theoretically plausible. Evolution 59: 691–695. [PubMed] [Google Scholar]
- Emerson, B. C., and N. Kolm, 2005. Species diversity can drive speciation. Nature 434: 1015–1017. [DOI] [PubMed] [Google Scholar]
- Farrell, M. J., and S. E. Finkel, 2003. The growth advantage in stationary-phase phenotype conferred by rpoS mutation is dependent on the pH and nutrient environment. J. Bacteriol. 185: 7044–7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel, S. E., 2006. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat. Rev. Microbiol. 4: 113–120. [DOI] [PubMed] [Google Scholar]
- Finkel, S. E., and R. Kolter, 1999. Evolution of microbial diversity during prolonged starvation. Proc. Natl. Acad. Sci. USA 96: 4023–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel, S. E., E. R. Zinser and R. Kolter, 2000. Long-term survival and evolution in the stationary phase, pp. 231–238 in Bacterial Stress Responses, edited by G. Storz and R. Hengge-Aronis. ASM, Washington, DC.
- Finkel, T., 2003. Oxidant signals and oxidative stress. Curr. Opin. Cell Biol. 15: 247–254. [DOI] [PubMed] [Google Scholar]
- Gerrish, P. J., and R. E. Lenski, 1998. The fate of competing beneficial mutations in an asexual population. Genetica 103: 127–144. [PubMed] [Google Scholar]
- Gillespie, J. H., 2001. Is the population size of a species relevant to its evolution? Evolution 55: 2161–2169. [DOI] [PubMed] [Google Scholar]
- Hedrick, P. W., 1986. Genetic polymorphism in heterogeneous envrionments: a decade later. Annu Review of Ecology and Systematics 17: 535–566. [Google Scholar]
- Hedrick, P. W., M. E. Ginevan and E. P. Ewing, 1976. Genetic polyorphism in a heterogeneous environment. Annu. Rev. Ecol. Syst. 7: 1–32. [Google Scholar]
- Hengge-Aronis, R., 2000. The general stress response in Escherichia coli, pp. 161–178 in Bacterial Stress Responses, edited by G. Storz and R. Hengge-Aronis. ASM, Washington, DC.
- Hill, W. G., and A. Robertson, 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269–294. [PubMed] [Google Scholar]
- Hillebrand, H., 2004. On the generality of the latitudinal diversity gradient. Am. Nat. 163: 192–211. [DOI] [PubMed] [Google Scholar]
- Jablonski, D., K. Roy and J. W. Valentine, 2006. Out of the tropics: evolutionary dynamics of the latitudinal gradient. Science 314: 102–106. [DOI] [PubMed] [Google Scholar]
- Kassen, R., A. Buckling, G. Bell and P. B. Rainey, 2000. Diversity peaks at intermediate productivity in a laboratory microcosm. Nature 406: 508–512. [DOI] [PubMed] [Google Scholar]
- MacArthur, R., and E. Pianka, 1966. On optimal use of a patchy environment. Am. Nat. 100: 603–609. [Google Scholar]
- Mittelbach, G. G., C. F. Steiner, S. M. Scheiner, K. L. Gross, H. L. Reynolds et al., 2001. What is the observed relationship between species richness and productivity? Ecology 82: 2381–2396. [Google Scholar]
- Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
- Nemoto, S., and T. Finkel, 2004. Aging and the mystery at Arles. Nature 429: 149–152. [DOI] [PubMed] [Google Scholar]
- Notley-McRobb, L., T. King and T. Ferenci, 2002. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress types. J. Bacteriol. 184: 806–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi, E., 2005. What determines species diversity? Science 309: 90. [DOI] [PubMed] [Google Scholar]
- Pinheiro, J., and D. Bates, 2000. Mixed-Effects Models in S and S-PLUS. Springer, New York.
- R Development Core Team, 2007. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna.
- Rangel, T. F. L. V. B., and J. A. F. Diniz-Filho, 2005. Neutral community dynamics, the mid-domain effect and spatial patterns in species richness. Ecol. Lett. 8: 783–790. [Google Scholar]
- Rosenzweig, M. L., 1992. Species diversity gradients: we know more and less than we thought. J. Mammal. 73: 715–730. [Google Scholar]
- Simpson, E. H., 1949. Measurement of diversity. Nature 163: 688. [Google Scholar]
- Stevens, M. H. H., and W. P. Carson, 1999. The significance of assemblage level thinning for species richness. J. Ecol. 87: 490–502. [Google Scholar]
- Stevens, M. H. H., O. L. Petchey and P. E. Smouse, 2003. Stochastic relations between species richness and the variability of species composition. Oikos 103: 479–488. [Google Scholar]
- Suding, K. N., S. L. Collins, L. Gough, C. Clark, E. E. Cleland et al., 2005. Functional- and abundance-based mechanisms explain diversity loss due to n fertilization. Proc. Natl. Acad. Sci. USA 102: 4387–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon, O., B. Toupance, H. Le Nagard, F. Taddei and B. Godelle, 1999. Mutators, population size, adaptive landscape and the adaptation of asexual populations of bacteria. Genetics 152: 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman, D., 1982. Resource Competition and Community Structure (Monographs in Population Biology). Princeton University Press, Princeton, NJ. [PubMed]
- VanderMeulen, M., A. Hudson and S. M. Scheiner, 2001. Three evolutionary hypotheses for the hump-shaped productivity-diversity curve. Evol. Ecol. Res. 3: 379–392. [Google Scholar]
- Waide, R. B., M. R. Willig, C. F. Steiner, G. Mittelbach, L. Gough et al., 1999. The relationship between productivity and species richness. Annu. Rev. Ecol. Syst. 30: 257–300. [Google Scholar]
- Willig, M. R., D. M. Kaufman and R. D. Stevens, 2003. Latitudinal gradients of biodiversity: pattern, process, scale, and synthesis. Annu. Rev. Ecol. Syst. 34: 273–309. [Google Scholar]
- Yeiser, B., E. D. Pepper, M. F. Goodman and S. E. Finkel, 2002. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc. Natl. Acad. Sci. USA 99: 8737–8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano, M. M., and R. Kolter, 1993. Escherichia coli mutants lacking Nadh Dehydrogenase-I have a competitive disadvantage in stationary phase. J. Bacteriol. 175: 5642–5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano, M. M., and R. Kolter, 1996. GASPing for life in stationary phase. Cell 86: 181–184. [DOI] [PubMed] [Google Scholar]
- Zambrano, M. M., D. A. Siegele, M. Almiron, A. Tormo and R. Kolter, 1993. Microbial competition—Escherichia coli mutants that take over stationary phase cultures. Science 259: 1757–1760. [DOI] [PubMed] [Google Scholar]