Abstract
SOD-1 and SOD-2 detoxify superoxide in the cytoplasm and mitochondria. We find that, although several long-lived mutants of Caenorhabditis elegans have increased SOD levels, this phenomenon does not correlate with life span or growth rate. Furthermore, although disruption of sod-1 or -2 expression produces numerous phenotypes, including increased sensitivity to paraquat and increased oxidative damage to proteins (except in daf-2 mutants), this fails to shorten the life span of these long-lived mutants. In fact, sod-1(RNAi) increases the life span of daf-2 mutants and sod-2(RNAi) that of clk-1 mutants. Our results suggest that increased superoxide detoxification and low oxidative damage are not crucial for the longevity of the mutants examined, with the possible exception of daf-2, where our results are inconclusive. These results are surprising because several of the long-lived mutants that we examined specifically affect mitochondrial electron transport, a process whose involvement in life-span determination is believed to be related to superoxide generation. We discuss the significance of our findings in light of the oxidative stress theory of aging.
A variety of Caenorhabditis elegans mutants display an increased, or shortened, life span compared to the wild type under standard conditions (Lakowski and Hekimi 1998; Guarente and Kenyon 2000; Hekimi et al. 2001b; Tissenbaum and Guarente 2002; Hekimi and Guarente 2003; Boehm and Slack 2005; Kenyon 2005). In life-span studies in a variety of organisms, it is frequently found that increased or decreased life span is associated with changes in the biology of reactive oxygen species (ROS) (Beckman and Ames 1998; Golden et al. 2002; Droge 2003; Hekimi and Guarente 2003; Balaban et al. 2005), and this is also true for a variety of C. elegans mutants (Vanfleteren and Braeckman 1999; Feng et al. 2001; Senoo-Matsuda et al. 2001; Hekimi and Guarente 2003; Shibata et al. 2003; de Castro et al. 2004; Kayser et al. 2004; Sedensky and Morgan 2006) and wild-type animals treated by RNA interference (RNAi) (Dillin et al. 2002; Lee et al. 2003). However, although these studies, as well as many other studies that have investigated the biochemical and molecular changes that accompany altered patterns of aging, provide strong arguments for the importance of ROS in life-span determination, it remains difficult to prove or disprove the hypothesis that ROS cause aging by damaging macromolecules. To date, there is conflicting evidence in favor of both views. For example, although overexpression of catalase in mouse mitochondria increases life span in some genetic backgrounds (Schriner et al. 2005), the life span of Sod2 +/− mice, which demonstrably suffer increased oxidative stress (Mansouri et al. 2006), is not shortened (Van Remmen et al. 2003), nor does transgenic overexpression of the cytoplasmic SOD1 in mice increase the life span of the transgenic animals (Huang et al. 2000).
One way to manipulate ROS levels in vivo, and to test their effects on the life span of intact organisms, is by altering the expression of genes that code for proteins involved in ROS production or detoxification. This strategy has been followed in a variety of organisms, in particular by knocking out, or overexpressing, detoxifying enzymes such as superoxide dismutases (SOD) (Phillips et al. 1989; Parkes et al. 1999; Melov et al. 2001; Sun et al. 2002; Duttaroy et al. 2003; Fabrizio et al. 2003; Harris et al. 2003). In C. elegans, there are five distinct genes that code for SODs: sod-2 and sod-3 encode MnSODs that are localized in the mitochondrial matrix (Hunter et al. 1997), sod-1 encodes the classical cytoplasm CuZnSOD (Giglio et al. 1994), sod-4 encodes a membrane-bound and extracellular CuZn SOD (Fujii et al. 1998), and sod-5 encodes a relatively uncharacterized, alternative cytoplasmic CuZnSOD (Jensen and Culotta 2005). We focus on sod-1 and sod-2 because they encode the primary and the best-characterized superoxide dismutases of the two categories: mitochondrial matrix MnSODs and CuZnSODs.
Most, or all, long-lived C. elegans mutants display other phenotypes in addition to increased life span (Hekimi et al. 2001a,b; Hekimi and Guarente 2003). To test whether the increased life span of these mutants could be due to low ROS levels, we have investigated whether any of the phenotypes of long-lived mutants, including their life span, were correlated with the expression of SODs or could be suppressed by knocking down the SODs by RNAi. We focused on strains carrying one or several mutations in four well-characterized genes whose alteration can result in increased longevity and whose interactions have been previously studied. daf-2 encodes an insulin-receptor-like tyrosine kinase (Kenyon et al. 1993); clk-1 encodes a ubiquinone biosynthetic enzyme (Ewbank et al. 1997; Stenmark et al. 2001) and clk-1 mutants display respiratory defects (Felkai et al. 1999; Miyadera et al. 2001; Braeckman et al. 2002a,b; Kayser et al. 2004); isp-1 encodes the “Rieske” iron–sulfur protein (Feng et al. 2001), a catalytic subunit of mitochondrial complex III; and ctb-1, a gene encoded by the mitochondrial genome, encodes the cytochrome b of complex III (Feng et al. 2001). Mutations in daf-2 and clk-1 act synergistically to induce a very long life span (Lakowski and Hekimi 1996). In contrast, mutations in isp-1 and daf-2 are nonadditive (Feng et al. 2001), suggesting that mutations in these genes affect life span by a mechanism that is similar, although loss-of-function mutations in daf-16, which abolish the increased life span of daf-2 mutants, do not abolish the increased life span of isp-1 (Feng et al. 2001) or isp-1 daf-2 mutants (J. Feng and S. Hekimi, unpublished observations). ctb-1(qm189) strongly suppresses many of the isp-1 phenotypes, including slow development and behavior, but does not suppress the increased life span (Feng et al. 2001).
It is often difficult to interpret the effect of life-span-shortening treatments because a deleterious treatment could shorten life span without acting on the actual aging process. However, it is possible to exclude a major role in life-span determination of a particular molecular process (here superoxide detoxification) when a treatment is well defined and its molecular and phenotypic effects can be directly observed, yet the process does not impinge significantly on life span. Here we show that the increased life spans of mutants that have altered mitochondrial function and/or increased resistance to oxidative stress are not affected by reductions in superoxide detoxification and concomitant increases in oxidative damage.
MATERIALS AND METHODS
Strains used:
The following strains were used: N2 (wild type), isp-1(qm150), isp-1(qm150); ctb-1(qm189), clk-1 (qm30), daf-2(e1370), daf-2(e1370); clk-1(qm30), daf-2(e1370); isp-1(qm150), daf-2 (e1370); isp-1(qm150); ctb-1(qm189), and mev-1(kn1). Worms were cultured at 20°.
Generation of sod-1 and sod-2 RNAi clones:
Full-length sod-1 and sod-2 cDNA sequences were obtained by RT–PCR and were cloned into the L4440 feeding vector (pPD129.36). The resulting plasmids were transformed into bacteria DH10b and then retransformed into the RNase III-deficient feeding strain HT115 (DE3) (Shibata et al. 2003). Primer sequences can be obtained upon request.
Feeding RNAi:
Single colonies of HT115 bacteria were inoculated in LB broth containing 50 μg/ml ampicillin (Amp) and grown overnight at 37°. Overnight cultures were diluted 1:100 and allowed to grow for 6–8 hr and then seeded directly onto NGM plates with Amp and 1 mm IPTG. Seeded plates were dried and induced overnight at room temperature. Animals that were grown on bacteria transformed with the empty vector were used as control. The technique used is essentially as described (Kamath et al. 2001).
Post-embryonic development:
L4 stage hermaphrodite worms (P0) were placed onto newly prepared (fresh) RNAi plates and were incubated at 20° for 24 hr. Then young adults were transferred to fresh RNAi plates and were incubated for 24–48 hr to lay eggs. Unstaged eggs were placed onto RNAi plates and left for 3 hr to hatch. Larvae (F1 progeny) that had hatched during that period were placed onto fresh RNAi plates (10/plate for a total of 50) at 20° and were monitored every 1.5 or 3 hr until maturity.
Life span:
The aging experiments were begun as in post-embryonic development assays. F1 worms were cultured on RNAi plates at 20° and examined every day until death. Animals were transferred to fresh RNAi plates twice each week.
Brood size:
F1 worms were placed singly onto fresh RNAi plates at 20° until they had matured and begun to lay eggs. The hermaphrodites then were transferred to RNAi plates daily and the progeny were counted. Worms that did not lay any eggs were defined as sterile.
Embryonic lethality:
Eggs produced by F1 worms during a limited time period (4–6 hr) were placed onto fresh RNAi plates and were monitored every day until hatching. The eggs that did not hatch were scored as dead embryos.
Paraquat resistance:
Groups of L4 larvae were grown on control as well as on sod-1 and sod-2 RNAi plates until their progeny had reached the L4 stage. A total of 50 of those were transferred onto NGM plates with 4 mm paraquat (PQ) and kept for 72 hr at 20°, at which time the number of surviving worms was counted.
Paraquat-induced damage to proteins:
L1 wild-type worms were transferred onto plates with different concentrations of paraquat (0.0, 0.1, 0.2, or 0.5 mm) and left to develop into adults. Their adult progeny was then harvested and used for protein extraction by freeze–thaw and dissolved in NET buffer. Protein concentrations were measured with the Bio-Rad (Hercules, CA) kit and then normalized to 5 μg/μl and used with the Oxyblot (Chemicon) kit and analyzed as described below.
Antibodies:
Whole cDNA sequences of sod-1 and sod-2 were amplified from a C. elegans cDNA library (Invitrogen, San Diego) and cloned into the pGEX-5X-1 vector (Invitrogen). Bacterially expressed GST-fusion proteins were extracted and injected into two rabbits to obtain polyclonal antibodies. The terminal bleed of each rabbit recognizes the bacterial antigen and, in worm extracts, a predominant band at the expected size (18.5 kDa for SOD-1 and 24.5 kDa for SOD-2), whose intensity was drastically reduced upon specific RNAi treatment (sod-1 or sod-2 RNAi).
Western blot analysis:
After RNAi treatment, 100 young adult worms of each genotype were picked, lysed in two times loading buffer, and electrophoresed on 12% SDS–polyacrylamide gels (SDS–PAGE), and then blotted onto nitrocellulose membrane (Bio-Rad). After applying primary antibody (1:1000, rabbit polyclonal antibody against worm SOD-1 or SOD-2) and secondary antibody (1:10,000 mouse anti-rabbit IgG, Invitrogen), the membranes were incubated with the ECL plus detection reagent (Amersham Biosciences) and scanned using a Typhoon trio plus scanner. Band densities were analyzed by ImageQuant TL V2003.03.
MnSOD activity assay:
A total of 100 μl of pellets of young adult worms were ground into powder with a mortar in liquid nitrogen and mixed with 100–150 μl of extraction buffer (50 mm phosphate buffer, pH 7.8) and then were centrifuged at 4°, 10,000 × g for 10 min. The supernatant was used for electrophoresis on nondenaturing polyacrylamide gels (7.5%). After electrophoresis, the gel was soaked in 1.23 mm nitroblue tetrazolium for 10 min in the presence of 4 mm potassium cyanide to inhibit CuZn SOD activity and then soaked in 36 mm phosphate buffer (pH 7.8) containing 28 μm riboflavin and 28 mm TEMED for another 10 min. The gel was illuminated on a light box for ∼10 min. The presence of the MnSOD proteins in the gel remained unstained, while the background of the gels was stained a bluish color.
O2 consumption experiment:
The rates of O2 consumption of the wild type and clk-1(qm30) and clk-1; sod-2(RNAi) and isp-1(qm150); ctb-1(qm189) were measured. Worms were grown on RNAi feeding bacteria from L3 to L4 larvae to young adult of the next generation. Young adult worms were collected and washed with M9 to be bacteria free. Oxygen consumption and protein quantification were measured as in Feng et al. (2001).
Protein damage measurement by Oxyblot:
One hundred adults from each strain or treatment or age class were picked, washed repeatedly, concentrated in 3 μl of M9 buffer, and frozen to −80° for storage. Before loading, 3 μl of lysis buffer and 4 μl 15% SDS were added into tubes. Samples were then treated by the freeze–thaw method to release protein. A total of 10 μl of DNP solution (Oxyblot) was added to the samples and kept at room temperature for 15 min, after which 7.5 μl of neutralization buffer (Oxyblot) was added. Samples (27.5 μl) were run in 7.5% SDS–PAGE gels, transferred to nitrocellulose (Bio-Rad), and blocked with 5% milk for 1 hr. Membrane was incubated with the first antibody (Oxyblot) (1:150) overnight at 4° and then for 1 hr with the second antibody (Oxyblot) (1:300) at room temperature. Membranes were incubated with the ECL plus detection reagent (Amersham Biosciences) and scanned using a Typhoon trio plus scanner. Band densities were analyzed by ImageQuant TL V2003.03. The densities of all bands in a given lane were added together and considered the Oxyblot value for the sample. Membranes were then incubated with 15% hydrogen peroxide for 30 min at room temperature and treated with a α-tubulin antibody to derive a density value for tubulin for each lane, with which an Oxyblot value normalized to tubulin was obtained. In Figure 3, the samples for which values are represented in a given panel were always run in the same gel and treated and analyzed together to reduce experimental variations. However, sometimes more than one sample of each genotype or condition was run on the same gel, which is why the sample size is not always identical for each genotype or condition. Each membrane contained at least two samples for each strain or treatment, including the control. The relative value for each sample, including for each control sample, was obtained by dividing the value of the sample by the average value for all control samples on the same blot, which is why the control bars in Figure 3 have SEM error bars.
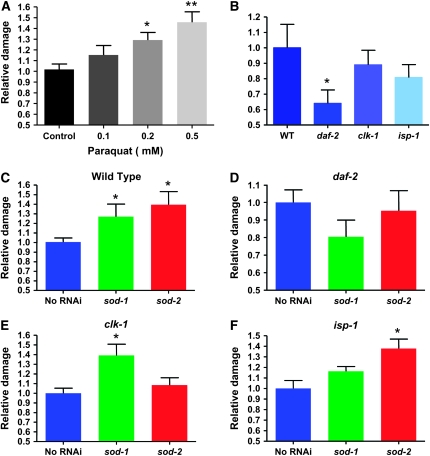
Figure 3.—
Oxidative damage to proteins after treatment with paraquat in mutant strains at various chronological ages and after RNAi against superoxide dismutases. Oxidative damage to proteins was measured by Oxyblot and is expressed as relative to control. The sample sizes given below in the legend indicate the number of independent experiments. Statistical significance is given relative to the wild type or no RNAi condition (*P ≤ 0.05; **P < 0.01, #P = 0.07). The error bars are SEM. (A) Treatment of wild-type worms with paraquat (n = 5); n represents repeats of the experiment. (B) The oxidative damage levels at the young adult stage in different strains [wild type (n = 7); daf-2 (n = 8); clk-1 (n = 7); isp-1 (n = 8)]. Although there is a tendency for less damage in all three long-lived mutant strains, it is significant only in daf-2 mutants. (C) Reduction of either SOD-1 or SOD-2 levels by RNAi leads to significant increases in oxidative damage in the wild type (n = 8; no RNAi n = 7). (D) Neither sod-1 nor sod-2 RNAi significantly affects protein damage levels in daf-2 mutants (n = 8). However, there is a tendency for a reduction of damage after sod-1 RNAi treatment. (E) Reduction of SOD-1 levels but not SOD-2 levels affects oxidative damage in clk-1 mutant (n = 7). (F) Reduction of both SOD-1 and SOD-2 levels increases oxidative damage in isp-1 mutants, although only the increase produced by SOD-2 is statistically significant (n = 7). (G) Reduction of both SOD-1 and SOD-2 levels increases oxidative stress in mev-1 (n = 3 for each condition). (H) Reduction of both SOD-1 and SOD-2 levels increase carbonyls in mutants to levels that are higher than in the wild type in a direct comparison (n = 7 except for isp-1 treatment with sod-2 RNAi for which n = 5). (I) The carbonyl levels in aged isp-1 mutants (13 days old; n = 10) are not reduced compared to wild type (n = 5).
Quantitative PCR measurements:
Worms from RNAi treatment plates were washed into a 1.5-ml tube with M9 when they reached the young adult stage. After three washes with 100 μl M9, all M9 was carefully removed. Total mRNA was extracted using the Trizol reagent (Invitrogen) and then quantified with a spectrophotometer (Beckman Coulter). One microgram of total mRNA was reverse transcribed to cDNA using the Omniscript RT kit (QIAGEN, Valencia, CA). One-fortieth of the reverse transcription product was used as template to perform real-time PCR using the Quantitect SYBR Grenn PCR kit (QIAGEN) and an icycler apparatus (Bio-Rad Version 4.006). Each sample was done in triplicate. Data were analyzed with icycler software (version: 3.0.6070).
Statistical analysis:
Mean post-embryonic rates were compared by using Student's t-test assuming unequal variances. Fertilities (self-brood size) were compared by using Student's t-test assuming equal variances. We performed log-rank statistics to determine if the adult life spans of the RNAi treatment groups were different from that of control groups. Embryonic viabilities and sensitivities to PQ were compared by a χ2 test (http://www.georgetown.edu/faculty/ballc/webtools/web_chi.html).
RESULTS
sod-1 and -2 expression in the wild type and after knockdown by RNAi:
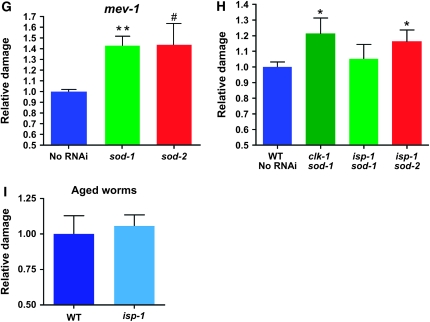
We have raised antisera specific for C. elegans SOD-1 and SOD-2 (Figure 1A). RNAi treatment against each of these genes almost completely abolished detection of the respective proteins by Western blotting but did not alter the level of expression of the other protein (Figure 1A). SOD-2 and SOD-3 are homologous proteins of similar size but the antiserum that we raised against SOD-2 recognizes SOD-3∷GFP only very weakly (not shown). However, sod-2 RNAi virtually abolishes not only SOD-2 protein expression, but also all MnSOD activity (Figure 1A). Furthermore, sod-2 RNAi severely reduces sod-3 mRNA levels in daf-2 and clk-1 mutants where they are elevated (see below). We conclude that our sod-2(RNAi) treatment also severely affects sod-3 expression. In any case, the sod-2 RNAi treatment appears to virtually abolish superoxide detoxification in the mitochondrial matrix of the wild type.
Figure 1.—
Expression of SOD-1 and SOD-2 in different genetic backgrounds and after RNAi treatments. (A) Western blot and MnSOD enzymatic activity blot of wild-type (N2) worm extracts after treatment with RNAi against sod-1 or sod-2 or without treatment. The labels on the left indicate the specificity of the antiserum used or the activity. The interruption of the band in the first two lanes of the activity blot is an unwanted technical artifact of the gel. The three lanes are from the same blot. The dotted lines have been added to clarify the lanes because of the artifact. The same worm extracts were used in all three Western blot panels. (B) Quantification of the levels of SOD-1 and SOD-2 in different genetic backgrounds using Western blotting. Amounts are expressed relative to the wild type; n = 5 repeats of the experiment; error bars are SEM. (C) Quantification of the levels of SOD-1 and SOD-2 after treatment with RNAi against the other SOD gene in four genetic backgrounds. Amounts are expressed relative to untreated controls, for each genotype; n = 5 repeats of the experiment; error bars are SEM.
SOD levels in long-lived mutants:
Using our antisera, we quantified the levels of SOD-1 and SOD-2 in a variety of long-lived, slow-growing mutants (Figure 1B). For this part of the study only, we also examined double mutants because they were known to differ dramatically from each other in life span or growth rate (Wong et al. 1995; Lakowski and Hekimi 1996; Feng et al. 2001), which could help in determining whether SOD levels could be uncoupled from either, or each, of the two parameters. Several mutant strains (isp-1, daf-2, daf-2 clk-1, daf-2 isp-1, and daf-2 isp-1; ctb-1) show increased levels of both proteins, but no strain shows a significant increase of only one of them. Also, no strain shows a significant decrease in SOD levels in comparison to the wild type. All the strains examined are both long lived and slow growing, but to various degrees compared to the wild type (Wong et al. 1995; Lakowski and Hekimi 1996; Feng et al. 2001). Increased SOD levels likely reduce superoxide levels, which could impact both of these phenotypes. However, we did not observe any correlation between increased SOD levels and life span or growth rate. For example, although isp-1 and isp-1; ctb-1 mutants have essentially the same life span (Feng et al. 2001), only isp-1 mutants have increased SOD levels (Figure 1B). Likewise, daf-2 clk-1 mutants display the same levels of SODs as daf-2 or isp-1, although their life span is substantially longer (Lakowski and Hekimi 1996; Feng et al. 2001). Similarly, for growth rate, we observe that, although clk-1 grows much more slowly than daf-2 (Wong et al. 1995; Lakowski and Hekimi 1996), daf-2 has much higher levels of SODs, and although daf-2; isp-1 mutants grow two times more slowly than daf-2 single mutants (Feng et al. 2001), they have similar SOD levels (Figure 1B).
Interactions of SOD-1 and -2 levels with each other:
The CuZn superoxide dismutase SOD-1 is expected to be expressed in the cytoplasm, but also in the mitochondrial intermembrane space (Okado-Matsumoto and Fridovich 2001; Sturtz et al. 2001; O'Brien et al. 2004). As hydrogen peroxide, which is the product of the reaction catalyzed by SODs, can cross membranes and affect cellular compartments distinct from that in which it was produced, we wondered whether the level of one type of SOD could affect the expression of the other type. We tested for this both in the wild type, as described above (Figure 1A), and in the mutants (Figure 1C). RNAi was as effective in reducing the level of the targeted SOD in the mutants as in the wild type (not shown). But we could detect no significant effects of RNAi on the levels of the untargeted SOD in the mutants or the wild type. The small effects of sod-1(RNAi) on SOD-2 levels appear minor, considering the much larger variations in SOD levels among genotypes (Figure 1B). These observations indicate that knocking down the expression of one sod gene does not induce the other and therefore suggests that superoxide detoxification is carried out independently in each cellular compartment.
Effect of sod RNAi on growth rate and other phenotypes in long-lived mutants:
We treated our collection of long-lived mutants with RNAi against sod-1 or sod-2 and examined the effect of the treatment on the length of post-embryonic development (Table 1), adult life span (Table 2), embryonic lethality (Table 3), and fertility (Table 4). In the absence of any treatment, all of the long-lived mutants display slow post-embryonic development and, except for daf-2, altered fertility. Some show increased embryonic lethality. Treatment with sod-1(RNAi) has only relatively mild effects, sometimes deleterious and sometimes slightly suppressive. For example, it appears to be deleterious for the fertility of the wild-type and daf-2 mutants, but slightly increases the fertility of clk-1 (Table 4). In fact, sod-1(RNAi) appears to be slightly suppressive for several phenotypes of clk-1 mutants, as noted previously (Shibata et al. 2003). However, in general, we believe that the relatively small magnitude of the effects observed here with sod-1(RNAi) precludes a clear interpretation of their meaning. On the other hand, sod-2(RNAi) appears to be clearly deleterious as it strongly slows down development of clk-1 and isp-1 (Table 1), dramatically increases embryonic lethality of these two mutants (Table 3), and decreases fertility, especially of isp-1 mutants (Table 4).
TABLE 1.
Effects of sod-1 and sod-2 RNAi on post-embryonic development rate
| Strain | Control | sod-1 RNAi | sod-2 RNAi |
|---|---|---|---|
| Wild type (N2) | 46.0 ± 1.9 | 46.6 ± 2.1 | 46.8 ± 2.1 |
| n = 146 | n = 134 | n = 148 | |
| daf-2(e1370) | 74.0 ± 3.5 | 76.1 ± 3.6 | 72.6 ± 2.7 |
| n = 135 | n = 120 | n = 133 | |
| isp-1(qm150) | 108.7 ± 7.6 | 100.2 ± 7.1 | 116.0 ± 6.7 |
| n = 138 | n = 148 | n = 144 | |
| clk-1(qm30) | 83.3 ± 4.7 | 74.4 ± 2.3 | 111.1 ± 15.3 |
| n = 129 | n = 148 | n = 81 |
Post-embryonic development rate is expressed in hours to reach adulthood after hatching. All values found for treated animals vs. untreated animals are significantly different at P < 0.05 using Student's t-test assuming unequal variances. The control plates contained bacteria with the empty vector.
TABLE 2.
Effects of sod-1 and sod-2 RNAi on adult life span
| Strain | Control | sod-1 RNAi | sod-2 RNAi |
|---|---|---|---|
| Wild type (N2) | 17.5 ± 5.6 | 16.2 ± 4.8 | 18.0 ± 5.8 |
| Maximum: 29 | Maximum: 28 | Maximum: 29 | |
| n = 150 | n = 142 | n = 150 | |
| P =0.008 | P = 0.236 | ||
| daf-2(e1370) | 33.0 ± 11.0 | 37.4 ± 11.3 | 33.5 ± 9.2 |
| Maximum: 53 | Maximum: 66 | Maximum: 52 | |
| n = 150 | n = 140 | n = 150 | |
| P = 0.0002 | P = 0.266 | ||
| isp-1(qm150) | 23.3 ± 8.2 | 22.1 ± 8.2 | 22.6 ± 7.9 |
| Maximum: 50 | Maximum: 50 | Maximum: 45 | |
| n = 260 | n = 227 | n = 267 | |
| P = 0.198 | P = 0.171 | ||
| clk-1(qm30) | 18.6 ± 6.4 | 18.7 ± 5.4 | 23.9 ± 7.7 |
| Maximum: 37 | Maximum: 37 | Maximum: 43 | |
| n = 150 | n = 150 | n = 150 | |
| P = 0.625 | P < 0.0001 | ||
| mev-1(kn-1) | 13.3 ± 3.8 | 14.3 ± 3.5 | 12.8 ± 3.1 |
| Maximum: 23 | Maximum: 23 | Maximum: 24 | |
| n = 63 | n = 67 | n = 55 | |
| P = 0.137 | P = 0.442 |
Adult life span is expressed in days. P-values are determined using the log-rank statistics. Values indicating a lack of significance are also shown.
TABLE 3.
Effects of sod-1 and sod-2 RNAi on embryonic lethality
| Strain | Control | sod-1 RNAi | sod-2 RNAi |
|---|---|---|---|
| Wild type (N2) | 3.9 | 2.9 | 1.9 |
| n = 178 | n = 173 | n = 213 | |
| daf-2(e1370) | 0 | 2.0* | 1.5 |
| n = 201 | n = 200 | n = 200 | |
| isp-1(qm150) | 6.3 | 8.1 | 30.6*** |
| n = 313 | n = 358 | n = 411 | |
| clk-1(qm30) | 2.5 | 5.8* | 50.3*** |
| n = 316 | n = 328 | n = 349 |
Embryonic lethality is expressed in percentages of unhatched eggs. Mutant embryonic lethalities were compared to the wild-type level by the χ2 nonparametric test. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
TABLE 4.
Effects of sod-1 and sod-2 RNAi on fertility
| Strain | Control | sod-1 RNAi | sod-2 RNAi |
|---|---|---|---|
| Wild type (N2) | 319.4 ± 30.4 | 267.0 ± 54.2** | 296.7 ± 59.7 |
| n = 20 | n = 20 | n = 20 | |
| daf-2(e1370) | 298.4 ± 23.5 | 266.8 ± 19.5** | 279.1 ± 15.4* |
| n = 15 | n = 15 | n = 15 | |
| isp-1(qm150) | 68.5 ± 21.4 | 66.8 ± 20.1 | 51.4 ± 30.7*** |
| n = 50 | n = 50 | n = 50 | |
| 5% sterile | |||
| clk-1(qm30) | 146.5 ± 54.5 | 195.5 ± 35.4* | 167.8 ± 25.0 |
| n = 10 | n = 10 | n = 10 |
Fertility is scored as self-brood size. P-values were determined by using Student's t-test assuming equal variances. *P < 0.05, **P < 0.01, and ***P < 0.001.
Effect of sod RNAi on sensitivity to paraquat:
No function other than superoxide detoxification has been reported for superoxide dismutases, which strongly suggests that any phenotypic effect of sod RNAi treatment on mutants is due to an increase in ROS resulting from poor superoxide detoxification. To investigate this further, we have examined how sod-1 or sod-2 RNAi affects survival after treatment with PQ, a superoxide generator that is frequently used to establish alterations in ROS resistance. We determined PQ resistance by treating worms for one generation with RNAi and then by testing the ability of adults to survive a relatively high concentration of PQ (4 mm) (Figure 2). We find that both sod-1 and sod-2 RNAi increase the sensitivity to killing by PQ of both the wild type and the mutants, with the exception of treatment of daf-2 with sod-2 RNAi, which has no effect. Furthermore, the mutants are more resistant than the wild type to killing by PQ in every condition, except for treatment of isp-1 with sod-1 RNAi, which is as effective as in the wild type. We conclude that the level of reduction of SOD-1 or SOD-2 expression by RNAi treatment effectively decreases superoxide detoxification. However, since we do not know the exact nature of the damage that kills the animals, the relative effects of sod-1 vs. sod-2 RNAi in this experiment does not tell us which treatment produces more overall oxidative stress or damage or how it might relate to aging. For the same reason, the increased resistance of the mutants to PQ confirms only the generally observed correlation between altered life span and altered resistance to imposed acute oxidative stress.
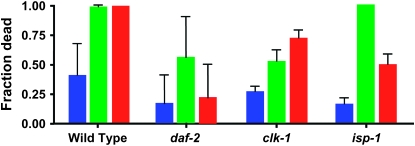
Figure 2.—
Sensitivity of the wild-type, daf-2, clk-1, and isp-1 mutants to PQ after sod RNAi treatment. The fraction of worms that die after 72 hr of treatment with 4 mm PQ was scored. Treatment of sod-1(RNAi) is shown in green and sod-2(RNAi) in red; no RNAi treatment is shown in blue. The experiment was repeated three times; error bars are SEM. The wild type and all three mutants were sensitized to PQ by RNAi treatments (all differences are significant at P < 0.01 by a χ2 test), except for daf-2, which is insensitive to sod-2(RNAi). Furthermore, after RNAi treatment, all three mutants are somewhat more resistant to PQ treatment than the wild type (P < 0.01; P < 0.025 for clk-1), except for isp-1, which is as sensitive to sod-1(RNAi) as the wild type.
Effect of life-span mutants and sod RNAi on ROS damage to proteins:
We have used a Western blotting method (Oxyblot; see materials and methods) to quantify oxidative damage to proteins (carbonyl formation) as the most sensitive method currently available for C. elegans to measure ROS damage (Shacter 2000; Dalle-Donne et al. 2003; Kayser et al. 2004; Yasuda et al. 2006). We have examined damage in response to increasing doses of PQ to establish that the method is capable of yielding a graded response (Figure 3A). Treatment with paraquat is well recognized as increasing oxidative stress. The 0.5-mm condition was nearly lethal (not shown). Note that Figure 3A shows the result of treating wild-type worms with PQ throughout development, which is a more severe treatment than growth on PQ only at the adult stage, as in the experiments of Figure 2, and thus requires lower doses of PQ. The results suggest that the Oxyblot measurements indeed measure oxidative stress and that a 1.5-fold increase corresponds to a degree of increase in damage that is close to the maximum compatible with survival. We have also examined the levels of carbonyls in mutants in comparison to the wild type (Figure 3B) and in response to sod RNAi treatments (Figure 3, C–F).
Figure 3B shows that in the absence of any treatment only daf-2 mutants have significantly reduced levels of oxidative damage. There is also a similar tendency for clk-1 and isp-1 mutants, but the effect is minor. Figure 3, C–F, shows that sod-1 RNAi increases damage in all strains except in daf-2 mutants where, surprisingly, it tends to lower damage. Possibly, in this mutant, the excess superoxide produced by the reduction in detoxification induces an overcompensating protective mechanism involving different detoxifying enzymes. It is also of note that the reduction in damage produced by sod-1 RNAi in daf-2 mutants is consistent with the slight increase in life span produced by this treatment (Figure 4). Figure 3, C–F, also shows that sod-2 RNAi increases damage significantly in the wild type and in isp-1 mutants, but is without effect in daf-2 and clk-1 mutants. In the case of clk-1, this might be the result of a form of developmental compensation (see below). It is important to note that the effects of sod-1 RNAi on clk-1 and of sod-2 RNAi on isp-1 increase damage ∼1.4-fold, while there appears to be only a minor reduction in damage in untreated clk-1 and isp-1 mutants compared to the wild type (Figure 3B). Thus, the level of damage in treated mutants must be similar to, or higher than, that in the wild type.
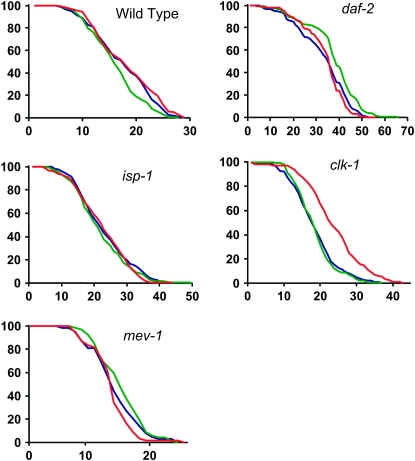
Figure 4.—
Changes in life span of the wild type and different mutants after RNAi treatments against sod-1 or sod-2. The x-axis is in days and the y-axis indicates the fraction of the worms in the sample that are still alive. Blue represents untreated worms, green represents worms treated by sod-1(RNAi), and red represents worms treated by sod-2(RNAi). The worms were checked every day. All statistical analyses are shown in Tables 1–4, and the results are discussed in detail in the text.
mev-1 encodes a subunit of mitochondrial complex II; mev-1 mutants have a short life span and are hypersensitive to paraquat (Ishii et al. 1998), which has suggested to previous researchers that their short life span was due to elevated mitochondrial oxidative stress. We have found that both sod-1 and sod-2 RNAi are capable of substantially increasing carbonyl damage in mev-1 mutants (Figure 3G). These results suggests that our RNAi treatments do strongly increase oxidative stress by a degree that is rather greater than that by which the mev-1 mutation increases oxidative stress.
In the experiments described in this section of the results, for each repeat of the experiments that compared RNAi-treated and mock-treated animals (Figure 3, C–G) all the samples were analyzed on the same gels to reduce experimental variation. However, we also wanted to make sure that we could relate the relative differences between treated and untreated animals to those differences that were observed between untreated wild-type and mutant animals (Figure 3B). Thus, for this, as a control, we prepared samples from clk-1 or isp-1 mutants treated with sod-1 or sod-2 RNAi and ran them together on the same gels with samples from untreated wild-type animals (Figure 3H). We found that, in both sod-1 and sod-2 RNAi-treated isp-1 and clk-1 mutants, the level of carbonyls was indeed as high or higher as in untreated wild-type animals (Figure 3H).
In the experiments described above, we treated young animals and compared their carbonyl levels. However, it is possible that protein oxidation changes with chronological age. To test this, we chose to examine and compare carbonyl levels in the wild type and isp-1 at a specific time: the time (13 days) at which 15% of the wild-type animals but none of the mutants had died (Figure 3I). We chose isp-1 because our experiments suggest that their carbonyl levels at a young age are not significantly different from those of the wild type (Figure 3B), yet were increased by sod RNAi treatments, albeit without effects on life span (see below). Furthermore, the robust difference in life span between the wild type and isp-1 allowed for measurement that indeed compared chronological with physiological age. We found that even at the time of 15% mortality for the wild type there was no difference in carbonyl levels between the wild-type and isp-1 mutants. Thus, no spontaneous reduction in oxidative damage to proteins in the mutants can explain why isp-1 mutants survive longer than the wild type.
Effect of sod RNAi on the life span of long-lived mutants:
Although we did not find any correlation between the levels of SODs and life span in the long-lived mutant strains that we have examined (Figure 1B), the reduction of a given SOD activity should produce an increase of superoxide in the compartment where the SOD is expressed. Indeed, this must be why these reductions induce various deleterious phenotypes (Tables 1–4) and/or reduce survival on PQ (Figure 2) and/or increase oxidative damage (Figure 3). Thus, these reductions in detoxification should shorten the life spans of these mutants if their long life spans were due to low oxidative stress. To test this hypothesis, we have examined the effects of sod RNAi on the life spans of the mutant strains, expecting to observe a total or partial suppression of their increased life spans. Surprisingly, this was not observed. Instead, we observed the following (Figure 4): (1) an almost complete absence of effect of sod-1(RNAi) except for a minor life-span-shortening effect in the wild type and an equally minor, albeit statistically significant, life-span-lengthening effect in daf-2 and (2) a life-span-lengthening effect of sod-2(RNAi) on clk-1.
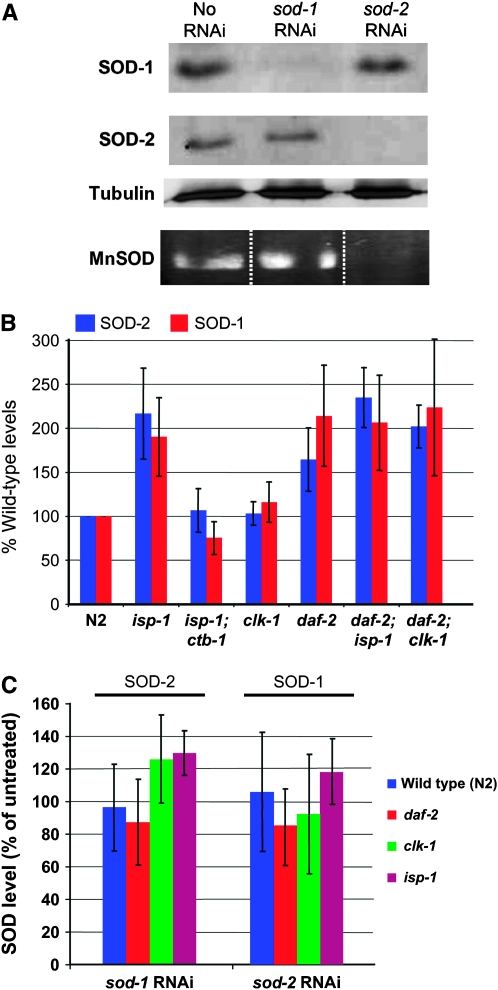
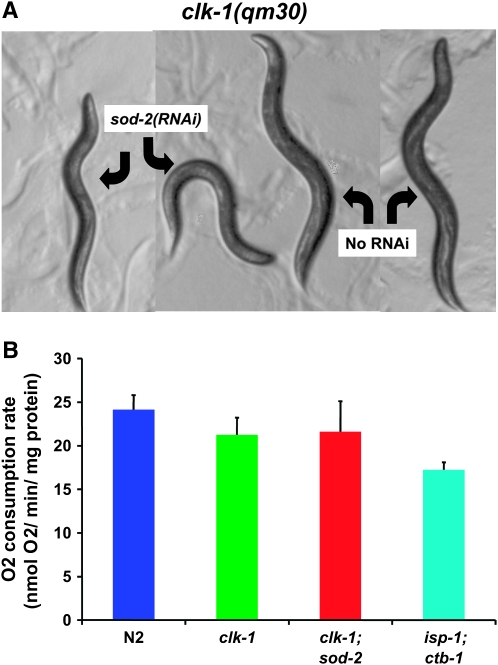
The life-span-lengthening effect of sod-2(RNAi) on clk-1 is accompanied by severely increased embryonic lethality and lengthening of post-embryonic development (Tables 1–4). In addition, the treated animals are significantly smaller than the untreated animals (Figure 5A). However, they show no reduction in oxygen consumption per milligram of protein (Figure 5B) and no elevation of the amount of oxidative damage per milligram of protein (Figure 3E). One possible explanation for the apparent paradox of a decrease in ROS detoxification resulting in an increase in life span is that the sod-2 RNAi treatment of clk-1 mutants damages their respiratory chain in some way. Indeed, the phenotype of these animals is similar to that observed when wild-type worms are treated by RNAi against subunits of respiratory chain complexes (Dillin et al. 2002; Lee et al. 2003). However, this also suggests that the increased longevity is not due to low oxidative damage, as oxidative damage is not decreased.
Figure 5.—
RNAi treatment against sod-2 alters the size and appearance of clk-1 mutants but not their oxygen consumption rates normalized to protein content. (A) Four-day-old adult clk-1(qm30) mutants after treatment by sod-2(RNAi) or without treatment. (B) Oxygen consumption of clk-1(qm30) worms treated by sod-2(RNAi) compared to untreated wild-type worms, clk-1(qm30) worms, and isp-1;ctb-1 worms (n = 3; error bars are SEM), which are known to have decreased oxygen consumption (Feng et al. 2001). sod-2(RNAi) treatment has no effect on clk-1 oxygen consumption, although the effect of the treatment on growth rate, size, life span, and the localization of SOD-2 in the mitochondrial matrix suggests the existence of a deleterious effect on mitochondrial function.
Interestingly, the slight life-span-lengthening effect of sod-1 RNAi on daf-2 is correlated by a paradoxical decrease of carbonyl levels brought about by this treatment (Figure 3D). One hypothesis to explain this effect is that, in response to reduced SOD-1 levels, there is overexpression of other protective or detoxifying activities that we do not monitor here.
Effect of sod RNAi on life span of mev-1:
In addition to testing the effect of sod RNAi on long-lived mutants, we tested the effect on the short-lived mutant mev-1 (Figure 4E). mev-1 encodes a subunit of the complex II of the respiratory chain and sustains increased oxidative stress (Ishii et al. 1998). We found that neither sod-1(RNAi) nor sod-2(RNAi) significantly altered the life span of the mutants. Yet we have shown that these treatments substantially increase oxidative stress as measured by carbonyl levels (Figure 3G). Although the short life of mev-1 mutants has sometimes been hypothesized to result from increased oxidative stress, there is no immediate demonstration of this. In fact, recent data very strongly indicate that a large part of the effect of mev-1 on life span is due to its effect on increasing programmed cell death through a direct effect on the ex pression of ced-9 (Senoo-Matsuda et al. 2001, 2003). Our findings suggest that the degree by which sod RNAi treatment increases oxidative stress (Figure 3E) in this mutant does not shorten its life span. This is consistent with the absence of a life-span-shortening effect of sod RNAi treatments on the wild type and suggests that, as in the case of the long-lived mutants, the respiratory defect of mev-1 (Senoo-Matsuda et al. 2001), rather than its increased oxidative stress, is the cause of its short life.
sod mRNA expression in daf-2 and clk-1 mutants:
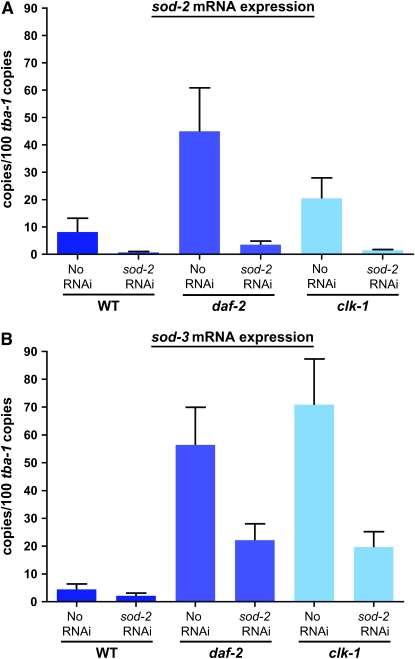
The carbonyl levels of daf-2 and clk-1 mutants are unaffected by treatment with sod-2, although the same treatment increases carbonyl levels in the wild type. This could be due to an increase in the expression of sod-3, whose protein levels we cannot directly measure because of the absence of a SOD-3-specific antiserum. To determine whether changes in sod-3 expression might be involved in this effect, we used quantitative RT–PCR to monitor the levels of sod-2 and sod-3 mRNAs after sod-2 RNAi treatment of the wild-type and daf-2 and clk-1 mutants (Figure 6). Both sod-2 and sod-3 mRNA levels in untreated daf-2 and clk-1 mutants are elevated compared to the wild type. Treatment with sod-2 RNAi reduces sod-2 mRNA to very low levels in all three backgrounds (Figure 6A). The same treatment also reduces sod-3 levels considerably in all backgrounds. This indicates that sod-2 RNAi treatment also affects the level of sod-3, likely because of the sequence homology, which is consistent with the absence of MnSOD activity after sod-2 treatment of the wild type (Figure 1A). Note, however, that an increased level of mRNA expression does not guarantee an increase in the level of protein present, as we do not observe an elevation of SOD-2 protein in clk-1 mutants (Figure 1B). Thus, increased levels of sod-2 and -3 expression might or might not participate in the somewhat lower levels of protein carbonyls observed in daf-2 mutants (Figure 3B). However, similar increases seem to have only a minimal effect on carbonyl levels in clk-1 mutants (Figure 3B). Furthermore, sod-2 RNAi has no effect on the life span of daf-2 (Figure 4). Thus, as the total level of expression of mitochondrial sod genes is reduced by sod-2 RNAi treatment (Figures 1 and 6), the levels observed without treatment are not necessary for the long life of the mutants. The situation is similar for clk-1, where the total level of sod-2 and -3 expression in response to sod-2 RNAi treatment is lower than without treatment and thus cannot explain the absence of an increase in carbonyls in response to sod-2 RNAi. Nor, of course, can it explain why sod-2 RNAi prolongs the life span of clk-1 (Figure 4).
Figure 6.—
mRNA levels by quantitative real-time PCR of sod-2 and sod-3 in the wild type as well as daf-2 and clk-1 mutants, with or without sod-2 RNAi treatment (n = 3 repeats; error bars are SEM). (A) sod-2 mRNA levels. (B) sod-3 mRNA levels.
DISCUSSION
Alterations in the regulation of SOD expression:
We have found increased expression of sod-1, -2, and -3 in several long-lived strains, but these changes did not correlate with life span, growth rate, embryonic lethality, or brood size as measured here (Tables 1–4 and Figure 4) or as published previously (Wong et al. 1995; Lakowski and Hekimi 1996; Feng et al. 2001). Furthermore, sod(RNAi) is equally effective in reducing SOD expression to background levels in all strains (data not shown), and we have not found a correlation between the level of sod's and the sensitivity of the growth rate or of the life span to sod(RNAi).
Testing the ROS toxicity theory of aging:
From the point of view of evolutionary theory, it is a reasonable hypothesis to speculate that aging is due to the accumulation of unrepaired damage, as there is an expected trade-off between reproduction and the costs associated with somatic maintenance (Hekimi et al. 2001b). Toxicity from the ROS produced in mitochondria is an obvious candidate for an important source of damage in this process (Sohal 2002; Droge 2003). In particular, aging appears to be strongly correlated with increases in oxidative stress and oxidative damage accumulation (Beckman and Ames 1998), in particular in the mitochondria (Van Remmen and Richardson 2001). Yet a causal role for ROS toxicity in bringing about aging remains a fundamentally unproven hypothesis.
To test this hypothesis, we have been analyzing mutant strains of C. elegans that display an increased life span. Beyond the initial hypothesis of an involvement of ROS in life-span determination, there were numerous reasons to suppose that the phenotypes of these strains, including their life span, are due to altered ROS metabolism. One of the main sources of cellular superoxide production is believed to be the bifurcating transfer of electrons from ubiquinone to the “Rieske” iron–sulfur protein and the cytochrome b of complex III of the mitochondrial respiratory chain (Raha and Robinson 2000). Two of the three long-lived mutants that we are analyzing are directly involved in this process: isp-1(qm150) is a point mutation in the iron–sulfur protein (Feng et al. 2001), and clk-1 affects the biosynthesis of ubiquinone (Miyadera et al. 2001). The third gene, daf-2, encodes an insulin-receptor-like kinase (Kimura et al. 1997) and affects a variety of processes in worms, including their resistance to oxidative stress (Honda and Honda 2002). The one short-lived mutant that we have also been studying, mev-1, is a point mutation in a subunit of mitochondrial complex II (Ishii et al. 1998).
Lack of support for the ROS toxicity theory of aging:
Strikingly, although several of the mutant strains tested have increased SOD levels (Figure 1B) and although the RNAi treatments are effective (Figure 1A), frequently appear deleterious (Tables 1–4 and Figure 2), and can be shown to increase measurable oxidative damage to proteins in the wild-type and mutant backgrounds (Figure 3), the RNAi treatments do not shorten the increased life span of the long-lived mutants (Figure 4) or worsen the short life span of mev-1 (Figure 4). Furthermore, we observe some paradoxical effects; for example, sod-1(RNAi) against daf-2 and sod-2(RNAi) against clk-1 increase life span. Thus, our results provide no substantial support for the hypothesis that the increased life span of most of the mutants that we have examined is due to low levels of ROS damage. Our results are consistent with observations using pharmacological SOD mimetics that were capable of protecting from experimental increases in oxidative stress, but were not capable of positively affecting wild-type life span in C. elegans (Keaney et al. 2004).
Our findings about the role that oxidative stress might or might not play in the long life span of daf-2 mutants are not conclusive. Indeed, daf-2 mutants are unaffected by our treatments except in a paradoxical fashion: sod-1 RNAi decreases oxidative damage and increases life span. Thus, we have not succeeded in uncoupling low oxidative stress from the increased life span of daf-2 mutants.
A superoxide-independent role for electron transport in life-span determination:
Electron transport in the mitochondrial respiratory chain generates superoxide at sites where electrons are transferred from prosthetic groups to ubiquinone and vice versa (Raha and Robinson 2000). Several of the mutants that we have examined (clk-1, isp-1, and mev-1) are closely involved in the process of electron transport (Ishii et al. 1998; Feng et al. 2001; Miyadera et al. 2001). Thus, it is likely that these mutants have altered ROS metabolism (Feng et al. 2001; Senoo-Matsuda et al. 2001; Shibata et al. 2003), as indicated by the various effects of SOD knockdowns on the mutant phenotypes of the long-lived mutants (Tables 1–4). Yet our results indicate that it is not a reduction of oxidative damage that is responsible for the observed increased life span. Our results therefore suggest that electron transport has a role in life-span determination that is independent of its role in the production of oxidative damage, as has been suggested previously on different grounds (Dillin et al. 2002; Hekimi and Guarente 2003). Our findings of the dramatic effect of sod-2(RNAi) on clk-1 (Figure 5) can also be interpreted in this light. That is, that the reduction of the activity of the mitochondrial matrix SOD-2 in the clk-1 background, with its altered quinone profile, damages the respiratory chain in a way that we do not yet understand, but that is favorable for life span. In fact, the phenotype of clk-1 mutants treated with sod-2(RNAi) is highly reminiscent of that obtained by knocking down subunits of mitochondrial complexes (Dillin et al. 2002; Lee et al. 2003), which also prolongs life span.
Oxidative stress and aging:
The possibility that the oxidative stress theory is not generally valid has been considered by others (Koc et al. 2004; Kujoth et al. 2005), but remains surprising (Hekimi and Guarente 2003). However, most evidence about oxidative stress and aging points to a general correlation and not necessarily to a causal relationship. A scientific theory has to be falsifiable to be meaningful. Thus, although a single study cannot address at once all the observations that previously appeared to be consistent with the theory, sound experimental results that are inconsistent with a theory represent an important challenge to that theory. For example, although for technical reasons we could examine only oxidative damage to proteins, but not to other macromolecules, no part of the oxidative stress theory of aging suggests that damage to proteins does not matter in bringing about aging or suggests how an increase of oxidative stress that is sufficient to damage proteins could fail to damage other macromolecules.
More generally, it is of interest that our results—which suggest that oxidative stress from mitochondrial respiration is likely not causal in aging in the mutants that we have examined—do not imply that oxidative stress does not play a role in producing the aged phenotype in these mutants or in any other organism. In fact, a sharp increase of oxidative damage with age is well documented in many organisms and tissues. For example, oxidative damage to DNA (Hamilton et al. 2001), measured as an increase in 8-hydroxguanosine, as well as oxidative damage to proteins (Yasuda et al. 1999; Levine and Stadtman 2001), measured as an increase in carbonyls, increase dramatically with age. Interestingly, the increase in carbonyls is most marked later in life, at a time when in fact the organisms are already aged (Levine and Stadtman 2001), which further implies that the increase itself cannot be causal in aging.
The higher oxidative damage of the aged phenotype suggests that aged organisms have higher level of ROS. This is supported by the finding that in Drosophila transgenic overexpression of superoxide dismutase can increase life span (Sun et al. 2002), especially in short-lived strains (Orr and Sohal 2003). However, this again does not indicate that oxidative stress is the cause of aging but only that oxidative damage is deleterious and that relieving it in part through a genetic or pharmacological intervention might improve the health of aged organisms. In summary, while our observations suggest that oxidative stress is not the cause of aging, observations of high oxidative stress and damage in old organisms, on the other hand, are consistent with the notion that increased oxidative stress is a consequence of aging.
Acknowledgments
We thank Jinliu Feng for unpublished results and Robyn Branicky for helpful comments on the manuscript. We gratefully acknowledge the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health, for strains. S.H. is Strathcona Professor of Zoology. This work was supported in part by a research contract from Chronogen.
References
- Balaban, R. S., S. Nemoto and T. Finkel, 2005. Mitochondria, oxidants, and aging. Cell 120: 483–495. [DOI] [PubMed] [Google Scholar]
- Beckman, K. B., and B. N. Ames, 1998. The free radical theory of aging matures. Physiol. Rev. 78: 547–581. [DOI] [PubMed] [Google Scholar]
- Boehm, M., and F. Slack, 2005. A developmental timing microRNA and its target regulate life span in C. elegans. Science 310: 1954–1957. [DOI] [PubMed] [Google Scholar]
- Braeckman, B. P., K. Houthoofd, K. Brys, I. Lenaerts, A. De Vreese et al., 2002. a No reduction of energy metabolism in Clk mutants. Mech. Ageing Dev. 123: 1447–1456. [DOI] [PubMed] [Google Scholar]
- Braeckman, B. P., K. Houthoofd and J. R. Vanfleteren, 2002. b Assessing metabolic activity in aging Caenorhabditis elegans: concepts and controversies. Aging Cell 1: 82–88; discussion 102–103. [DOI] [PubMed] [Google Scholar]
- Dalle-Donne, I., R. Rossi, D. Giustarini, A. Milzani and R. Colombo, 2003. Protein carbonyl groups as biomarkers of oxidative stress. Clin. Chim. Acta 329: 23–38. [DOI] [PubMed] [Google Scholar]
- de Castro, E., S. Hegi de Castro and T. E. Johnson, 2004. Isolation of long-lived mutants in Caenorhabditis elegans using selection for resistance to juglone. Free Radic. Biol. Med. 37: 139–145. [DOI] [PubMed] [Google Scholar]
- Dillin, A., A. L. Hsu, N. Arantes-Oliveira, J. Lehrer-Graiwer, H. Hsin et al., 2002. Rates of behavior and aging specified by mitochondrial function during development. Science 298: 2398–2401. [DOI] [PubMed] [Google Scholar]
- Droge, W., 2003. Oxidative stress and aging. Adv. Exp. Med. Biol. 543: 191–200. [DOI] [PubMed] [Google Scholar]
- Duttaroy, A., A. Paul, M. Kundu and A. Belton, 2003. A Sod2 null mutation confers severely reduced adult life span in Drosophila. Genetics 165: 2295–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewbank, J. J., T. M. Barnes, B. Lakowski, M. Lussier, H. Bussey et al., 1997. Structural and functional conservation of the Caenorhabditis elegans timing gene clk-1. Science 275: 980–983. [DOI] [PubMed] [Google Scholar]
- Fabrizio, P., L. L. Liou, V. N. Moy, A. Diaspro, J. S. Valentine et al., 2003. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics 163: 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felkai, S., J. J. Ewbank, J. Lemieux, J. C. Labbe, G. G. Brown et al., 1999. CLK-1 controls respiration, behavior and aging in the nematode Caenorhabditis elegans. EMBO J. 18: 1783–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, J., F. Bussiere and S. Hekimi, 2001. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell 1: 633–644. [DOI] [PubMed] [Google Scholar]
- Fujii, M., N. Ishii, A. Joguchi, K. Yasuda and D. Ayusawa, 1998. A novel superoxide dismutase gene encoding membrane-bound and extracellular isoforms by alternative splicing in Caenorhabditis elegans. DNA Res. 5: 25–30. [DOI] [PubMed] [Google Scholar]
- Giglio, A. M., T. Hunter, J. V. Bannister, W. H. Bannister and G. J. Hunter, 1994. The copper/zinc superoxide dismutase gene of Caenorhabditis elegans. Biochem. Mol. Biol. Int. 33: 41–44. [PubMed] [Google Scholar]
- Golden, T. R., D. A. Hinerfeld and S. Melov, 2002. Oxidative stress and aging: beyond correlation. Aging Cell 1: 117–123. [DOI] [PubMed] [Google Scholar]
- Guarente, L., and C. Kenyon, 2000. Genetic pathways that regulate ageing in model organisms. Nature 408: 255–262. [DOI] [PubMed] [Google Scholar]
- Hamilton, M. L., H. Van Remmen, J. A. Drake, H. Yang, Z. M. Guo et al., 2001. Does oxidative damage to DNA increase with age? Proc. Natl. Acad. Sci. USA 98: 10469–10474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, N., V. Costa, M. MacLean, M. Mollapour, P. Moradas-Ferreira et al., 2003. Mnsod overexpression extends the yeast chronological (G(0)) life span but acts independently of Sir2p histone deacetylase to shorten the replicative life span of dividing cells. Free Radic. Biol. Med. 34: 1599–1606. [DOI] [PubMed] [Google Scholar]
- Hekimi, S., and L. Guarente, 2003. Genetics and the specificity of the aging process. Science 299: 1351–1354. [DOI] [PubMed] [Google Scholar]
- Hekimi, S., C. Benard, R. Branicky, J. Burgess, A. K. Hihi et al., 2001. a Why only time will tell. Mech. Ageing Dev. 122: 571–594. [DOI] [PubMed] [Google Scholar]
- Hekimi, S., J. Burgess, F. Bussiere, Y. Meng and C. Benard, 2001. b Genetics of life span in C. elegans: molecular diversity, physiological complexity, mechanistic simplicity. Trends Genet. 17: 712–718. [DOI] [PubMed] [Google Scholar]
- Honda, Y., and S. Honda, 2002. Oxidative stress and life span determination in the nematode Caenorhabditis elegans. Ann. NY Acad. Sci. 959: 466–474. [DOI] [PubMed] [Google Scholar]
- Huang, T. T., E. J. Carlson, A. M. Gillespie, Y. Shi and C. J. Epstein, 2000. Ubiquitous overexpression of CuZn superoxide dismutase does not extend life span in mice. J. Gerontol. A Biol. Sci. Med. Sci. 55: B5–B9. [DOI] [PubMed] [Google Scholar]
- Hunter, T., W. H. Bannister and G. J. Hunter, 1997. Cloning, expression, and characterization of two manganese superoxide dismutases from Caenorhabditis elegans. J. Biol. Chem. 272: 28652–28659. [DOI] [PubMed] [Google Scholar]
- Ishii, N., M. Fujii, P. S. Hartman, M. Tsuda, K. Yasuda et al., 1998. A mutation in succinate dehydrogenase cytochrome b causes oxidative stress and ageing in nematodes. Nature 394: 694–697. [DOI] [PubMed] [Google Scholar]
- Jensen, L. T., and V. C. Culotta, 2005. Activation of CuZn superoxide dismutases from Caenorhabditis elegans does not require the copper chaperone CCS. J. Biol. Chem. 280: 41373–41379. [DOI] [PubMed] [Google Scholar]
- Kamath, R. S., M. Martinez-Campos, P. Zipperlen, A. G. Fraser and J. Ahringer, 2001. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2: RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser, E. B., M. M. Sedensky and P. G. Morgan, 2004. The effects of complex I function and oxidative damage on life span and anesthetic sensitivity in Caenorhabditis elegans. Mech. Ageing Dev. 125: 455–464. [DOI] [PubMed] [Google Scholar]
- Keaney, M., F. Matthijssens, M. Sharpe, J. Vanfleteren and D. Gems, 2004. Superoxide dismutase mimetics elevate superoxide dismutase activity in vivo but do not retard aging in the nematode Caenorhabditis elegans. Free Radic. Biol. Med. 37: 239–250. [DOI] [PubMed] [Google Scholar]
- Kenyon, C., 2005. The plasticity of aging: insights from long-lived mutants. Cell 120: 449–460. [DOI] [PubMed] [Google Scholar]
- Kenyon, C., J. Chang, E. Gensch, A. Rudner and R. Tabtiang, 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464. [DOI] [PubMed] [Google Scholar]
- Kimura, K. D., H. A. Tissenbaum, Y. Liu and G. Ruvkun, 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277: 942–946. [DOI] [PubMed] [Google Scholar]
- Koc, A., A. P. Gasch, J. C. Rutherford, H. Y. Kim and V. N. Gladyshev, 2004. Methionine sulfoxide reductase regulation of yeast life span reveals reactive oxygen species-dependent and -independent components of aging. Proc. Natl. Acad. Sci. USA 101: 7999–8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth, G. C., A. Hiona, T. D. Pugh, S. Someya, K. Panzer et al., 2005. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309: 481–484. [DOI] [PubMed] [Google Scholar]
- Lakowski, B., and S. Hekimi, 1996. Determination of life-span in Caenorhabditis elegans by four clock genes. Science 272: 1010–1013. [DOI] [PubMed] [Google Scholar]
- Lakowski, B., and S. Hekimi, 1998. The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 95: 13091–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. S., R. Y. Lee, A. G. Fraser, R. S. Kamath, J. Ahringer et al., 2003. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 33: 40–48. [DOI] [PubMed] [Google Scholar]
- Levine, R. L., and E. R. Stadtman, 2001. Oxidative modification of proteins during aging. Exp. Gerontol. 36: 1495–1502. [DOI] [PubMed] [Google Scholar]
- Mansouri, A., F. L. Muller, Y. Liu, R. Ng, J. Faulkner et al., 2006. Alterations in mitochondrial function, hydrogen peroxide release and oxidative damage in mouse hind-limb skeletal muscle during aging. Mech. Ageing Dev. 127: 298–306. [DOI] [PubMed] [Google Scholar]
- Melov, S., S. R. Doctrow, J. A. Schneider, J. Haberson, M. Patel et al., 2001. Life span extension and rescue of spongiform encephalopathy in superoxide dismutase 2 nullizygous mice treated with superoxide dismutase-catalase mimetics. J. Neurosci. 21: 8348–8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyadera, H., H. Amino, A. Hiraishi, H. Taka, K. Murayama et al., 2001. Altered quinone biosynthesis in the long-lived clk-1 mutants of Caenorhabditis elegans. J. Biol. Chem. 276: 7713–7716. [DOI] [PubMed] [Google Scholar]
- O'Brien, K. M., R. Dirmeier, M. Engle and R. O. Poyton, 2004. Mitochondrial protein oxidation in yeast mutants lacking manganese-(MnSOD) or copper- and zinc-containing superoxide dismutase (CuZnSOD): evidence that MnSOD and CuZnSOD have both unique and overlapping functions in protecting mitochondrial proteins from oxidative damage. J. Biol. Chem. 279: 51817–51827. [DOI] [PubMed] [Google Scholar]
- Okado-Matsumoto, A., and I. Fridovich, 2001. Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu,Zn-SOD in mitochondria. J. Biol. Chem. 276: 38388–38393. [DOI] [PubMed] [Google Scholar]
- Orr, W. C., and R. S. Sohal, 2003. Does overexpression of Cu,Zn-SOD extend life span in Drosophila melanogaster? Exp. Gerontol. 38: 227–230. [DOI] [PubMed] [Google Scholar]
- Parkes, T. L., A. J. Hilliker and J. P. Phillips, 1999. Motorneurons, reactive oxygen, and life span in Drosophila. Neurobiol. Aging 20: 531–535. [DOI] [PubMed] [Google Scholar]
- Phillips, J. P., S. D. Campbell, D. Michaud, M. Charbonneau and A. J. Hilliker, 1989. Null mutation of copper/zinc superoxide dismutase in Drosophila confers hypersensitivity to paraquat and reduced longevity. Proc. Natl. Acad. Sci. USA 86: 2761–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raha, S., and B. H. Robinson, 2000. Mitochondria, oxygen free radicals, disease and ageing. Trends Biochem. Sci. 25: 502–508. [DOI] [PubMed] [Google Scholar]
- Schriner, S. E., N. J. Linford, G. M. Martin, P. Treuting, C. E. Ogburn et al., 2005. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science 308: 1909–1911. [DOI] [PubMed] [Google Scholar]
- Sedensky, M. M., and P. G. Morgan, 2006. Mitochondrial respiration and reactive oxygen species in C. elegans. Exp. Gerontol. 41: 957–967. [DOI] [PubMed] [Google Scholar]
- Senoo-Matsuda, N., K. Yasuda, M. Tsuda, T. Ohkubo, S. Yoshimura et al., 2001. A defect in the cytochrome b large subunit in complex II causes both superoxide anion overproduction and abnormal energy metabolism in Caenorhabditis elegans. J. Biol. Chem. 276: 41553–41558. [DOI] [PubMed] [Google Scholar]
- Senoo-Matsuda, N., P. S. Hartman, A. Akatsuka, S. Yoshimura and N. Ishii, 2003. A complex II defect affects mitochondrial structure, leading to ced-3- and ced-4-dependent apoptosis and aging. J. Biol. Chem. 278: 22031–22036. [DOI] [PubMed] [Google Scholar]
- Shacter, E., 2000. Quantification and significance of protein oxidation in biological samples. Drug Metab. Rev. 32: 307–326. [DOI] [PubMed] [Google Scholar]
- Shibata, Y., R. Branicky, I. O. Landaverde and S. Hekimi, 2003. Redox regulation of germline and vulval development in Caenorhabditis elegans. Science 302: 1779–1782. [DOI] [PubMed] [Google Scholar]
- Sohal, R. S., 2002. Oxidative stress hypothesis of aging. Free Radic. Biol. Med. 33: 573–574. [DOI] [PubMed] [Google Scholar]
- Stenmark, P., J. Grunler, J. Mattsson, P. J. Sindelar, P. Nordlund et al., 2001. A new member of the family of di-iron carboxylate proteins. Coq7 (clk-1), a membrane-bound hydroxylase involved in ubiquinone biosynthesis. J. Biol. Chem. 276: 33297–33300. [DOI] [PubMed] [Google Scholar]
- Sturtz, L. A., K. Diekert, L. T. Jensen, R. Lill and V. C. Culotta, 2001. A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J. Biol. Chem. 276: 38084–38089. [DOI] [PubMed] [Google Scholar]
- Sun, J., D. Folk, T. J. Bradley and J. Tower, 2002. Induced overexpression of mitochondrial Mn-superoxide dismutase extends the life span of adult Drosophila melanogaster. Genetics 161: 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum, H. A., and L. Guarente, 2002. Model organisms as a guide to mammalian aging. Dev. Cell 2: 9–19. [DOI] [PubMed] [Google Scholar]
- Vanfleteren, J. R., and B. P. Braeckman, 1999. Mechanisms of life span determination in Caenorhabditis elegans. Neurobiol. Aging 20: 487–502. [DOI] [PubMed] [Google Scholar]
- Van Remmen, H., and A. Richardson, 2001. Oxidative damage to mitochondria and aging. Exp. Gerontol. 36: 957–968. [DOI] [PubMed] [Google Scholar]
- Van Remmen, H., Y. Ikeno, M. Hamilton, M. Pahlavani, N. Wolf et al., 2003. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol. Genomics 16: 29–37. [DOI] [PubMed] [Google Scholar]
- Wong, A., P. Boutis and S. Hekimi, 1995. Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics 139: 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda, K., H. Adachi, Y. Fujiwara and N. Ishii, 1999. Protein carbonyl accumulation in aging dauer formation-defective (daf) mutants of Caenorhabditis elegans. J. Gerontol. A Biol. Sci. Med. Sci. 54: B47–B51; discussion B52–B43. [DOI] [PubMed] [Google Scholar]
- Yasuda, K., T. Ishii, H. Suda, A. Akatsuka, P. S. Hartman et al., 2006. Age-related changes of mitochondrial structure and function in Caenorhabditis elegans. Mech. Ageing Dev. 127: 763–770. [DOI] [PubMed] [Google Scholar]