Abstract
The alternative oxidase transfers electrons from ubiquinol to molecular oxygen, providing a mechanism for bypassing the later steps of the standard cytochrome-mediated electron transport chain. The enzyme is found in an array of organisms and in many cases is known to be produced in response to perturbations of the standard chain. Alternative oxidase is encoded in the nucleus but functions in the inner mitochondrial membrane. This implies the existence of a retrograde regulation pathway for communicating from the mitochondrion to the nucleus to induce alternative oxidase expression. Previous studies on alternative oxidase in fungi and plants have shown that a number of genes are required for expression of the enzyme, but the identity of these genes has remained elusive. By gene rescue we have now shown that the aod-2 and aod-5 genes of Neurospora crassa encode transcription factors of the zinc-cluster family. Electrophoretic mobility shift assays show that the DNA-binding domains of the AOD2 and AOD5 proteins act in tandem to bind a sequence element in the alternative oxidase gene promoter that is required for expression. Both proteins contain potential PAS domains near their C terminus, which are found primarily in proteins involved in signal transduction.
THE alternative oxidase is found in green plants, many fungi, some protists, some bacteria, and a few animals (Vanlerberghe and Mcintosh 1997; Joseph-Horne et al. 2001; Stenmark and Nordlund 2003; Veiga et al. 2003; McDonald and Vanlerberghe 2004, 2005; Chaudhuri et al. 2006). The enzyme catalyzes the transfer of electrons directly from ubiquinol to oxygen, allowing electrons to bypass the final steps of the cytochrome-mediated electron transport chain. Alternative oxidase does not pump protons so that use of the alternate pathway leaves only complex I as a site of proton gradient formation for ATP synthesis while additional energy is dissipated as heat.
A number of functions have been suggested for the alternative oxidase. For example, in plants the enzyme may prevent the accumulation of reactive oxygen species, prevent programmed cell death, and/or ensure proper energy and biosynthetic balance during growth (Maxwell et al. 1999; Hansen et al. 2002; Moore et al. 2002; Robson and Vanlerberghe 2002; Vanlerberghe et al. 2002; Sieger et al. 2005; Umbach et al. 2005; Ho et al. 2007). The best-defined function of alternative oxidase is in certain plant species where the heat generated via the alternative pathway of electron transport is used to volatilize aromatic compounds that attract insect pollinators (Vanlerberghe and McIntosh 1997). In Trypanosoma brucei, the enzyme is the exclusive terminal oxidase in mitochondria of the bloodstream form of this parasitic protozoan and may be essential for its survival (Helfert et al. 2001; Chaudhuri et al. 2006). It has been suggested that fungi or animals that encounter inhibitors of complex III or IV in nature may utilize alternative oxidase to escape the effects of these compounds (Lambowitz and Zannoni 1978; Joseph-Horne et al. 2001; McDonald and Vanlerberghe 2004).
Alternative oxidase expression has been studied in various organisms, but perhaps most extensively in plants where the enzyme is encoded by multigene families (Considine et al. 2002). Individual genes within the family are expressed differentially in response to a variety of stresses or developmental signals (reviewed in Vanlerberghe and McIntosh 1997; Arnholdt-Schmitt et al. 2006; Clifton et al. 2006; Rhoads et al. 2006; Rhoads and Subbaiah 2007). For at least some genes in some species, it has been shown that expression likely involves control at the level of transcription, although additional controls at the level of mRNA stability and translation may also exist (Yu et al. 2001; Thirkettle-Watts et al. 2003; Dojcinovic et al. 2005; Zarkovic et al. 2005). In T. brucei, the level of alternative oxidase is controlled post-transcriptionally in a developmentally regulated fashion (Chaudhuri et al. 2002). In fungi, the enzyme is generally induced by conditions that impair the function of the normal electron transport chain and/or increase reactive oxygen species levels, but constitutive expression has also been reported. Regulation of expression may occur at both the transcriptional and the post-transcriptional levels in fungi (Yukioka et al. 1998; Huh and Kang 1999; Borghouts et al. 2001; Huh and Kang 2001; Tanton et al. 2003).
Alternative oxidase is encoded in the nucleus, synthesized on cytosolic ribosomes, imported into mitochondria, and assembled into the inner mitochondrial membrane. Since most organisms have been found to produce the enzyme in response to inhibition of the electron transport chain, it is assumed that one or more pathways of retrograde regulation must exist to communicate signals that originate in the mitochondrion to the nucleus for expression of the gene. Examination of mutants has been used in an attempt to elucidate the number and type of factors required for alternative oxidase expression. Specific deletion of the Candida albicans SLN1 gene, which encodes a histidine kinase, resulted in slight alterations in expression of alternative oxidase (Huh and Kang 2001), although it was not established whether these effects were direct or indirect. More broadly based mutant screens have been performed in Arabidopsis thaliana and Neurospora crassa. In A. thaliana, several genes were found that are required to induce the alternative oxidase, most likely at the level of transcription, in response to either antimycin A or monofluroacetate, an inhibitor of the Kreb's cycle (Zarkovic et al. 2005). In N. crassa, alternative oxidase is encoded by the alternative oxidase (aod-1) gene. Under normal growth conditions, alternative oxidase is not detectable, but when oxidative phosphorylation is impaired by mutations or inhibitors, expression of the aod-1 gene is induced (Bertrand et al. 1983; Tanton et al. 2003). In the 1980s, mutant screens revealed that the aod-2 gene was required for expression of aod-1 (Bertrand et al. 1983; Lambowitz et al. 1989). Additional genes (aod-4, aod-5, aod-6, and aod-7) that may be involved in a signal transduction pathway for alternative oxidase expression in N. crassa have been discovered more recently (Descheneau et al. 2005). To date, the nature of the products encoded by genes uncovered in mutant screens in plants or fungi remains unknown.
The N. crassa aod-1 gene promoter contains an alternative oxidase induction motif (AIM) that is required for efficient expression of the gene (Chae et al. 2007). The AIM contains two directly repeated CGG triplets separated by 7 bp, both copies of which are required for induction. This motif structure is typical of sequence elements recognized by transcription factors of the zinc-cluster family (Macpherson et al. 2006). Here we describe the identification of the N. crassa aod-2 and aod-5 gene products as zinc-cluster transcription factors that control the expression of the aod-1 gene via interaction with the AIM sequence.
MATERIALS AND METHODS
Strains, cosmids, plasmids, and oligonucleotides:
The strains used in this study are listed in Table 1. Cosmids were isolated from the pMOcosX libraries (Kelkar et al. 2001), which were obtained from the Fungal Genetics Stock Center. Plasmid pAO2-2 contains the coding sequence for the AOD2 protein, plus 135 bp upstream and 480 bp downstream. The sequence was generated by PCR using primers Orf52-5′ and Orf52-3′ and cloned into the NotI restriction sites of pBSSKII. Plasmid pAO2XBen-4 is derived from pAO2-2 but contains an additional 324 bp of upstream sequence and a benomyl resistance gene. pAO2XBen-4 was used to produce mutant versions of the aod-2 gene. Plasmid pXba-11 containing the aod-5 coding sequence and flanking regions was produced by cloning a 3.4-kb XbaI fragment from cosmid X14F7 into pMOcosX. The aod-5 plasmid for site-directed mutagenesis was pBNA5-2, which contains the 3.4-kb XbaI fragment and a benomyl resistance gene cloned into pBSSKII. Mutant derivatives of plasmids are described in the results and were produced by site-directed mutagenesis as described previously (Chae et al. 2007). Mutations generated in aod-2 or aod-5 plasmids were confirmed by DNA sequencing. For mutant versions of genes that were inefficient in rescuing the aod-2 or aod-5 phenotype, the entire gene was sequenced to confirm that no PCR errors had been introduced. Oligonucleotides used in this study are listed in supplemental Table 1 at http://www.genetics.org/supplemental/.
TABLE 1.
Strains used in this study
| Strain | Genotype | Origin, source, or reference |
|---|---|---|
| NCN251 (also called 74A) | A | FGSC no. 2489 |
| NCN233 | pan-2 A | Nargang lab |
| 7064 | aod-2 nic-1 al-2 a | H. Bertrand |
| CNA33 | aod-2 pan-2 | Cross of NCN233 × 7064 |
| EN14-34 | aod-2 ad-2 al-2 A (also carries pBAT reporter system) | Descheneau et al. (2005) |
| PL40-23 | aod-5 pan-2 A | Descheneau et al. (2005) |
FGSC, Fungal Genetics Stock Center.
Neurospora transformations:
Spheroplasts of CNA33 and PL40-23 for transformation with cosmids and plasmids were generated as described (Drygas et al. 1989) after germination for 3 hr at 30°. Rescue of the aod-2 and aod-5 mutations was achieved by including antimycin A in both the plates and top agar at a final concentration of 0.25–0.50 μg/ml, depending on the lot of antimycin A used. Plasmids were also transformed into CNA33 or PL40-23 using electroporation (Tanton et al. 2003). When plasmid tranformants were selected on the basis of benomyl resistance, both plates and top agar contained benomyl at a concentration of 1 μg/ml.
Respiration:
Oxygen consumption by N. crassa mycelium was measured using an oxygen electrode. Alternative oxidase was induced by growth in the presence of chloramphenicol as described (Tanton et al. 2003).
Production of AOD2 and AOD5 DNA-binding domains:
The N-terminal regions of the AOD2 and AOD5 proteins were produced in the Escherichia coli Rosetta (DE3) strain following cloning into plasmid pET-26b (Novagen, Mississauga, Ontario). The desired fragments for cloning were produced by PCR using plasmids containing cloned cDNAs as templates. For aod-2, primers MCHA53 and MCHA55 were used to amplify the first 351 bp of the coding sequence. MCHA53 introduced an NdeI restriction site at the 5′-end that included the start codon, and MCHA55 inserted a SalI restriction site immediately following base pair 351 of the aod-2 coding sequence. The restriction sites were used to clone the fragment into the NdeI and XhoI restriction sites of pET-26b to give plasmid p55-4a, which expresses the DNA-binding domain of AOD2 plus a C-terminal hexahistidinyl tag encoded on the vector. For aod-5, primers MCHA56 and MCHA58 were used to amplify the first 426 bp of the coding sequence, which introduced NdeI and XhoI sites on the 5′- and 3′-ends, respectively. The fragment was cloned into pET-26b to yield plasmid p58-1, which expresses the DNA-binding domain of AOD5 plus a C-terminal hexahistidinyl tag encoded on the vector. DNA sequencing confirmed that no PCR errors had been introduced into either plasmid.
Plasmids p55-4a and p58-1 were separately transformed into Rosetta (DE3) cells for expression of the fragments. The strain was also transformed with pET-26b to give a control “empty vector” strain. Growth of strains and production of soluble cell supernatants using lysozyme and sonication were according to the supplier's instructions (Novagen) except that ZnCl2 was included in the growth medium at a concentration of 1 mm and in the lysis buffer at 10 μm. The cell lysate supernatants were stored at 4° at a concentration of 1 μg protein/μl. Although the fragments produced contained histidine tags, which could have been used for purification, it was found that the cell lysate supernatants could be used directly in electrophoretic mobility shift assay (EMSA) experiments.
EMSA:
Hybridization of single-stranded oligonucleotides was used to generate wild-type or mutant double-stranded molecules with seven nucleotide overhangs for use in labeling reactions. Annealing reactions contained complementary oligonucleotides (20 nmol each) and were assembled in 500 μl of annealing buffer (10 mm Tris–HCl, pH 7.9, 50 mm NaCl, 10 mm MgCl2, 1 mm dithiothreitol). The mixture was incubated at 95° for 10 min in an aluminum block and cooled slowly to room temperature to give double-stranded DNA stocks. For labeling, the 5′ overhangs were filled in using Klenow enzyme, dATP, dGTP, dTTP, and [α-32P]dCTP (3000 Ci/mmol). Following 1 hr labeling at 37°, the probe was precipitated with ethanol, dried, and resuspended in annealing buffer at a concentration of 10,000 counts/min/μl.
Binding reactions were done in 15-μl volumes and contained 10 mm HEPES, pH 8.0, 1 mm EDTA, 10 mm KCl, 10% glycerol, 10 μm ZnCl2, 1 mm dithiothreitol, 1 μg of sonicated salmon sperm DNA, and 10,000 counts/min of labeled probe. Protein (5 μg) was added directly from bacterial cell lysate supernatant fractions. When two lysates were added to a single binding reaction, 2.5 μg of protein from each was used. Following a 30-min incubation at room temperature, 1 μl of a 0.2% (w/v) bromophenol blue stock was added. The reactions were then loaded onto a 6% polyacrylamide (79:1 acrylamide:bis-acrylamide) gel made in 0.5× TBE (90 mm Tris, 90 mm boric acid, 2 mm EDTA, pH 8.2) that had been prerun at 100 V for 2 hr at 4°. The samples were electrophoresed for 3 hr under the same conditions. The gels were dried under vacuum and exposed to X-ray film.
Quantitative PCR:
Strain NCN233 was grown for 12 hr in liquid Vogel's medium in the absence of antimycin A or for 22 hr in the presence of antimycin A (0.5 μg/ml). Two separate flasks were grown for each condition to provide biological replicates. Mycelium was harvested by vacuum filtration and immediately frozen in liquid nitrogen. Total RNA was isolated using the QIAGEN (Mississauga, Ontario) RNeasy plant mini kit according to the supplier's instructions. The integrity of RNA samples was confirmed by examination on an Agilent 2100 Bioanalyzer using an RNA Nano Chip (Agilent, Santa Clara, CA). First-strand cDNA synthesis was done with SuperscriptIII reverse transcriptase (Invitrogen, Burlington, ON) according to the instructions of the supplier. The cDNAs were stored at −80°.
Primers for PCR were developed for aod-1, aod-2, aod-5, and tom40 using primer express software version 3.0 (Applied Biosystems, Foster City, CA). The primer sets chosen gave products of ≤151 bp in length, and one primer of each set was developed across an exon/exon boundary to avoid amplification of any contaminating genomic DNA. Primers for aod-2 and aod-5 were chosen in regions that did not encode the DNA-binding domain of zinc-cluster proteins since a large family of proteins in N. crassa contain this domain (Borkovich et al. 2004). Quantitative PCR (qPCR) was performed using the standard curve method for absolute quantification with an Applied Biosystems 7500 Fast Real Time PCR system according to the instructions of the manufacturer. Standard curves generated for all genes tested using quantified plasmids containing double-stranded cDNA clones had R2 values of >0.999. By convention, we refer to “levels of transcript” for each gene in the text and figures, even though we have not examined reverse transcription efficiencies.
RESULTS
Identification and properties of the aod-2 and aod-5 gene products:
Cultures of N. crassa grown in chloramphenicol have a reduced ability to translate mitochondrial gene products, which results in a decrease of electron transport complexes and oxidative phosphorylation. Under such conditions, wild-type cells induce alternative oxidase but strains containing mutations in the aod-2 or aod-5 genes are unable to induce expression. Furthermore, aod-2 and aod-5 mutant strains are unable to grow in the presence of antimycin A because the drug effectively eliminates electron transport through complex III of the standard electron transport chain and, without alternative oxidase, no respiration is possible (Descheneau et al. 2005). We exploited this trait to isolate the aod-2 and aod-5 genes using a rescue approach.
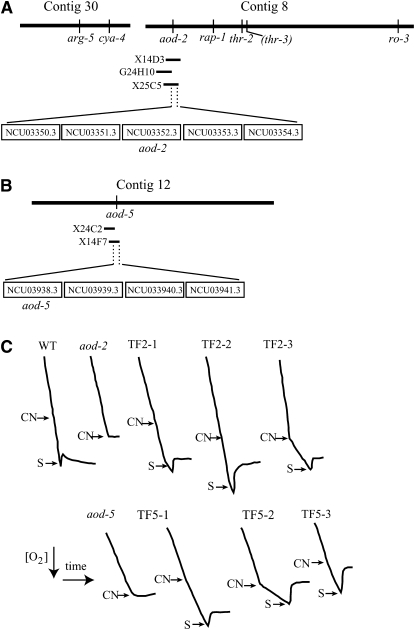
The aod-2 gene was mapped previously to chromosome II near arg-5 and thr-3 in the vicinity of the centromere (Bertrand et al. 1983). We chose 94 cosmids from the pMOcosX libraries (Kelkar et al. 2001) that provided coverage of ∼1.75 Mb in this region of the chromosome. All cosmids were from physical contigs 8, 30, and 33 as defined in assembly 7 (version 3) of the N. crassa sequencing project (Galagan et al. 2003). Isolated cosmids were pooled in sets of two and used to transform spheroplasts of the aod-2 mutant strain CNA33. The transformation mix was plated onto medium containing antimycin A so that only transformants that have a restored ability to induce the alternative oxidase would grow. In 4–5 days, colonies were visible from a single pool of transforming DNA containing cosmids G24H10 and X14D3. A subsequent round of transformation using the individual cosmids showed that X14D3 was able to rescue the aod-2 mutant (Figure 1A). This cosmid contained 10 predicted open reading frames (ORFs) that potentially represented the aod-2 gene, but 4 of these could be eliminated because they were also contained on G24H10, which was unable to rescue. Contig 8 was then reexamined for additional cosmids that contained portions of the sequence of X14D3. Cosmid X25C5 partially overlapped and was found to rescue aod-2 in transformation experiments. This eliminated one additional ORF as a possible aod-2 gene (Figure 1A). To test the remaining candidate ORFs individually, we designed primers to generate PCR products for each remaining ORF that included ∼500 bp upstream and 500 bp downstream of the predicted coding sequences. The first products obtained were for ORFs NCU03352.3 and NCU03350.3. These were cloned into plasmid pBSSKII and used for transformation of CNA33. The plasmid containing NCU03352.3 (pAO2-2) was able to rescue the aod-2 mutation.
Figure 1.—
Identification of aod-2 and aod-5. (A) Contigs 30 (404,010 bp) and 8 (971,569 bp), found on N. crassa chromosome II, are shown at the top. These contigs contained the arg-5 and thr-3 genes previously found to be closely linked to aod-2. The thr-3 gene, in parentheses, has not been identified in the N. crassa sequence and its position near thr-2 is estimated from previous mapping studies (Perkins et al. 2001). The sites of various other genes whose locations have been defined are indicated on the contigs. The position of the three cosmids discussed in the text and the original candidate aod-2 ORFs are shown. NCU03352.3 was determined to be the aod-2 gene. aod, alternative oxidase; arg, arginine; cya, cytochrome aa3; rap, ribosome-associated protein; thr, threonine. (B) Contig 12 (879,620 bp) of chromosome VI. The position of the two cosmids discussed in the text and the potential aod-5 ORFs are shown. NCU03938.3 was identified as aod-5. (C) Respiration tracings of controls, mutant strains, and transformants. The aod-2 mutant strain CNA33 was transformed with plasmid pAO2-2, which contains the PCR product encompassing NCU03352.3. The aod-5 mutant strain PL40-23 was transformed with pXba-11, which contains the XbaI fragment, including NCU03938.3. NCN251 is a wild-type (WT) control. Conidia of each strain were inoculated into medium containing chloramphenicol (2 mg/ml) and grown for 16 hr. One milliliter of the culture was then transferred to a chamber of the oxygen respirometer containing 2 ml of fresh Vogel's medium, and oxygen consumption was measured. The tracings show the decrease in oxygen with time. The presence of alternative oxidase is indicated by continued oxygen consumption following addition of KCN (CN), which inhibits complex IV of the cytochrome-mediated electron transport chain. Three transformants of aod-2 (TF2-1, -2, -3) and three of aod-5 (TF5-1, -2, -3) are shown. For strains that continued to respire following the addition of KCN, salicylhydroxamic acid (S), which inhibits alternative oxidase, was added.
A similar approach was taken to identify aod-5, which had been mapped to chromosome VI (Descheneau et al. 2005). Strain PL40-23 was transformed with cosmids found on chromosome VI from contigs 34, 16, 22, and 12 as defined in assembly 7, version 3 of the genome sequence. Cosmid X14F7 of contig 12 was shown to rescue the aod-5 mutant while an overlapping cosmid, X24C2, could not (Figure 1B). ORF NCU03938.3, which occurred on a 3.4-kb XbaI restriction fragment from this cosmid, was identified as the aod-5 gene.
Rescued aod-2 and aod-5 transformants were examined for their ability to induce alternative oxidase. Transformants were isolated from antimycin A-containing plates and purified through one round of single-colony isolation on the same medium. Cells were then grown in medium containing chloramphenicol and examined for the presence of alternative oxidase using an oxygen respirometer. It was found that KCN was unable to stop respiration in about half the transformants examined, indicating that these strains had recovered the ability to induce alternative oxidase (Figure 1C). We assume that transformants that did not display alternative oxidase activity (not shown) were not stably transformed.
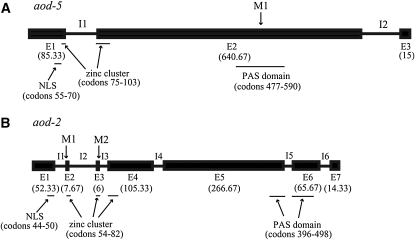
For AOD5, the N. crassa genome project predicted that the 740 residue NCU03938.3 protein was a zinc-cluster transcription factor because it contains the canonical sequence CX2CX6CX5-12CX2CX6-8C near its N terminus (Figure 2A). These proteins form an abundant class of transcription factors in fungi (Macpherson et al. 2006), including N. crassa (Borkovich et al. 2004). We also confirmed the position of predicted introns by sequencing an aod-5 cDNA produced by RT–PCR. The nature of the AOD2 protein identified as NCU03352.3 was not immediately obvious. However, inspection and analysis of the sequence strongly suggested that the ORF predicted from the sequencing project (assembly 7, version 3) had excluded a small exon (exon 3 in Figure 2B) that would complete the coding of the zinc-binding region of a zinc-cluster transcription factor of 517 amino acids (Figure 2B). We have confirmed the presence of the missing exon as part of the coding sequence by sequencing a wild-type cDNA clone generated by RT–PCR. We have corrected the information in the N. crassa sequencing project via the community annotation service. Thus, both aod-2 and aod-5 encode zinc-cluster transcription factors.
Figure 2.—
Structure of the aod-2 and aod-5 genes. (A) The aod-5 gene. Exons (E) are shown as solid boxes and introns (I) as solid lines. The number of codons in each exon is in parentheses. Sites within exons that contain motifs identified as a possible nuclear localization signal (NLS), zinc cluster, and PAS domain are indicated by thin lines below the gene. Above the gene, M1 indicates the site of the nonsense mutation in strain PL40-23. (B) The aod-2 gene. As described for A, except that M1 indicates the site of the original aod-2 mutation, which changes the codon for the first Cys residue of the zinc cluster to a Tyr residue. M2 shows the position of the mutation in strain EN14-34 that affects the splice junction immediately following exon 3.
To confirm that the predicted proteins represented AOD2 and AOD5, we wanted to show that mutant strains contained mutations in the corresponding gene. Genomic DNA was prepared from strain CNA33, which contains the original aod-2 mutation; from EN14-34, which contains a more recently isolated aod-2 mutant allele (Descheneau et al. 2005); and from PL40-23, which contains the aod-5 mutant allele. PCR products that contained ORF NCU03352.3 or NCU03938.3, and their respective flanking regions, were generated and sequenced directly. The original aod-2 mutation was shown to change a Cys codon that is part of the zinc-binding region of the protein at position 54 to a Tyr codon (TGC to TAC) while the other mutant allele of aod-2 contained a mutation (GT to AT) affecting the consensus splice site immediately following exon 3 (Figure 2B). The mutant aod-5 gene contained a mutation that generates a stop codon at residue 512 (TGG to TGA) of the protein (Figure 2A).
Further analysis of the predicted amino acid sequences revealed that each protein contained a predicted nuclear localization signal prior to the zinc-binding domain (Figure 2). Although zinc-cluster proteins typically contain a coiled-coil region following the zinc-cluster domain to facilitate formation of homo- or heterodimers (Macpherson et al. 2006), we could not detect strong evidence for a coiled coil in AOD2 or AOD5, although it has been noted that these regions are often difficult to identify in this class of proteins (Schjerling and Holmberg 1996). Comparisons to the Pfam database (Finn et al. 2006) revealed the presence of a possible PAS domain in the C-terminal region of both proteins (Figure 2). These domains are known to act as sensors of an array of biological signals, including changes in light, oxygen concentration, redox potential, and cellular energy (Taylor and Zhulin 1999; Hefti et al. 2004). The domains are also often involved in the dimerization of proteins.
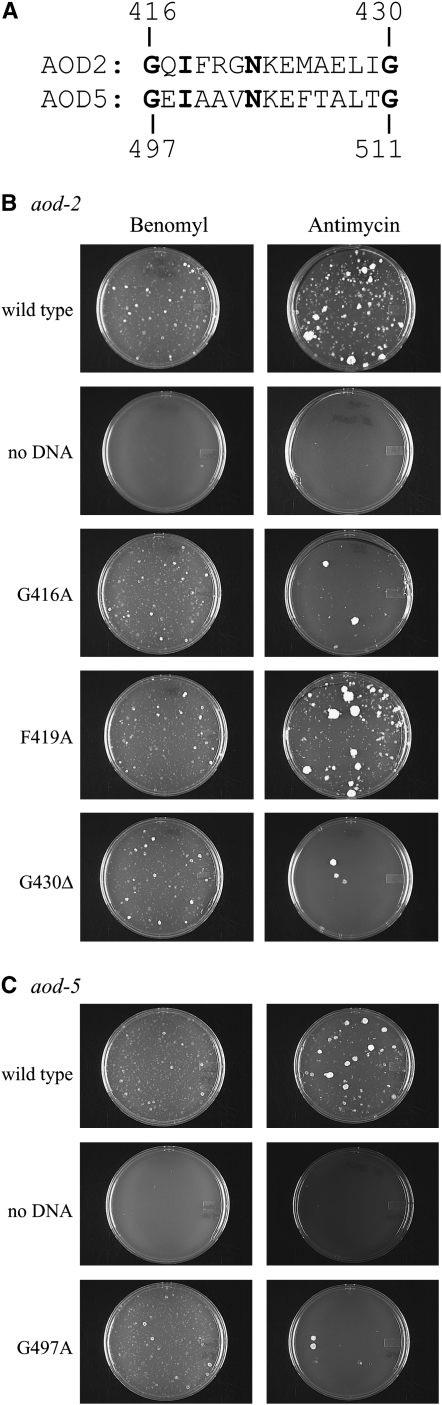
We wanted to examine the importance of residues in the possible PAS domains. These domains average ∼100–130 amino acid residues in length (Taylor and Zhulin 1999; Gilles-Gonzalez and Gonzalez 2004; Hefti et al. 2004) and are thought to give rise to a characteristic conserved structure despite poor conservation of the primary sequence of the motif. However, many PAS domains do contain short regions in which a few residues are relatively well conserved. For example, the sequence G X1 I X3 N X7 G appears in many PAS domains (Taylor and Zhulin 1999; Hefti et al. 2004) and this motif is completely conserved in both AOD2 and AOD5 (Figure 3A). For aod-2, mutations affecting each of the two codons for the Gly residues in this motif were constructed: G416A and G430Δ. The mutant alleles were transformed into CNA33 and examined for their ability to restore growth on plates containing antimycin A. Both mutations greatly reduced the ability of the aod-2 gene to enable growth of CNA33 transformants on medium containing antimycin A while a mutation of a nonconserved residue in the same region (F419A) had little effect on the ability to rescue (Figure 3B). For aod-5, we determined the effect of changing the first Gly residue of the motif, G497A. This mutation drastically reduced the ability of the cloned gene to rescue the aod-5 mutant (Figure 3C). These results support the prediction that the C-terminal region of both proteins contains a functional PAS domain and that the conserved Gly residues are important for its function. For both aod-2 and aod-5, mutations that severely reduced the transformation efficiency still produced occasional colonies on the antimycin plates. The reasons for this are not known but could be due to rare localized homologous recombination events that restore a functional copy of the gene. Alternatively, ectopic integrants at certain loci might be highly expressed. It is conceivable that mutant proteins present at high levels might provide enough function for rescue.
Figure 3.—
Effect of mutations in the potential PAS domain of aod-2 and aod-5. (A) Sequence of a region in AOD2 and AOD5 that contains four conserved residues (shown in boldface type) in a region of the possible PAS domain. Numbers indicate the position of the residues in each protein. (B) Plasmids containing wild-type (pAOX2Ben-4) or mutated versions of the aod-2 gene were used to transform the aod-2 mutant strain CNA33. A portion of the transformation mixture was plated on benomyl-containing medium as a control for transformation efficiency. Another portion was plated on medium containing antimycin A to determine if the mutated versions could rescue the inability of the aod-2 mutant strain to grow in the presence of the drug. (C) As in B, except plasmids containing either a wild-type or a G497A mutant version of the aod-5 gene were used to transform strain PL40-23.
Homologs of AOD2 and AOD5:
BLAST searches revealed homologs of the AOD2 protein in several fungal species whose genomes have been sequenced such as Chaetomium globosum, Giberella zeae, Magnaporthe grisea, and others. The Saccharomyces cerevisiae protein most closely related to AOD2 was Rds2p, also known as Ypl133cp (supplemental Figure 1 at http://www.genetics.org/supplemental/). Rds2p is best known as a regulator of drug sensitivity, but deletions of the gene also prevent growth on nonfermentable carbon sources (Akache et al. 2001; Akache and Turcotte 2002), suggesting a role in the yeast respiratory pathway and a possible functional relationship to AOD2. Homologs for AOD5 were also identified in other fungi. The closest S. cerevisiae homolog was Ybr239cp (supplemental Figure 2), a protein of unknown function. It should also be noted that AOD2 and AOD5 are homologous to each other (supplemental Figure 3).
Binding to the AIM sequence:
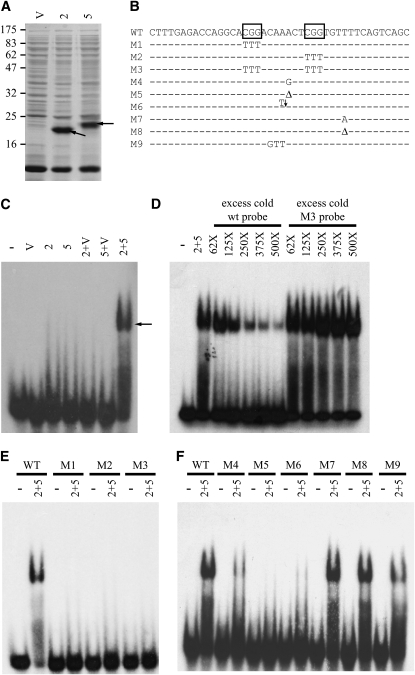
We recently described an AIM in the aod-1 promoter region that contained repeated CGG triplets in an arrangement characteristic of those bound by zinc-cluster transcription factors (Chae et al. 2007). Thus, the AOD2 and AOD5 proteins were logical choices for binding to this site of the aod-1 promoter. We created plasmid constructs expressing the N-terminal 117 residues of aod-2 and 142 residues of aod-5. The protein fragments produced from these constructs contain the zinc-cluster DNA-binding domains of each protein. Following expression in bacterial cells, the AOD2 and AOD5 fragments were abundant in the bacterial cell lysate supernatant fractions (Figure 4A). Aliquots of these fractions were used directly in EMSAs with a 32P-labeled probe containing the wild-type AIM sequence (Figure 4B). Neither the AOD2 nor the AOD5 N-terminal fragments bound efficiently to the wild-type probe, although trailing in the lanes containing either of these protein fragments suggested possible inefficient binding (Figure 4C). However, when both fragments were added to the same binding reaction, an obvious mobility shift of the wild-type probe was observed (Figure 4C). This binding is effectively competed by unlabeled wild-type probe, but unlabeled probe lacking both of the CGG repeats, which are predicted to act as the binding sites, did not compete (Figure 4D). Similarly, labeled probes in which either or both of the CGGs have been replaced with the sequence TTT do not bind the AOD2 and AOD5 protein fragments (Figure 4E). We also examined the importance of the 7-bp sequence that separates the two CGGs (Figure 4F). When substitutions of either 1 or 3 bp were made in the region between the CGG repeats, binding was decreased but not eliminated. On the other hand, deletion or insertion of a single base pair resulted in loss of binding. Alterations outside the CGG repeats did not affect binding.
Figure 4.—
The DNA-binding domains of both AOD2 and AOD5 are required for binding the AIM sequence. (A) Coomassie-blue-stained gel showing bacterial lysate supernatants prepared from cells expressing the N-terminal fragments of AOD2 (2) or AOD5 (5). The relevant bands are indicated by arrows. Lysate prepared from cells carrying the empty cloning vector (V) is shown as a control. Each lane contains 40 μg of cell lysate supernatant protein. (B) Probes prepared for EMSA binding reactions containing the wild-type (WT) AIM sequence and mutant derivatives (M1–M9). Boxes designate the CGG repeats thought to represent the binding sites for transcriptional activators (Chae et al. 2007). Bases indicated below the wild-type sequence show the replacements made in the individual mutant probes. Δ, a deletion of a single base. The arrow and T for M6 indicate addition of a single T into the sequence at that site. (C) Aliquots of the lysates in A were incubated in binding reactions with 32P-labeled probe containing the wild-type AIM sequence. The minus sign indicates a lane containing only the free probe and the specific lysate supernatant(s) used in each reaction is indicated above the other lanes. The binding reactions were electrophoresed on polyacrylamide gels that were then dried and exposed to X-ray film. The arrow indicates probe bound by the AOD2 plus AOD5 N-terminal fragments. (D) Binding reactions containing 32P-labeled wild-type probe were incubated with aliquots of the lysates containing the AOD2 plus AOD5 fragments as in the last lane in C. As indicated, aliquots of either unlabeled (cold) wild-type probe or unlabeled mutant M3 probe were included in binding reactions at increasing concentrations. The reactions were processed as for C. (E) Labeled probes of either the wild-type (WT) sequence or containing specific mutations (M1–M3) in the AIM sequence were generated to determine the specificity of binding at the site. Binding reactions contained the AOD2 plus AOD5 lysates and were processed as in C. (F) As in E, except probes were either wild type or mutants M4–M9.
Transcript levels:
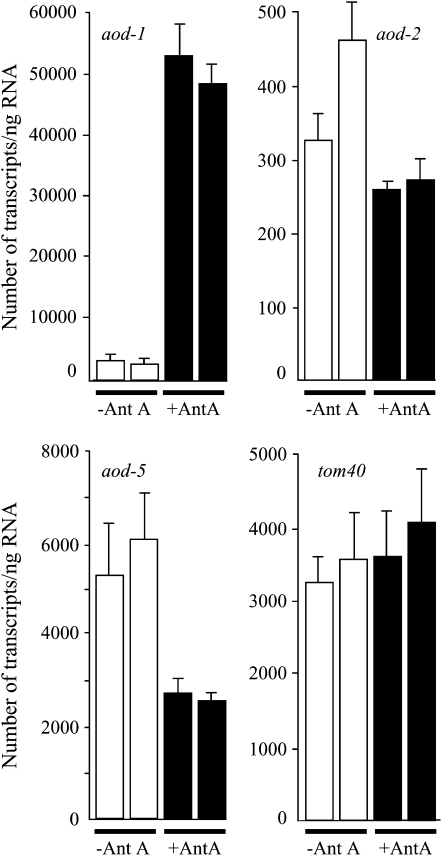
We wanted to determine if the transcript levels of aod-2 and/or aod-5 were more abundant in cells grown under conditions that induce aod-1 transcription. RNA was isolated from a wild-type strain grown in either the absence or the presence of antimycin A and qPCR was used to examine the abundance of the aod-1, aod-2, and aod-5 transcript levels under each condition. The levels of tom40 transcripts were examined as an endogenous control. Tom40 is the major component of the outer membrane translocase that initiates the process of importing proteins into mitochondria (Neupert and Herrmann 2007). The level of aod-1 transcript found under normal growth conditions was increased by ∼17-fold in cells grown in the presence of antimycin A whereas the levels of the tom40 transcript remained virtually unchanged under the two conditions (Figure 5). Transcripts of both aod-2 and aod-5 appeared to be slightly reduced in cells grown in the presence of antimycin A compared to noninduced cells. The number of transcripts predicted for aod-2 is extremely low compared to the other transcripts examined. However, we cannot formally rule out the possibility that the apparent low levels of aod-2 transcripts are due to inefficient reverse transcription of the aod-2 message.
Figure 5.—
qPCR. Cultures of the wild-type strain NCN233 were grown either for 12 hr under normal conditions or for 22 hr in the presence of antimycin A (0.5 μg/ml). Two cultures were grown under each condition to provide biological replicates. Poly(A)–RNA was isolated and converted to cDNA, and qPCR was performed. For each gene examined, the value for each biological replicate grown without (open bars) or with (solid bars) antimycin A is given. Each of the four bars in one graph represents the mean of four to six determinations. Error bars show 1 SD.
DISCUSSION
The aod-2 gene was originally identified as necessary for aod-1 expression >20 years ago, but the gene product has remained unidentified. Here we have shown that aod-2 encodes a transcription factor of the zinc-cluster family. The cloned gene rescues the phenotype of aod-2 cells with respect to their ability to grow in the presence of antimycin A. Similarly, we have identified the more recently described aod-5 gene as a transcription factor of the same family. Conceivably, our gene rescue approach could have identified second-site suppressors of the alternative oxidase-deficient phenotype. However, the fact that the mutations that defined each gene were found within the predicted coding sequences effectively argues against this possibilty.
The results of our EMSAs show that AOD2 and AOD5 act synergistically to bind the AIM in a sequence-specific fashion. Each of the CGG repeats in the AIM, and proper spacing between them, is required for binding. This agrees well with our previous observations that the CGG repeats are required for expression of alternative oxidase (Chae et al. 2007). We have not identified a dimerization domain in the truncated proteins used in the EMSA experiments, but the observation that the closest yeast homologs of these proteins, Rds2p and Ybr239cp, interact in a yeast two-hybrid assay (Ito et al. 2001) supports the notion that AOD2 and AOD5 interact with each other. It is also possible that binding of two independent proteins at individual sites of the AIM sequence results in a synergistic increase in binding efficiency. In N. crassa, there is evidence for both a low level of constitutive transcription of aod-1 and an induced high level of transcription of the gene when electron transport is inhibited (Tanton et al. 2003). The observation that neither aod-2 nor aod-5 mutants produce aod-1 mRNA under inducing conditions (Descheneau et al. 2005) and the fact that the AOD2 and AOD5 proteins cooperate to bind the AIM sequence, which is required for expression of alternative oxidase (Chae et al. 2007), strongly suggest that these proteins are directly required for inducing the aod-1 gene.
Although it is possible that post-transcriptional control mechanisms could prevent translation of the aod-2 and aod-5 mRNAs, the existence of transcripts from both genes in cultures grown under noninducing conditions suggests that the proteins preexist and can rapidly respond to changing conditions. There are different possibilities for the mechanism of transduction of the activating signal from mitochondria to the aod-1 promoter. For example, AOD2 and/or AOD5 may be directly affected by changes that occur in the cell as a result of inefficient oxidative phosphorylation. Alternatively, other factors might communicate the signal to AOD2/AOD5 as part of a larger signal transduction cascade. The finding that other genes are involved in controlling alternative oxidase expression (Descheneau et al. 2005) argues for a more complex pathway.
The possible existence of PAS domains in the AOD2 and AOD5 proteins suggests a mechanism whereby either or both proteins could respond to different conditions in the cell that are relevant to alternative oxidase induction. PAS domains are known to bind cofactors such as FAD, which enables the detection of redox levels (Taylor and Zhulin 1999). However, the putative PAS domains in AOD2 and AOD5 do not appear to be closely related to those known to bind FMN or FAD (Huala et al. 1997; Crosson and Moffat 2001). PAS domains are also involved in homo- and heterodimerization of proteins. Since the protein fragments used in our EMSAs did not contain the putative PAS domains, they are obviously not essential for heterodimerization of the proteins in these assays. However, formation of dimers via PAS domains between AOD2 and/or AOD5, or even additional proteins, could also affect activity and/or localization of the proteins and respond to different conditions in the cell.
Another mechanism for controlling the activity or localization of transcription factors in response to redox conditions in the cell is via changes in the oxidation state of Cys residues (Buchanan and Balmer 2005; Hansen et al. 2006). For example, the Yap1 transcription factor of S. cerevisiae regulates the expression of several antioxidant defense genes when reactive oxygen species are encountered. The active form of the protein occurs when a nuclear export signal is masked due to the intramolecular oxidation of Cys residues in the C terminus of the protein. When the disulfide bonds are reduced, the export signal is exposed and the protein can be exported to the cytoplasm (Wood et al. 2004). Both AOD2 and AOD5 contain five Cys residues in addition to those associated with the zinc-binding region of the protein. It is conceivable that the redox state of these residues could affect the function or localization of one or both of these proteins. Future work will be aimed at identifying other components required for aod-1 expression and determining the mechanism by which AOD2 and AOD5 are activated to induce transcription of aod-1.
Acknowledgments
We are grateful to Gary Eitzen for advice on protein expression systems and to Troy Locke for advice on qPCR. A.T.T. was supported by graduate scholarships from the National Sciences and Engineering Research Council (NSERC) and the Alberta Heritage Foundation for Medical Research (AHFMR). C.C.L. was supported by summer studentships from NSERC and AHFMR. This work was supported by a Discovery Grant from NSERC to F.E.N.
References
- Akache, B., and B. Turcotte, 2002. New regulators of drug sensitivity in the family of yeast zinc cluster proteins. J. Biol. Chem. 277: 21254–21260. [DOI] [PubMed] [Google Scholar]
- Akache, B., K. Wu and B. Turcotte, 2001. Phenotypic analysis of genes encoding yeast zinc cluster proteins. Nucleic Acids Res. 29: 2181–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnholdt-Schmitt, B., J. H. Costa and D. F. de Melo, 2006. AOX: A functional marker for efficient cell reprogramming under stress? Trends Plant Sci. 11: 281–287. [DOI] [PubMed] [Google Scholar]
- Bertrand, H., A. Argan and N. A. Szakacs, 1983. Genetic control of the biogenesis of cyanide insensitive respiration in Neurospora crassa, pp. 495–507 in Mitochondria, edited by R. J. Schweyen, K. Wolf and F. Kaudewitz. Walter de Gruyter, Berlin.
- Borghouts, C., A. Werner, T. E. Elthon and H. D. Osiewacz, 2001. Copper-modulated gene expression and senescence in the filamentous fungus Podospora anserina. Mol. Cell. Biol. 21: 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich, K. A., L. A. Alex, O. Yarden, M. Freitag, G. E. Turner et al., 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68: 1–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan, B. B., and Y. Balmer, 2005. Redox regulation: a broadening horizon. Annu. Rev. Plant Biol. 56: 187–220. [DOI] [PubMed] [Google Scholar]
- Chae, M. S., C. C. Lin, K. E. Kessler, C. E. Nargang, L. L. Tanton et al., 2007. Identification of an alternative oxidase induction motif in the promoter region of the aod-1 gene in Neurospora crassa. Genetics 175: 1597–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri, M., R. Sharan and G. C. Hill, 2002. Trypanosome alternative oxidase is regulated post-transcriptionally at the level of RNA stability. J. Eukaryot. Microbiol. 49: 263–269. [DOI] [PubMed] [Google Scholar]
- Chaudhuri, M., R. D. Ott and G. C. Hill, 2006. Trypanosome alternative oxidase: from molecule to function. Trends Parasitol. 22: 484–491. [DOI] [PubMed] [Google Scholar]
- Clifton, R., A. H. Millar and J. Whelan, 2006. Alternative oxidases in Arabidopsis: a comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim. Biophys. Acta 1757: 730–741. [DOI] [PubMed] [Google Scholar]
- Considine, M. J., R. C. Holtzapffel, D. A. Day, J. Whelan and A. H. Millar, 2002. Molecular distinction between alternative oxidase from monocots and dicots. Plant Physiol. 129: 949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosson, S., and K. Moffat, 2001. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. USA 98: 2995–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descheneau, A. T., I. A. Cleary and F. E. Nargang, 2005. Genetic evidence for a regulatory pathway controlling alternative oxidase production in Neurospora crassa. Genetics 169: 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dojcinovic, D., J. Krosting, A. J. Harris, D. J. Wagner and D. M. Rhoads, 2005. Identification of a region of the Arabidopsis AtAOX1a promoter necessary for mitochondrial retrograde regulation of expression. Plant Mol. Biol. 58: 159–175. [DOI] [PubMed] [Google Scholar]
- Drygas, M. E., A. M. Lambowitz and F. E. Nargang, 1989. Cloning and analysis of the Neurospora crassa gene for cytochrome c heme lyase. J. Biol. Chem. 264: 17897–17907. [PubMed] [Google Scholar]
- Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich et al., 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34: D247–D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read et al., 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422: 859–868. [DOI] [PubMed] [Google Scholar]
- Gilles-Gonzalez, M.-A., and G. Gonzalez, 2004. Signal transduction by heme-containing PAS-domain proteins. J. Appl. Physiol. 96: 774–783. [DOI] [PubMed] [Google Scholar]
- Hansen, J. M., Y.-M. Go and D. P. Jones, 2006. Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annu. Rev. Pharmacol. Toxicol. 46: 215–234. [DOI] [PubMed] [Google Scholar]
- Hansen, L. D., J. N. Church, S. Matheson, V. W. McCarlie, T. Thygerson et al., 2002. Kinetics of plant growth and metabolism. Thermochim. Acta 388: 415–425. [Google Scholar]
- Hefti, M. H., K.-J. Francoijs, S. C. de Vries, R. Dixon and J. Vervoort, 2004. The PAS fold. A redefinition of the PAS domain based upon structural prediction. Eur. J. Biochem. 271: 1198–1208. [DOI] [PubMed] [Google Scholar]
- Helfert, S., A. M. Estevez, B. Bakker, P. Michels and C. Clayton, 2001. Roles of triosephosphate isomerase and aerobic metabolism in Trypanosoma brucei. Biochem. J. 357: 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, L. H. M., E. Giraud, R. Lister, D. Thirkettle-Watts, J. Low et al., 2007. Characterization of the regulatory and expression context of an alternative oxidase gene provides insights into cyanide-insensitive respiration during growth and development. Plant Physiol. 143: 1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huala, E., P. W. Oeller, E. Liscum, I.-S. Han, E. Larsen et al., 1997. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science 278: 2120–2123. [DOI] [PubMed] [Google Scholar]
- Huh, W., and S. Kang, 1999. Molecular cloning and functional expression of alternative oxidase in Candida albicans. J. Bacteriol. 181: 4098–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W., and S. Kang, 2001. Characterization of the gene family encoding alternative oxidase from Candida albicans. Biochem. J. 356: 595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori et al., 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98: 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph-Horne, T., D. W. Holloman and P. M. Wood, 2001. Fungal respiration: a fusion of standard and alternative components. Biochim. Biophys. Acta 1504: 179–195. [DOI] [PubMed] [Google Scholar]
- Kelkar, H. S., J. Griffith, M. E. Case, S. F. Covert, R. D. Hall et al., 2001. The Neurospora crassa genome: cosmid libraries sorted by chromosome. Genetics 157: 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz, A. M., and D. Zannoni, 1978. Cyanide-insensitive respiration in Neurospora: genetic and biophysical approaches, pp. 283–291 in Plant Mitochondria, edited by G. Ducet and C. Lance. Elsevier/North-Holland Biomedical Press, Amsterdam.
- Lambowitz, A. M., J. R. Sabourin, H. Bertrand, R. Nickels and L. McIntosh, 1989. Immunological identification of the alternative oxidase of Neurospora crassa mitochondria. Mol. Cell. Biol. 9: 1362–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson, S., M. Larochelle and B. Turcotte, 2006. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 70: 583–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell, D. P., Y. Wang and L. McIntosh, 1999. The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. USA 96: 8271–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, A. E., and G. C. Vanlerberghe, 2004. Branched mitochondrial electron transport in the animalia: presence of alternative oxidase in several animal phyla. IUBMB Life 56: 333–341. [DOI] [PubMed] [Google Scholar]
- McDonald, A. E., and G. C. Vanlerberghe, 2005. Alternative oxidase and plastoquinol terminal oxidase in marine prokaryotes of the Sargasso Sea. Gene 349: 15–24. [DOI] [PubMed] [Google Scholar]
- Moore, A. L., M. S. Albury, P. G. Crichton and C. Affourtit, 2002. Function of the alternative oxidase: Is it still a scavenger? Trends Plant Sci. 7: 478–481. [DOI] [PubMed] [Google Scholar]
- Neupert, W., and J. M. Herrmann, 2007. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76: 723–749. [DOI] [PubMed] [Google Scholar]
- Perkins, D. D., A. Radford and M. S. Sachs, 2001. The Neurospora Compendium: Chromosomal Loci. Academic Press, San Diego.
- Rhoads, D. M., and C. C. Subbaiah, 2007. Mitochondrial retrograde regulation in plants. Mitochondrion 7: 177–194. [DOI] [PubMed] [Google Scholar]
- Rhoads, D. M., A. L. Umbach, C. C. Subbaiah and J. N. Siedow, 2006. Mitochondrial reactive oxygen species: contribution to oxidative stress and interorganellar signaling. Plant Physiol. 141: 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson, C. A., and G. C. Vanlerberghe, 2002. Transgenic plant cells lacking mitochondrial alternative oxidase have increased susceptibility to mitochondria-dependent and -independent pathways of programmed cell death. Plant Physiol. 129: 1908–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjerling, P., and S. Holmberg, 1996. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 24: 4599–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieger, S. M., B. K. Kristensen, C. A. Robson, S. Amirsadeghi, E. W. Y. Eng et al., 2005. The role of alternative oxidase in modulating carbon use efficiency and growth during macronutrient stress in tobacco cells. J. Exp. Bot. 56: 1499–1515. [DOI] [PubMed] [Google Scholar]
- Stenmark, P., and P. Nordlund, 2003. A prokaryotic alternative oxidase present in the bacterium Novosphingobium aromaticivorans. FEBS Lett. 552: 189–192. [DOI] [PubMed] [Google Scholar]
- Tanton, L. L., C. E. Nargang, K. E. Kessler, Q. Li and F. E. Nargang, 2003. Alternative oxidase expression in Neurospora crassa. Fungal Genet. Biol. 39: 176–190. [DOI] [PubMed] [Google Scholar]
- Taylor, B. L., and I. B. Zhulin, 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63: 479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirkettle-Watts, D., T. C. McCabe, R. Clifton, C. Moore, P. M. Finnegan et al., 2003. Analysis of the alternative oxidase promoters from soybean. Plant Physiol. 133: 1158–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach, A. L., F. Fiorani and J. N. Siedow, 2005. Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol. 139: 1806–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe, G. C., and L. McIntosh, 1997. Alternative oxidase: from gene to function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 703–734. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe, G. C., C. A. Robson and J. Y. Yip, 2002. Induction of mitochondrial alternative oxidase in response to a cell signal pathway down-regulating the cytochrome pathway prevents programmed cell death. Plant Physiol. 129: 1829–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga, A., J. D. Arrabaca, F. Sansonetty, P. Ludovico, M. Corte-Real et al., 2003. Energy conversion coupled to cyanide-resistant respiration in the yeasts Pichia membranifaciens and Debaryomyces hansenii. FEMS Yeast Res. 3: 141–148. [DOI] [PubMed] [Google Scholar]
- Wood, M. J., G. Storz and N. Tjandra, 2004. Structural basis for redox regulation of Yap1 transcription factor localization. Nature 430: 917–921. [DOI] [PubMed] [Google Scholar]
- Yu, J., R. Nickels and L. McIntosh, 2001. A genome approach to mitochondrial-nuclear communication in Arabidopsis. Plant Physiol. Biochem. 39: 345–353. [Google Scholar]
- Yukioka, H., S. Inagaki, R. Tanaka, K. Katoh, N. Miki et al., 1998. Transcriptional activation of the alternative oxidase gene of the fungus Magnaporthe grisea by a respiratory-inhibiting fungicide and hydrogen peroxide. Biochim. Biophys. Acta 1442: 161–169. [DOI] [PubMed] [Google Scholar]
- Zarkovic, J., S. L. Anderson and D. M. Rhoads, 2005. A reporter gene system used to study developmental expression of alternative oxidase and isolate mitochondrial retrograde regulation mutants in Arabidopsis. Plant Mol. Biol. 57: 871–888. [DOI] [PubMed] [Google Scholar]