Abstract
The S locus of Nicotiana alata encodes a polymorphic series of ribonucleases (S-RNases) that determine the self-incompatibility (SI) phenotype of the style. The pollen product of the S locus (pollen S) in N. alata is unknown, but in species from the related genus Petunia and in self-incompatible members of the Plantaginaceae and Rosaceae, this function has been assigned to an F-box protein known as SLF or SFB. Here we describe the identification of 10 genes (designated DD1–10) encoding SLF-related proteins that are expressed in N. alata pollen. Because our approach to cloning the DD genes was based on sequences of SLFs from other species, we presume that one of the DD genes encodes the N. alata SLF ortholog. Seven of the DD genes were exclusively expressed in pollen and a low level of sequence variation was found in alleles of each DD gene. Mapping studies confirmed that all 10 DD genes were linked to the S locus and that at least three were located in the same chromosomal segment as pollen S. Finally, the different topologies of the phylogenetic trees produced using available SLF-related sequences and those produced using S-RNase sequences suggests that pollen S and the S-RNase have different evolutionary histories.
SELF-INCOMPATIBILITY (SI) is a genetic mechanism found in many flowering plants that acts to reduce the negative fitness effects associated with inbreeding (de Nettancourt 2001). In the best-understood examples, SI is controlled by a single genetic S locus with many S alleles: styles reject pollen grains when both express the same S specificity. Although SI systems have evolved independently several times during the diversification of flowering plants (Matton et al. 1994), in three families—the Solanaceae, Plantaginaceae, and Rosaceae—the SI phenotype of the style (but not of pollen) is controlled by an extracellular ribonuclease known as the S-RNase (Anderson et al. 1986; McClure et al. 1989; Sassa et al. 1993; Xue et al. 1996), a feature that suggests a single evolutionary origin for RNase-based SI systems (Igic and Kohn 2001). Mechanistically, S-RNases are thought to act as cytotoxins that can enter both compatible and incompatible pollen tubes (Luu et al. 2000; Goldraij et al. 2006), retarding the growth of incompatible pollen tubes possibly by degrading the limited amount of ribosomal RNA that they contain (McClure et al. 1990).
The S-locus pollen factor, known as pollen S, until recently was unknown. The first indication of its identity came from Lai et al. (2002), who sequenced a 63-kb region of Antirrhinum hispanicum (Plantaginaceae) genomic DNA that contained the S-RNase gene and who found within this sequence an F-box gene specifically expressed in pollen. F-box genes were soon identified at the S loci of Petunia (Solanaceae) (Qiao et al. 2004a; Y. Wang et al. 2004; Tsukamoto et al. 2005), Prunus, Pyrus, and Malus (Rosaceae) (Entani et al. 2003; Ushijima et al. 2003; Sassa et al. 2007), and collectively these genes are now called the S locus F-box genes (abbreviated as SLF for the Solanaceae and Plantaginaceae and as SFB for the Rosaceae). Functionally, F-box proteins are a component of one type of E3 ubiquitin (Ub)-protein ligase, the enzyme responsible for transferring Ub from the E2 Ub-conjugating enzyme to free Lys residues on a selected protein target (Vierstra 2003). This association with protein ubiquitination has led to a model in which compatible pollen tubes are protected from S-RNase cytotoxicity by a SLF-mediated mechanism involving Ub attachment and degradation (Lai et al. 2002; Qiao et al. 2004b).
In each family, evidence exists to show that SLF/SFB encodes pollen S. In the Rosaceae, for example, where producing transgenic plants is difficult, statistical tests of amino acid variation between different Prunus SFB alleles have identified two short regions of sequence that are under balancing selection (Ikeda et al. 2004; Nunes et al. 2006). SI is one of the best-documented examples of balancing selection and the way in which it affects nucleotide sequence variation has previously been used to identify genes involved in SI (Takebayashi et al. 2003). Also consistent with a role for SFB in SI are truncations or deletions of this gene in self-compatible cultivars of fruit trees such as Prunus avium (sweet cherry) and Prunus mume (Japanese apricot) that carry mutations specifically affecting the SI phenotype of pollen (Ushijima et al. 2004; Sonneveld et al. 2005). For the Solanaceae and Plantaginaceae, transgenic experiments have been used to show that SLF encodes a protein that behaves in the manner expected of pollen S (Qiao et al. 2004a; Sijacic et al. 2004). A SLF transgene does not cause pollen from a transgenic plant to express a new S-allele identity, but instead causes a breakdown of SI in pollen if the SLF transgene is derived from an S allele that differs from the plant's own S alleles (Qiao et al. 2004a; Sijacic et al. 2004). Thus, the SLF transgene derived from the Petunia inflata S2 allele (PiSLF2) caused loss of SI expression in pollen carrying either the S1 or S3 allele but not in pollen carrying the S2 allele (Sijacic et al. 2004). The ability of two different pollen S alleles present in the same pollen grain to “competitively interact” with each other, and thus allow the pollen tube to grow through an otherwise incompatible style, is well known, having previously been observed in tetraploids derived from self-incompatible diploids (e.g., Chawla et al. 1997). SI plants with pollen-part mutations (pollen-part mutants, PPMs) are another example of competitive interaction between pollen S alleles. PPMs are diploid plants with an extra copy of the S locus that is present either as a small additional chromosome called a centric fragment, as a translocated fragment attached to a nonhomologous (i.e., non-S-bearing) chromosome, or as a fragment attached to a homologous (S-bearing) chromosome via unequal exchange (Golz et al. 1999, 2001).
Genes related to SLF/SFB but with no known role in SI also occur in plant genomes, including those of self-compatible plants lacking an RNase-based SI mechanism (L. Wang et al. 2004). These genes, referred to here as SLF-like genes, are members of a large family of F-box protein genes found only in plants (L. Wang et al. 2004). Except for SLF/SFB, functions for most members of this family are unknown, even though it includes ∼13% of the F-box protein genes (92/694) in Arabidopsis thaliana (L. Wang et al. 2004). Phylogenetic analysis divides the SLF-like gene family into five different classes (A, B, C, M, and S), with all SLF/SFB genes and some SLF-like genes belonging to the S class (L. Wang et al. 2004). The presence of SLF-like genes can complicate the process of identifying SLF/SFB genes, especially when SLF-like genes are pollen expressed and linked to the S locus (Entani et al. 2003; Ushijima et al. 2003; Zhou et al. 2003; Y. Wang et al. 2004; Sassa et al. 2007).
In this article we describe an analysis of the pollen-expressed S-class SLF-like genes from the solanaceous self-incompatible plant Nicotiana alata. A motivation for this study was to broaden the range of solanaceous plants for which information on SLF genes is available, as all existing sequences are from Petunia (Qiao et al. 2004a; Sijacic et al. 2004; Tsukamoto et al. 2005). Using degenerate primers and a reverse transcriptase PCR (RT–PCR) approach with N. alata pollen RNA as the template, full-length sequences for 10 S-class genes were obtained. Because potentially 1 of these 10 genes is the N. alata SLF ortholog, we gave them the temporary names DD1–10. Of the 10 DD genes, 7 were exclusively expressed in pollen. Polymorphisms in 7 DD genes were used to show linkage to the S locus: placement of individual DD genes with regard to the S-RNase gene and surrounding S-locus markers was done using the PPM plants described in Golz et al. (2001). Finally, phylogenetic analysis using DD and other S-class sequences suggests that the SLFs in the Solanaceae and Plantaginaceae do not have a long history of co-evolving with the S-RNase gene. We discuss the implications that this has for identifying pollen S in these families and for understanding the evolutionary history of the pollen and stylar genes of the S locus.
MATERIALS AND METHODS
Plant material:
SI lines of N. alata homozygous for S1, S2, S3, S6, or S7 were maintained as described in Anderson et al. (1986). The collection of N. alata PPMs homozygous for the S6 allele and with mapped duplications containing the S3 allele have been described previously (Golz et al. 1999, 2001). N. alata families segregating for four S alleles were produced by germinating S1S3 × S6S7 seed and S2S3 × S6S7 seed. S genotypes were determined by PCR amplification of S-RNase genes as described by Li et al. (2000).
Primer design and PCR:
The primers D2 5′-YTIATIGGICCITGYRAYGG-3′ (forward) and D4 5′-CICCRTAYTSIWTCATNAYCC-3′ (reverse) were designed on the basis of regions conserved in SLF genes from the Solanaceae and Plantaginaceae. PCR was performed with ∼50 ng of genomic DNA or 1 μl of cDNA template. Reactions were carried out in a final volume of 20 μl of 1× PCR buffer (Invitrogen, San Diego) containing template DNA, 0.2–0.5 μm of each primer, 0.2 mm dNTPs, 1.5 mm MgCl2, and 2 units of Taq polymerase (Invitrogen) on a GeneAmp 2700 thermal cycler (Applied Biosystems, Foster City, CA). Cycling conditions were as follows: 94° for 2 min and then 32 cycles of 94°at 30 sec, 45°–65° at 30 sec, and 72° at 60 sec. Rapid amplification of cDNA ends (RACE) PCR was performed using the Smart RACE kit (CLONTECH), as described in the manufacturer's instructions.
Cloning and DNA sequencing:
PCR products were purified using the QIAquick kit (QIAGEN, Valencia, CA) and ligated into the pGEM-T easy vector (Promega, Madison, WI). Electrocompetent Escherichia coli (DH10B) cells were transformed and recombinant clones were selected for a PCR-based screen using standard vector specific primers (T7/Sp6). Resulting products were digested with 1 unit of either HaeIII or RsaI (Promega) and separated by gel electrophoresis. Plasmid DNA from clones with unique restriction digestion patterns was isolated and sequenced commercially (Macrogen, Seoul). Sequences of the N. alata DD genes have been deposited in GenBank with the accession nos. EF420251–EF4202510. Protein domain analysis was done using the SMART search tool (Letunic et al. 2006) (http://smart.embl-heidelberg.de/). All evolutionary analyses were performed using the Phylip package of programs (Felsenstein 2004). DNA alignments were generated using TRANALIGN (http://bioweb.pasteur.fr/docs/EMBOSS/tranalign), based on ClustalW (v1.8) (Higgins and Sharp 1988) protein alignments of the corresponding sequences. Maximum-parsimony (MP) trees were estimated from the DNA alignment using DNAPARS with default settings. Neighbor-joining (Saitou and Nei 1987) distance trees were generated using NEIGHBOR, with a DNA distance matrix computed using DNADIST. Bootstrap analyses (Felsenstein 1985) were carried out using 1000 bootstrap pseudoreplicates.
DNA blot analysis:
Genomic DNA was extracted from leaf material using the plant DNAeasy kit (QIAGEN). Leaf DNA (10 μg) was digested to completion with EcoRI (Promega), fractionated on an agarose gel, and transferred to Hybond N+ (Amersham, Piscataway, NJ) membrane using the alkaline blot procedure described by the manufacturer. DNA fragments were radiolabeled with [32P]dCTP using the Primagene kit (Promega). Hybridizations were performed overnight at 65° in 5× SSPE, 5× Denhardt's solution, 0.5% SDS, and 100 μg/ml denatured herring sperm. After hybridization membranes were washed twice in 2× SSC and 0.1% SDS and exposed to film.
RT–PCR analysis:
Total RNA was prepared using Trizol reagent (Invitrogen). DNA contamination was removed by treating isolated RNA (1 μg) with 2 units of DNase I (Invitrogen). First-strand cDNA synthesis was carried out using an oligo(dT17) primer and 200 units of Superscript III (Invitrogen). Sequences of the gene-specific primers used in this experiment are shown in supplemental Table 1 at http://www.genetics.org/supplemental/.
RESULTS
Cloning and sequence analysis of candidate S-class SLF-like protein genes from N. alata pollen:
Partial cDNAs for S-class SLF-like genes were isolated from N. alata pollen by a degenerate PCR approach. Primers D2 and D4 were designed to bind to sequences conserved in Antirrhinum and Petunia SLFs. Pollen RNAs isolated from N. alata plants homozygous for the S1, S2, S3, S6, or S7 allele were reverse transcribed and used as templates. PCR products were cloned and sequenced, and conceptual translations of all six frames were compared to GenBank using BlastP. After several rounds of screening, 10 partial cDNAs with high amino acid sequence similarity to Petunia SLF and SLF-like sequences were identified and given the provisional names DD1–DD10. Sequence differences between the 10 partial cDNAs were 30–40% and there was ≤5% difference in the sequence of the same DD cDNA amplified from different pollen RNA templates (data not shown). As the DD cDNAs represented candidates for N. alata SLF, RACE was used to obtain additional 5′ and 3′ sequence information. Full-length sequences for DD1 and DD5–8 were obtained by RACE using S2S2 pollen cDNA as the template. Full-length sequences for DD2 (S6S6), DD3/DD4 (S1S1), and DD9 (S3S3) were obtained from the indicated pollen cDNAs, and a near full-length sequence for DD10 (missing the initiator Met codon and 5′-UTR) was obtained from S6S6 pollen cDNA.
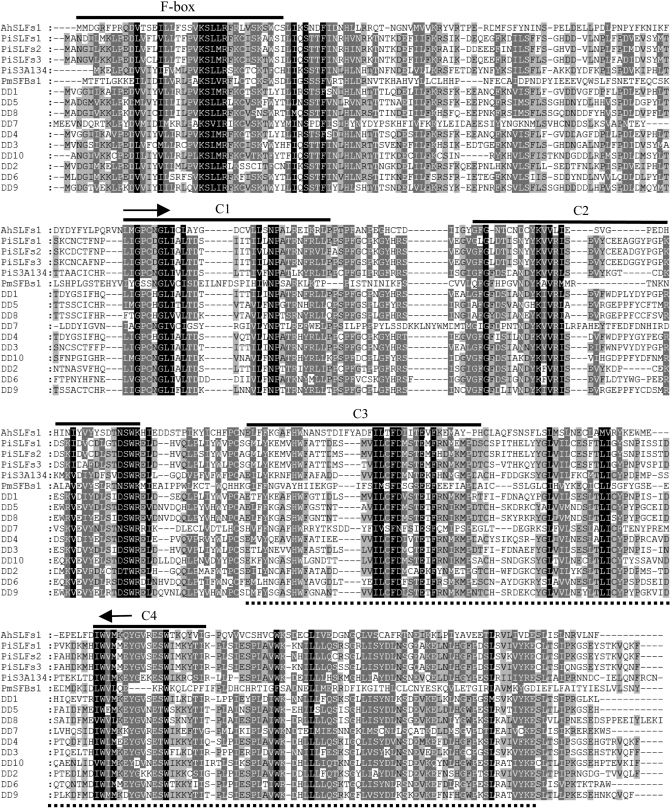
Figure 1 shows conceptual translations of the 10 DD cDNAs aligned with representative SLF and SLF-like proteins from P. inflata (Solanaceae), A. hispanicum (Plantaginaceae) and P. mume (Rosaceae). DD5 and DD8 were the most closely related of the N. alata DD proteins with 88% amino acid similarity (78% identity), and DD3 and DD7 were the least related with 50% amino acid similarity (35% identity). The protein similarity between the DD sequences and P. inflata SLF-S3 ranged from 53% (DD7) to 80% (DD3). SMART and PFAM analysis identified an N-terminal F-box domain in all N. alata DD proteins, consistent with them being members of the F-box protein superfamily. Other F-box proteins have diverse regions outside the F-box domain that bind to specific substrate proteins (Gagne et al. 2002). Potential substrate-binding motifs present in the DD proteins include the related F-box-associated domains type 1 and type 3 (FBA_1, PF07734; FBA_3, PF08268) (Figure 1). L. Wang et al. (2004) incorporated these domains into the four conserved C-terminal motifs (C1–C4) used to define the S-class SLF-like proteins. C1–C4 are present in the SLF/SFB proteins and in all 10 DD proteins (Figure 1).
Figure 1.—
Amino acid sequences encoded by the 10 N. alata DD cDNAs aligned with representative SLF-like sequences from A. hispanicum, P. inflata, and P. mume. Primer binding sites for degenerate primers D2 and D4 are indicated by arrows. The F-box and four conserved motifs identified in L. Wang et al. (2004) are indicated by solid lines. The C-terminal F-box associated domain (FBA 1 and FBA 3) is indicated by the dashed line below the alignment. Accession numbers of sequences from species other than N. alata are AhSLFS1 (AJ515535), PiSLFS1 (AY500390), PiSLFS2 (AY136628), PiSLFS3 (AY500392), PiS3A134 (AY363975), and PmSFBS1 (BAD08320).
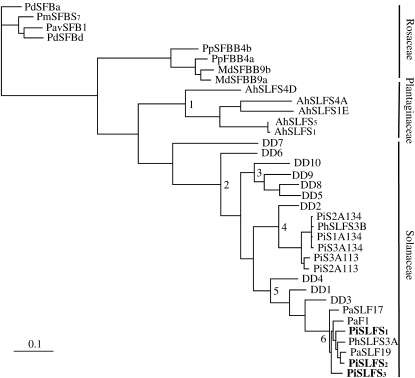
Nucleotide sequences of the N. alata DD genes were aligned with sequences of the SLF and S-class SLF-like genes from P. inflata, Petunia axillaris, and Petunia hybrida (Solanaceae) and A. hispanicum (Plantaginaceae). Sequences of SFBs from species in the Rosaceae were also included. Figure 2 shows the MP tree produced from the DNA alignment. Bootstrap analysis provides strong support for almost all nodes, and distance trees generated using the same alignment had an identical topology (results not shown). The Solanaceae SLF and SLF-like sequences are in a single cluster, as are the Antirrhinum sequences, with nodes at the base of the Solanaceae and Plantaginaceae clusters (nodes 1 and 2) having strong bootstrap support (100%). DD7 and DD6 are at the base of the Solanaceae cluster and sisters to three well-supported smaller clusters (nodes 3–5). The P. inflata SLF sequences, PiSLF-S1, PiSLF-S2, and PiSLF-S3, group together in a cluster (node 6) with PaF1, an SLF-like sequence from P. axillaris (Tsukamoto et al. 2005). These sequences are sisters to the Nicotiana sequences DD1, DD3, and DD4 (node 5). The second small cluster contains P. hybrida and P. inflata SLF-like sequences and DD2 (node 4), and the third cluster comprises only DD sequences (node 3).
Figure 2.—
Maximum-parsimony tree (5087 steps) derived from aligned SLF/SFB and SLF-like DNA sequences. The three Petunia sequences identified with pollen S are indicated by boldface type. Bootstrap values (1000 pseudoreplicates) for numbered nodes are 1 = 100%, 2 = 100%, 3 = 96%, 4 = 100%, 5 = 99%, 6 = 100%. Sequences in the tree and their accession numbers are PaSLFS17 (AY766153), PaSLFS19 (AY766154), PhSLFS3A (AY639403), PhSLFS3B (AY639402), PaF1 (AY766155), PiS3A113 (AY363972), PiS1A134 (AY363973), PiS2A134 (AY363974), PiS2A113 (AY363971), AhSLFS5 (AJ515536), AhSLF-S1E (AJ515535), AhSLF-S4D (AJ515534), AhSLFS4A (AJ515534), PmSFBS7 (AB092622), PdSFBa (AB092966), MdSFBB9b (AB270794), MdSFBB9a (AB270793), PpFBB4a (AB270797), PpSFBB4b (AB270798), PavSFB1 (PavSFB1), and PbSFBb (AB081648).
Expression of the DD genes in various N. alata S genotypes and tissues:
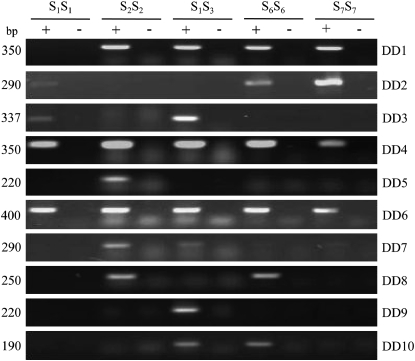
To determine whether expression of a DD gene was restricted to pollen of a particular S genotype, PCR experiments were done with gene-specific primers and S1S1, S2S2, S1S3, S6S6, and S7S7 pollen cDNA templates (Figure 3). In each case, DNA sequence analysis and/or restriction enzyme digests were used to confirm that the correct products had been amplified (data not shown), and pollen RNA (−RT control) was used to show that the products were derived from cDNA and not from genomic DNA. Transcripts for 2 of the 10 DD genes were amplified from all five pollen templates tested (DD4 and DD6) and transcripts for another 6 DD genes (DD1–DD3, DD7, DD8, and DD10) were amplified from between two and four templates. For example, DD1 transcripts were present in all pollen cDNAs except S1S1, and DD2 transcripts were detectable in S1S1, S6S6, and S7S7 cDNAs, but not in S2S2 and S1S3 cDNAs. DD3 was detected in S1S1 and S1S3 pollen cDNAs but could not be amplified from S3S3 cDNA (data not shown). The remaining two DD genes were more restricted in their expression and transcripts were detectable in only a single cDNA type: the S2S2 template for DD5 and the S1S3 template for DD9.
Figure 3.—
RT–PCR was carried out using primers specific for each DD gene shown on the right, with pollen cDNA templates of the indicated S genotype shown at the top of the gel. “No reverse transcriptase controls” are indicated by the minus sign and the size of each PCR product in base pairs is shown at the left of each gel.
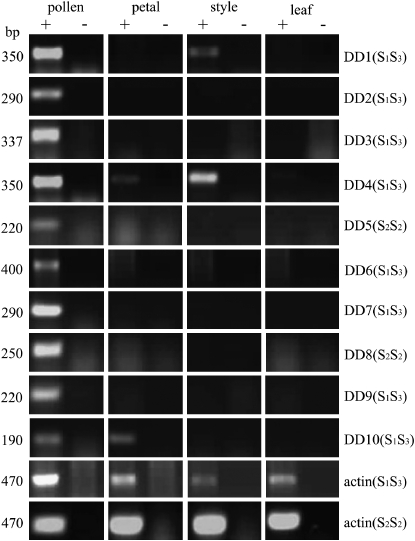
The tissue-specific expression patterns of the DD genes were investigated by RT–PCR using cDNA synthesized from N. alata pollen, style, leaf, and petal RNA. Figure 4 shows that DD2, DD3, and DD5–9 were expressed only in pollen, that DD1 and DD4 were expressed in pollen and style, and that DD10 was expressed in pollen and petal. The DD genes were not expressed at detectable levels in nonfloral tissues such as leaf or root (Figure 4 and data not shown).
Figure 4.—
Spatial expression pattern of DD genes. RT–PCR was performed on N. alata tissue indicated at the top of each gel. Primers used were specific for the gene shown at the right, and the cDNA genotype tested is indicated in parentheses. “No reverse transcriptase controls” are indicated by the minus sign, and actin-specific primers were used to confirm the presence of template. PCR product sizes in base pairs are indicated at the left.
Determining linkage between the DD genes and the S locus:
Evidence of linkage between the S locus and individual DD genes was obtained using two families of N. alata plants in which four S alleles were segregating. S genotypes of progeny plants resulting from a S1S3 × S6S7 cross or a S2S3 × S6S7 cross were determined using allele-specific PCR of the S-RNase gene (results not shown). Cleavable amplified polymorphic sequence markers were designed for eight of the N. alata DD genes to test for cosegregation with an individual S-RNase allele (e.g., S3). Analysis of 18–20 plants detected no recombination between the DD marker and S3-RNase (supplemental Table 2 at http://www.genetics.org/supplemental/), indicating that these DD genes are <5 cM from the S locus.
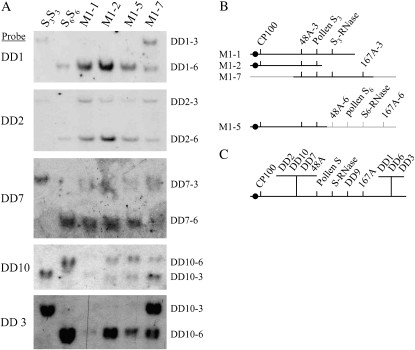
Mapping of the DD genes was also done using a collection of PPM N. alata plants that are homozygous for the S6 allele and that carry chromosomal duplications of varying lengths containing an S3 allele (dS3). The duplications are present in the genomes of these plants as small extra chromosomes (centric fragments), segments that have been added to other chromosomes (translocations), or segments that are linked to an S6 allele via an unequal crossover (Figure 5B). This approach to mapping genes to the N. alata S locus is quicker than the standard linkage-based approach (supplemental Table 2 at http://www.genetics.org/supplemental/) and provides information about relative gene order, but is suitable only for genes with S3- and S6-associated polymorphisms. Suitable polymorphisms were detected for DD1–3, DD6, DD7, DD9, and DD10. DNA blot analysis indicated that the DD genes were all single-copy sequences in the N. alata genome (Figure 5A and data not shown).
Figure 5.—
Linkage of the DD genes to the S locus. (A) Representative Southern blots using probes specific for the indicated DD gene. The source of genomic DNA is shown at the top of each lane and the different alleles are indicated at the right. (B) Maps of dS3 in each of the indicated PPMs. A solid dot indicates the centromere. All PPMs used in this experiment have the genotype S6S6dS3. (C) Map of the N. alata S locus showing relative placement of the seven DD genes mapped using this approach. Placement of genes in the segment between CP100 and S-RNase with respect to each other is arbitrary.
Figure 5 illustrates how the PPMs were used to determine map locations for the DD genes. Four PPM lines were selected for this analysis (M1-1, M1-2, M1-5, and M1-7) with Figure 5B showing the previously described order of markers (CP100, 48A, S-RNase, and 167A) on dS3 in each line. CP100, 48A, and S-RNase all map without recombination to the S locus and 167A is 0.9 cM away (Li et al. 2000). On blots of genomic DNA from S3S3, S6S6, M1-1, M1-2, M1-5, and M1-7 (Figure 5A), the DD1 probe detected a restriction fragment length polymorphism between S3S3 (DD1–3) and S6S6 (DD1–6). As expected, DD1–6 was present in all M1 plants (S6S6dS3) but DD1–3 was present only in M1-7, placing DD1 to the right of marker 167A (Figure 5C). Similarly, the DD2 probe hybridized to DD2–3 and DD2–6 in S3S3 and S6S6, respectively (Figure 5A). DD2–3 was present in all M1 plants, placing this gene within the S locus in a chromosomal segment delimited by the left border of the translocation in M1-7 and the right border of the unequal exchange in M1-5 (Figure 5C). The other genes known to lie in this segment are 48A and pollen S. Figure 5C shows DD2 grouped with 48A on the centromeric side of the S locus, although the placement of these markers with respect to each other and pollen S is arbitrary. Figure 5C summarizes the PPM mapping data. DD7 and DD10 are in the same chromosomal segment as DD2, DD9 lies between the S-RNase and 167A, and DD1, DD3, and DD6 are farther from the S locus than 167A (supplemental Figure 6 at http://www.genetics.org/supplemental/).
DISCUSSION
This article describes the identification and initial characterization of 10 genes expressed in N. alata pollen (DD1–10) that code for S-class SLF-like proteins, a class that includes all the known SLFs of Antirrhinum and Petunia, the SFBs of Prunus, and other clearly related proteins in these and other species with no obvious role in SI (L. Wang et al. 2004). The DD genes were all present as single-copy sequences and the 10 DD genes appear to represent the full complement of S-class S SLF-like genes expressed in N. alata pollen that can be recovered using the RT–PCR-based approach described here. Because none of the DD genes could unambiguously be identified as encoding the N. alata ortholog of SLF (see below), we propose to continue using this provisional naming scheme until such time as functions are identified and individual DD genes can be given more descriptive names.
A surprising aspect of this work was the linkage detected in small-scale experiments between 8 of the 10 DD genes (DD3–DD10) and the S locus (supplemental Table 2 at http://www.genetics.org/supplemental/). This potentially indicated the existence of a single cluster of DD genes. Clusters of F-box protein genes have been described in Arabidopsis by Gagne et al. (2002), where 35.9% of the 694 F-box protein genes are in arrays of two to seven members. Instead of a single cluster of DD genes, it could equally be true that there are many separate SLF-like genes, all subject to the suppressed recombination affecting the S locus and precluding the production of well-resolved genetic maps (Li et al. 2000; Wang et al. 2003). The relative order of seven of the DD genes with respect to each other and the surrounding S locus markers, however, could be determined using a small number of PPMs with duplicated S3 allele segments of differing sizes (Golz et al. 2001). This analysis showed that four DD genes (DD2, DD7, DD9, and DD10) exist as single genes (or possibly as a cluster of a few genes) on either side of the S-RNase gene and within the S locus. Three of the DD genes (DD1, DD3, and DD6) were farther from the S-RNase gene than the marker 167A and accordingly must be at least 0.9 cM from the S locus. In total, all 10 DD genes were linked to the S locus, although a lack of suitable polymorphisms for DD4, DD5, and DD8 meant that the position of these genes relative to other S-locus markers and each other could not be determined.
Even though all the class S SLF-like genes identified in this study were expressed in N. alata pollen, they clearly cannot all be involved in SI. Some, for example, were expressed in tissues other than pollen—specifically DD1 and DD3, which were also expressed in styles, and DD10, which had some expression in petals. Since all SLFs characterized to date are expressed only in pollen, these genes can be excluded from consideration as potential SLF orthologs. The remaining seven DD genes, however, appear to be expressed only in pollen and thus warrant further study. Although all the DD genes are at or near the N. alata S locus, fine-scale mapping indicates that only three—DD2, DD7, and DD10—are in the same region of the chromosome as pollen S. Since expression in petal means that DD10 can be excluded, only DD2 and DD7 can still be considered possible SLF orthologs, with the status of DD4, DD5, and DD8 being uncertain.
Interestingly, of these five “candidate” genes, DD5 transcripts were amplified only from S2S2 pollen cDNA, and DD2, DD7, and DD8 transcripts were amplified only from two of the five pollen cDNA templates tested (Figure 3 and data not shown). DD4 transcripts were amplified from all five pollen templates. While this could potentially reflect a level of S-allele specificity, amplification of transcripts from some cDNA templates but not others was also seen with DD genes that are clearly not pollen S (e.g., DD9 was amplified only from S1S3 pollen cDNA).
Both the number of S-class SLF-like genes at or near the S locus and their pollen expression are worthy of comment. No such clustering of S-class SLF-like genes is apparent in Arabidopsis, where the 7 genes of this type are distributed across three of the five chromosomes (L. Wang et al. 2004). Predominant or exclusive expression of 10 S-class SLF-like genes in N. alata pollen also contrasts with the situation in Arabidopsis, where six of the seven genes are constitutively expressed and only one, AtSFL79, is strictly pollen expressed (L. Wang et al. 2004). SLF-like genes, however, have been found at or near the S locus in the Rosaceae (e.g., see Entani et al. 2003; Ushijima et al. 2003; Sassa et al. 2007), the Plantaginaceae (Zhou et al. 2003), and in the solanaceous species P. inflata (Wang et al. 2003). In each case, these S-locus-linked F-box protein genes are predominantly or exclusively expressed in pollen, just like the DD genes. We speculate that the number of S-class SLF-like genes at the S locus and their pollen expression is not due to chance but is an evolutionary outcome arising from the state of permanent heterozygosity that SI imposes.
In a process analogous to that leading to a loss of active genes on the male-determining Y chromosome of animals (see Skaletsky et al. 2003), permanent heterozygosity at the S locus will reduce the effectiveness of selection against deleterious mutations occurring in genes with unrelated functions that are embedded within it (Uyenoyama 1997, 2005). As each individual S allele experiences a small effective population size, deleterious mutations in genes tightly linked to the S locus are highly likely to replace functional wild-type alleles, a process that should, over time, lead to the degeneration of most genes within the S-locus region except those with SI-specific functions. Any gene expressed in a haploid cell like a pollen grain will escape this effect because there are no genes associated with a second S allele to provide in trans the functions lost by mutation. Hence these genes will still be subject to selection against deleterious mutations, especially if their products contribute to the reproductive fitness of pollen. Selection may also favor expanding the number of these genes at the S locus through gene duplication in cases where higher expression levels enhance male fitness. Thus, this evolutionary scenario suggests that the cluster of DD genes at the N. alata S locus arises because enforced heterozygosity favors genes that contribute to male fitness and that through pollen expression avoid gene erosion processes.
The phylogenetic tree in Figure 2 highlights several puzzling aspects of the S-class SLF-like proteins. Most notably, the SLF/SFB proteins of the Plantaginaceae, Solanaceae, and Rosaceae do not form a monophyletic clade to the exclusion of other S-class SLF-like proteins with no role in SI, such as many of the DD proteins. Contrast can be made here to trees of S-RNases and the related S-like RNases, which are not involved in SI. Phylogenetic analysis places S-RNases and S-like RNases into one of three classes: a single monophyletic clade that contains all S-RNases from the Solanaceae, Plantaginaceae, and Rosaceae and two separate clades of S-like RNases (Igic and Kohn 2001; Steinbachs and Holsinger 2002). The apparent homology of the S-RNases leads to the conclusion that the three known RNase-based SI systems share a single evolutionary origin. In Figure 2, however, S-class SLF-like proteins are grouped not according to function but on the basis of the taxonomic relationships of the organisms from which they were derived. Thus the conclusion that the S-RNases of the Solanaceae, Rosaceae, and Plantaginaceae are homologous is not borne out by this analysis of the SLFs and SFBs. Assuming that the SLFs and SFBs are involved in SI, then the most parsimonious interpretation of Figure 2 is that their encoding genes have been independently recruited to roles in RNase-based SI in the Rosaceae, Plantaginaceae, and Solanaceae. Lack of homology implies that different mechanisms may be used to achieve the rejection of incompatible pollen in each family.
A second notable aspect of the phylogenetic reconstruction in Figure 2 is the well-resolved clade arising at node 5 that contains all the Petunia SLFs, proteins such as PaF1 (P. axillaris F-box protein 1) that are presumed not to be at the Petunia S locus (Tsukamoto et al. 2005), and two DD proteins (DD1 and DD3) that are not considered candidates for the N. alata SLF ortholog because their encoding genes are at least 0.9 cM from the S locus (Figure 5). Numerous phylogenetic analyses of the solanaceous S-RNases place these sequences in a well-resolved monophyletic clade that excludes S-like RNases (e.g., Igic and Kohn 2001; Steinbachs and Holsinger 2002). Any RNase sequence falling into the S-RNase clade not involved in self-incompatibility (e.g., one obtained from a self-compatible plant) is presumed to be derived from a nonfunctional S-RNase (relic S-RNases; Golz et al. 1998). For the clade in Figure 2 to be a clade of solanaceous SLFs resembling the clade of solanaceous S-RNases, DD1, DD3, and PaF1 must all be relic SLFs. Since these proteins are from self-incompatible species, it therefore becomes problematic to assign the name SLF to any protein sequence within this clade unless its function in SI has been verified experimentally. It is also notable that four of the remaining five SLF candidates—DD2, DD5, DD7, and DD8—are not in this SLF-containing clade.
The final puzzling feature of the tree is the shortness of terminal branches for SLFs in the Plantaginaceae and Solanaceae, reflecting the low levels of sequence polymorphism among SLF alleles in these two families. Notably, this feature is not seen with SFBs from the Rosaceae. The four Antirrhinum SLF alleles are identical at 97% or more of their amino acid positions and the two most divergent Petunia SLF alleles, PiSLF-S3 and PaSLF-S17, are 87% identical. These percentages of identity values are closer to those of genes with functions unrelated to self-incompatibility that also lie within the S locus (such as 48A; Takebayashi et al. 2003) than they are to the S-RNases, where pairwise amino acid identities of 40–50% are common (Clark and Kao 1994). The pairwise amino acid identity in partial cDNAs of DD1, DD2, DD4, DD6, DD7, DD8, and DD10 from different S-allele backgrounds ranges from 1 to 5%.
SI imposes a very intense form of balancing selection on the S locus, as a consequence of which high numbers of S-allele lineages are maintained over long periods of time (reviewed by Clark and Kao 1994; Richman 2000). The effect that balancing selection has had on the phylogenetic history of the S-RNase gene, particularly on extending the phylogenies of allelic lineages over time spans that are vastly longer than those expected under neutrality, has been demonstrated many times (see Ioerger et al. 1990; Richman et al. 1996). In the Rosaceae, identifying SFBs as the pollen S factor is based in part on agreement between the expected phylogeny of a gene under balancing selection and the observed SFB phylogenies (Ikeda et al. 2004; Nunes et al. 2006; Sassa et al. 2007). Conversely, genes have also been excluded from consideration as pollen S if their phylogeny did not show the expected features (e.g., Takebayashi et al. 2003).
Little evidence of this predicted evolutionary history is seen in the Petunia and Antirrhinum SLFs, however: the clades containing these sequences are marked by short terminal branches that quickly converged on the common ancestor of all sampled lineages. Because in the Rosaceae SFBs, sites identified as positively selected largely overlap with regions of highest sequence variability (Ikeda et al. 2004; Nunes et al. 2006), we visually inspected an alignment of SLF proteins from the Solanaceae for highly variable sites (i.e., sites occupied by many different amino acids; see supplemental Figure 7 at http://www.genetics.org/supplemental/). As only one such site was found, it is probable that the sequences also lack positively selected sites likely to be responsible for defining pollen specificities.
The marked lack of concordance between S-RNase trees, which are largely consistent with well-established theories as to how genes evolve under balancing selection (Clark and Kao 1994), and the S-class SLF-like tree for the Solanaceae leads to the unsettling conclusion that the SLFs and S-RNases are not stably co-evolving partners, which previously had been assumed would be true of the pollen and stylar factors of the S locus. Two possible explanations of this situation exist: either there is no obligate relationship between the S-RNase and a particular S-class F-box protein or the SLFs do not fulfill the role of pollen S in the Solanaceae and Plantaginaceae.
Although phylogenetic reconstruction of the S-class SLF-like proteins provides little indication that the SLFs have a long history of evolving under balancing selection, other evidence is consistent with the expected behavior of pollen S. Apart from their genetic location and expression pattern, this evidence includes the binding of recombinant SLF proteins to S-RNases and the pollination behavior of transgenic plants that express SLFs in their pollen (e.g., see Qiao et al. 2004b; Sijacic et al. 2004). It should, however, be noted that so far it has not been possible to demonstrate allele specificity for the SLFs (McClure 2004). Recombinant SLF proteins bind S-RNases nonspecifically, and SLF expression in transgenic plants results in a loss of SI rather than in the gain of a new S-allele specificity. Since other proteins can bind S-RNases (Sims and Ordanic 2001), and since mutations in other genes can lead to self-compatible phenotypes (e.g., Ai et al. 1991), it remains formally possible that the properties currently used to define pollen S in the Solanaceae and Plantaginaceae are not exclusive to this factor.
The alternative explanation is that the functions of pollen S are transient and able to be performed by any one of the many paralagous S-class SLF-like genes within the S locus. Sassa et al. (2007) recently made a similar suggestion on the basis of their observation that the S locus in Malus and Pyrus (Rosaceae) contained two or more copies of the SFB gene (“SFB brothers” or SFBBs). If paralagous SLF genes act as proposed, then pollen S lineages might be expected to turn over far more rapidly than S-RNase lineages. Although this suggestion currently lacks experimental support, it is obvious that identifying which, if any, of the DD genes codes for pollen S in N. alata will help refine ideas as to why the SLF and S-RNase genes have such remarkably divergent evolutionary histories.
Acknowledgments
We thank Bruce McGinness for assistance in the glass house and Poh Ling Koh for experimental help. This work is supported by a grant from the Australian Research Council.
References
- Ai, Y. J., E. Kron and T. H. Kao, 1991. S-alleles are retained and expressed in a self-compatible cultivar of Petunia hybrida. Mol. Gen. Genet. 230: 353–358. [DOI] [PubMed] [Google Scholar]
- Anderson, M. A., E. C. Cornish, S.-L. Mau, E. G. Williams, R. Hoggart et al., 1986. Cloning of cDNA for a stylar glycoprotein associated with expression of self-incompatibility in Nicotiana alata. Nature 321: 38–44. [Google Scholar]
- Chawla, B., R. Bernatzky, W. Liang and M. Marcotrigiano, 1997. Breakdown of self-incompatibility in tetraploid Lycopersicon peruvianum: inheritance and expression of S-related proteins. Theor. Appl. Genet. 95: 992–996. [Google Scholar]
- Clark, A. G., and T. Kao, 1994. Self-incompatibility: theoretical concepts and evolution, pp. 220–242 in Genetic Control of Self-Incompatibility and Reproductive Development in Flowering Plants, edited by E. G. Williams, R. B. Knox and A. E. Clarke. Kluwer, Dordrecht, The Netherlands.
- de Nettancourt, D., 2001. Incompatibility and Incongruity in Wild and Cultivated Plants. Springer, New York.
- Entani, T., M. Iwano, H. Shiba, F.-S. Che, A. Isogai et al., 2003. Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: identification of a pollen-expressed F-box gene with allelic diversity. Genes Cells 8: 203–213. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 2004. PHYLIP (Phylogeny Inference Package), Version 3.6. University of Washington, Seattle.
- Gagne, J. M., B. P. Downes, S. H. Shiu, A. M. Durski and R. D. Vierstra, 2002. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldraij, A., K. Kondo, C. B. Lee, C. N. Hancock, M. Sivaguru et al., 2006. Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature 439: 805–810. [DOI] [PubMed] [Google Scholar]
- Golz, J. F., A. E. Clarke, E. Newbigin and M. Anderson, 1998. A relic S-RNase is expressed in the styles of self-compatible Nicotiana sylvestris. Plant J. 16: 591–599. [DOI] [PubMed] [Google Scholar]
- Golz, J. F., V. Su, A. E. Clarke and E. Newbigin, 1999. A molecular description of mutations affecting the pollen component of the Nicotiana alata S locus. Genetics 152: 1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golz, J. F., H. Y. Oh, V. Su, M. Kusaba and E. Newbigin, 2001. Genetic analysis of Nicotiana pollen-part mutants is consistent with the presence of an S-ribonuclease inhibitor at the S locus. Proc. Natl. Acad. Sci. USA 98: 15372–15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, D. G., and P. M. Sharp, 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73: 237–244. [DOI] [PubMed] [Google Scholar]
- Igic, B., and J. R. Kohn, 2001. Evolutionary relationships among self-incompatibility RNases. Proc. Natl. Acad. Sci. USA 98: 13167–13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, K., B. Igic, K. Ushijima, H. Yamane, N. R. Hauck et al., 2004. Primary structural features of the S haplotype-specific F-box protein, SFB, in Prunus. Sex. Plant Reprod. 16: 235–243. [Google Scholar]
- Ioerger, T. R., A. G. Clark and T. H. Kao, 1990. Polymorphism at the self-incompatibility locus in Solanaceae predates speciation. Proc. Natl. Acad. Sci. USA 87: 9732–9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, Z., W. Ma, B. Han, L. Liang, Y. Zhang et al., 2002. An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol. Biol. 50: 29–42. [DOI] [PubMed] [Google Scholar]
- Letunic, I., R. R. Copley, B. Pils, S. Pinkert, J. Schultz et al., 2006. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34: D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.-H., N. Nass, M. Kusaba, P. N. Dodds, N. Treloar et al., 2000. A genetic map of the Nicotiana alata S locus that includes three pollen-expressed genes. Theor. Appl. Genet. 100: 956–964. [Google Scholar]
- Luu, D. T., X. Qin, D. Morse and M. Cappadocia, 2000. S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility. Nature 407: 649–651. [DOI] [PubMed] [Google Scholar]
- Matton, D. P., N. Nass, A. E. Clarke and E. Newbigin, 1994. Self-incompatibility: how plants avoid illegitimate offspring. Proc. Natl. Acad. Sci. USA 91: 1992–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure, B., 2004. S-RNase and SLF determine S-haplotype-specific pollen recognition and rejection. Plant Cell 16: 2840–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure, B. A., V. Haring, P. R. Ebert, M. A. Anderson, R. J. Simpson et al., 1989. Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature 342: 955–957. [DOI] [PubMed] [Google Scholar]
- McClure, B. A., J. E. Gray, M. A. Anderson and A. E. Clarke, 1990. Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature 347: 757–760. [Google Scholar]
- Nunes, M. D. S., R. A. M. Santos, S. M. Ferreira, J. Vieira and C. P. Vieira, 2006. Variability patterns and positively selected sites at the gametophytic self-incompatibility pollen SFB gene in a wild self-incompatible Prunus spinosa (Rosaceae) population. New Phytol. 172: 577–587. [DOI] [PubMed] [Google Scholar]
- Qiao, H., F. Wang, L. Zhao, J. Zhou, Z. Lai et al., 2004. a The F-box protein AhSLF-S2 controls the pollen function of S-RNase-based self-incompatibility. Plant Cell 16: 2307–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, H., H. Wang, L. Zhao, J. Zhou, J. Huang et al., 2004. b The F-box protein AhSLF-S2 physically interacts with S-RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. Plant Cell 16: 582–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman, A., 2000. Evolution of balanced genetic polymorphism. Mol. Ecol. 9: 1953–1963. [DOI] [PubMed] [Google Scholar]
- Richman, A. D., M. K. Uyenoyama and J. R. Kohn, 1996. Allelic diversity and gene genealogy at the self-incompatibility locus in the Solanaceae. Science 273: 1212–1216. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and M. Nei, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sassa, H., H. Hirano and H. Ikehashi, 1993. Identification and characterization of stylar glycoproteins associated with self-incompatibility genes of Japanese pear, Pyrus serotina Rehd. Mol. Gen. Genet. 241: 17–25. [DOI] [PubMed] [Google Scholar]
- Sassa, H., H. Kakui, M. Miyamoto, Y. Suzuki, T. Hanada et al., 2007. S-locus F-box brothers: multiple and pollen-specific F-box genes with S haplotype-specific polymorphisms in apple and Japanese pear. Genetics 175: 1869–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijacic, P., X. Wang, A. L. Skirpan, Y. Wang, P. E. Dowd et al., 2004. Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429: 302–305. [DOI] [PubMed] [Google Scholar]
- Sims, T. L., and M. Ordanic, 2001. Identification of a S-ribonuclease-binding protein in Petunia hybrida. Plant Mol. Biol. 47: 771–783. [DOI] [PubMed] [Google Scholar]
- Skaletsky, H., T. Kuroda-Kawaguchi, P. J. Minx, H. S. Cordum, L. Hillier et al., 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423: 825–837. [DOI] [PubMed] [Google Scholar]
- Sonneveld, T., K. R. Tobutt, S. P. Vaughan and T. P. Robbins, 2005. Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. Plant Cell 17: 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbachs, J. E., and K. E. Holsinger, 2002. S-RNase-mediated gametophytic self-incompatibility is ancestral in eudicots. Mol. Biol. Evol. 19: 825–829. [DOI] [PubMed] [Google Scholar]
- Takebayashi, N., P. B. Brewer, E. Newbigin and M. K. Uyenoyama, 2003. Patterns of variation within self-incompatibility loci. Mol. Biol. Evol. 20: 1778–1794. [DOI] [PubMed] [Google Scholar]
- Tsukamoto, T., T. Ando, H. Watanabe, E. Marchesi and T. H. Kao, 2005. Duplication of the S-locus F-box gene is associated with breakdown of pollen function in an S-haplotype identified in a natural population of self-incompatible Petunia axillaris. Plant Mol. Biol. 57: 141–153. [DOI] [PubMed] [Google Scholar]
- Ushijima, K., H. Sassa, A. M. Dandekar, T. M. Gradziel, R. Tao et al., 2003. Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 15: 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima, K., H. Yamane, A. Watari, E. Kakehi, K. Ikeda et al., 2004. The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J. 39: 573–586. [DOI] [PubMed] [Google Scholar]
- Uyenoyama, M. K., 1997. Genealogical structure among alleles regulating self-incompatibility in natural populations of flowering plants. Genetics 147: 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyenoyama, M. K., 2005. Evolution under tight linkage to mating type. New Phytol. 165: 63–70. [DOI] [PubMed] [Google Scholar]
- Vierstra, R. D., 2003. The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8: 135–142. [DOI] [PubMed] [Google Scholar]
- Wang, L., L. Dong, Y. Zhang, W. Wu, X. Deng et al., 2004. Genome-wide analysis of S locus F-box-like genes in Arabidopsis thaliana. Plant Mol. Biol. 56: 929–945. [DOI] [PubMed] [Google Scholar]
- Wang, Y., X. Wang, A. G. McCubbin and T. H. Kao, 2003. Genetic mapping and molecular characterization of the self-incompatibility (S) locus in Petunia inflata. Plant Mol. Biol. 53: 565–580. [DOI] [PubMed] [Google Scholar]
- Wang, Y., T. Tsukamoto, K. W. Yi, X. Wang, S. Huang et al., 2004. Chromosome walking in the Petunia inflata self-incompatibility (S-) locus and gene identification in an 881-kb contig containing S2-RNase. Plant Mol. Biol. 54: 727–742. [DOI] [PubMed] [Google Scholar]
- Xue, Y., R. Carpenter, H. G. Dickinson and E. S. Coen, 1996. Origin of allelic diversity in Antirrhinum S locus RNases. Plant Cell 8: 805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., F. Wang, W. Ma, Y. Zhang, B. Han et al., 2003. Structural and transcriptional analysis of S-locus F-box genes in Antirrhinum. Sex. Plant Reprod. 16: 165–177. [Google Scholar]