Abstract
Hybridization between plant species can induce speciation as well as phenotypic novelty and heterosis. Hybrids also can show genome rearrangements and gene expression changes compared with their parents. Here we determined the allelic variation in gene expression in Populus trichocarpa × Populus deltoides F1 hybrids. Among 30 genes analyzed in four independently formed hybrids, 17 showed >1.5-fold expression biases for one of the two alleles, and there was monoallelic expression of one gene. Expression ratios of the alleles differed between leaves and stems for 10 genes. The results suggest differential regulation of the two parental alleles in the hybrids. To determine if the allelic expression biases were caused by hybridization we compared the ratios of species-specific transcripts between an F1 hybrid and its parents. Thirteen of 19 genes showed allelic expression ratios in the hybrid that were significantly different from the ratios of the parental species. The P. deltoides allele of one gene was silenced in the hybrid. Modes of gene regulation were inferred from the hybrid–parent comparisons. Cis-regulation was inferred for 6 genes, trans-regulation for 1 gene, and combined cis- and trans-regulation for 9 genes. The results from this study indicate that hybridization between plant species can have extensive effects on allelic expression patterns, some of which might lead to phenotypic changes.
PLANT hybridization is a common process in nature and it plays a vital role in plant breeding. Hybridization can generate phenotypic novelty including a broad array of new and sometimes transgressive phenotypes (Rieseberg et al. 1999, 2003b). Hybridization also can lead to speciation, adaptive evolution, and ecological innovations (Rieseberg 1997; Rieseberg et al. 2003a; Arnold 2004; Hegarty and Hiscock 2005). Interspecific crosses in plants often generate hybrids that exhibit heterosis compared to their parents. Considerable changes have been observed in the genomes of some hybrids. Interspecific hybridization may lead to chromosomal rearrangements (Rieseberg et al. 1996; Shaked et al. 2001), transposable element mobilization (Liu and Wendel 2000; Shan et al. 2005), and DNA methylation changes (Salmon et al. 2005). Interspecific hybridization provides a vast reservoir of new alleles for gene evolution. Allelic variation resulting from interspecific hybridization can potentially contribute to phenotypic variation. For example, the complementation and interaction of different alleles in hybrids are hypothesized to be a component of the genetic basis for heterotic phenotypes (Birchler et al. 2003, 2006; Guo et al. 2004; Song et al. 2004; Springer and Stupar 2007a).
Hybridization between two plant species can result in changes in gene expression (reviewed in Adams 2007). Up- and downregulation of expression in hybrids compared to their parents has been shown in interspecific triploid Senecio hybrids (Hegarty et al. 2005, 2006). Intraspecific hybridization between two cultivars, ecotypes, or accessions also can result in up- or downregulation of gene expression, as shown in recent studies of diploid and triploid maize hybrids (Auger et al. 2005; Guo et al. 2006; Stupar and Springer 2006; Swanson-Wagner et al. 2006; Meyer et al. 2007; Springer and Stupar 2007b; Stupar et al. 2007; Uzarowska et al. 2007) in diploid wheat and rice hybrids (Wu et al. 2003; Bao et al. 2005; Wang et al. 2006, and in hybrids between ecotypes of Arabidopsis thaliana (Vuylsteke et al. 2005). Hybridization also can affect expression of individual alleles, although few studies have assayed allelic expression variation in diploid hybrids. Allelic expression differences of nonimprinted autosomal genes have been reported in interspecific hybrids of Drosophila (Wittkopp et al. 2004), as well as intraspecific F1 hybrids of mice (Cowles et al. 2002) and Saccharomyces cerevisiae (Ronald et al. 2005). Recent studies of intraspecific maize hybrids have shown unequal expression of parental alleles, including silencing of one allele (Guo et al. 2003, 2004; Stupar and Springer 2006; Springer and Stupar 2007b; Stupar et al. 2007). A study of an Adh gene in interspecific cotton F1 hybrids revealed organ-specific allelic silencing of this gene (Adams and Wendel 2005). Despite recent progress allelic variation in gene expression remains poorly investigated for interspecific plant hybrids.
Allelic variation in gene expression may arise from cis- or trans-regulatory factors (Wittkopp et al. 2004). Cis-regulators are genetically tightly linked to a gene and influence transcription in an allele-specific manner. In contrast, trans-regulators are located elsewhere in the genome and modify gene expression by interacting with cis-regulators. Following hybridization, genes under pure cis-regulation tend to show additive expression patterns, whereas those under trans-regulation can display either additive or nonadditive expression, depending on whether a dosage effect exists (Stupar and Springer 2006; Springer and Stupar 2007a). Cis- or trans-regulation can be inferred by comparing the ratios of species-specific transcripts between the F1 hybrids and the parental species (Wittkopp et al. 2004). Genes with strict cis-regulation have the same bias of expression of two alleles in both the hybrid and the parents. Genes with strict trans-regulation display allelic bias in the parents but are expected to have equal levels of allelic expression in the hybrid. While pure cis-effects imply the preservation of parental regulatory function, differential expression between parents and hybrid due to trans-effects are caused by hybridization that brings two genomes together, allowing both alleles to be exposed to a common set of trans-elements.
Populus hybrids provide a promising plant system to study interspecific hybridization and its genetic and molecular consequences. There are 30–40 different Populus species worldwide, including the common North American species Populus trichocarpa (black cottonwood), P. deltoides (eastern cottonwood), P. nigra (Lombardy poplar), and P. tremuloides (aspen). Populus has become a model system for research on wood-forming plants. P. trichocarpa is the first (and currently only) tree for which the genome has been sequenced (Tuskan et al. 2006) and it is one of only four flowering plant species with a sequenced genome at the present time (the others being A. thaliana, Oryza sativa, and Vitis vinifera). As a sustainable source for paper fiber and biofuel, Populus hybrids are important economic plants. Populus hybrids often show strong heterosis (Bradshaw and Stettler 1995; Li et al. 1998), and the study of molecular responses to hybridization may provide insights into heterosis of Populus hybrids.
Here we studied the allelic variation in gene expression levels using P. trichocarpa × P. deltoides interspecific F1 hybrids. The allele-specific expression for 30 genes in four independently formed hybrids was assayed. To investigate whether there are organ-specific differences in allelic expression, both leaves and stems were examined for the same genes. To determine if biased allelic expression was the result of hybridization or reflects differing expression levels in the parents, we compared the ratio of species-specific transcripts in an F1 hybrid vs. that in its parents, and the results have implications for cis- and trans-gene regulation.
MATERIALS AND METHODS
Plant materials:
To survey allelic expression levels for 30 different genes, plant tissues were collected from four P. trichocarpa × P. deltoides F1 hybrids (11-BULH-4-1, 9-KTWD-4, 7-IRVC-3-1, and 7-IRVD-5-1), described in (Gilchrist et al. 2006). These cottonwood hybrids originally came from central British Columbia and were planted at the University of British Columbia (UBC) Botanical Garden. Young leaves and stems from all four hybrids were collected in October 2005 and June 2006. For each tissue sample, two replicates were collected at the same time. All harvested tissue samples were frozen immediately in liquid nitrogen and stored at −80° until use. For the analysis of 30 genes in four hybrids, the genes 4CL3, CHI, LFY, MP, and NPR1 were assayed using the plant material collected in October 2005, and the other genes were assayed using the June 2006 material.
The analysis of cis- and trans-regulatory variation was conducted using a P. trichocarpa × P. deltoides F1 hybrid and its parental clones, P. trichocarpa accession Nisqually-1 and P. deltoides accession ILL 101. The hybrid was originally from a plantation of P. trichocarpa × P. deltoides F1 hybrids that were derived from the same cross by Dan Carson from Scott Paper in Harrison Mills, British Columbia. The maternal parent P. trichocarpa was planted at the UBC Botanical Garden and the paternal parent P. deltoides was provided by Carl Douglas. Cuttings of all three genotypes were grown under common greenhouse conditions at the UBC Horticultural Greenhouse for several months before tissue sampling. Young leaves from the hybrid and the two parents were collected at the same time during May 2007. For each tissue sample, three replicates were harvested and frozen immediately in liquid nitrogen and stored at −80° until use.
Sequence database searches:
Gene sequences were obtained from NCBI and the whole-genome shotgun sequence database of the P. trichocarpa Nisqually-1 genome that was available from the Joint Genome Institute (http://genome.jgi-psf.org/poplar0/poplar0.home.html). Primers for PCR (supplemental Table 1 at http://www.genetics.org/supplemental/) were designed using Primer Premier 5.0 to amplify both genomic DNAs and cDNAs.
Extraction of nucleic acids and synthesis of cDNA:
DNAs were extracted by using QIAGEN (Valencia, CA) DNeasy plant mini kit. Total RNA extraction was performed as described previously (Adams et al. 2003). DNA and RNA concentrations and purities were measured by using a NanoDrop spectrophotometer. RNAs were treated with DNaseI (New England Biolabs, Beverly, MA) before reverse transcription. Single-stranded cDNA was synthesized from 500 ng of total RNA using M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. As controls for DNA contamination, reactions were also performed without reverse transcriptase at the same time. For the cis- and trans-regulatory variation analysis, mixed cDNAs were synthesized from equal mixes of the two parent RNAs.
Genotyping:
Genes of interest were PCR amplified from genomic DNAs of the hybrid poplars. The PCR products were sequenced using Big Dye Terminator 3.1 sequencing chemistry (Applied Biosystems, Foster City, CA) by the Nucleic Acids Protein Service (NAPS) unit at the University of British Columbia. Single nucleotide polymorphisms (SNPs) in exon regions were identified and selected for allele-specific expression analysis. Common SNPs were selected for the four hybrid poplars from central British Columbia. The same SNPs were selected for the cis- and trans-regulatory variation analysis if they also existed in the hybrid synthesized by Scott Paper; otherwise other SNPs were selected for this hybrid.
Single-base primer extension assay:
DNA and cDNA segments surrounding the SNPs present in the hybrid poplars were PCR amplified. cDNAs from equally mixed parental RNAs were also PCR amplified. Following PCR thermal cycling, unincorporated primers and dNTPs were removed by adding 1.67 units of shrimp alkaline phosphatase (SAP) (Fermentas, Burlington, Ontario) and 1 unit of Exonuclease I (Fermentas) to each 5-μl PCR product. Reactions were mixed briefly and incubated at 37° for 60 min then 80° for 15 min. The PCR products were then subjected to a primer extension assay (SNaPshot, Applied Biosystems) using extension primers designed to anneal to the amplified DNA adjacent to the SNP site (supplemental Table 2 at http://www.genetics.org/supplemental/). Primer extension reactions were carried out in a total volume of 10 μl containing 0.5 μl ABI Prism SNaPshot multiplex kit mix (Applied Biosystems), 0.2 μm extension primer, 2 μl of PCR product, and 6.5 μl of deionized water. Thermal cycling conditions for extension reactions were carried out with the following program: 2 min at 94°, and 25 cycles consisting of 10 sec at 96°, 5 sec at 50°, and 30 sec at 60°. After cycling, the unincorporated fluorescent ddNTPs (dideoxynucleotide triphosphates) were removed by adding 1 unit of SAP and incubating for 60 min at 37°, followed by 15 min at 65° for enzyme inactivation. The resulting primer extension products were analyzed on an ABI 3730 capillary electrophoresis DNA instrument, using GeneMapper 3.7 software (Applied Biosystems) according to the manufacturer's protocol. The expression percentages of the two alleles were measured by comparing the peak heights. Since differing fluorophores may influence the incorporation and migration rates of four types of ddNTPs, the peak heights are not always identical between two alleles of equal abundance (Pinsonneault et al. 2004). Therefore, allelic ratios of genomic DNAs assumed to be present in equal amounts (ratio = 1) were used to normalize allelic ratios of cDNA samples (Pinsonneault et al. 2004; Wang et al. 2005). Genes showing expression of only one allele were further examined with direct sequencing of the RT–PCR products to confirm the monoallelic expression patterns.
Statistical analyses:
Standard errors of replicates were calculated. Two-tailed homoscedastic variance t-tests (P = 0.05) were performed with Microsoft Excel to test whether the expression differed between leaves and stems. Two-tailed homoscedastic variance t-tests (P = 0.05) were also used to assess the difference between the allelic expression ratio in the F1 hybrid vs. that in mixed parental RNA compared with a 50:50 value.
RESULTS
Gene selection and identification of SNPs:
Genes assayed in this study, listed in Table 1, were selected on the basis of the following criteria:
Twenty-seven genes had sequence homology to genes important for plant growth and development, or other known functions. Selected genes covered various gene categories.
Four genes, MATE, HAT22, Unknown1, and Unknown2, located next to the already selected genes DXPS, KNAT1, ISP, and ADK, were chosen to compare allelic expression patterns between physically adjacent gene pairs.
Only genes that are single copy or that were easily distinguishable from other paralogs in sequence were selected.
The genes were expressed in at least one of the two organ types, leaves and stems.
Marker SNPs were present between the two alleles in at least two of the hybrids.
TABLE 1.
List of Populus genes surveyed for allelic expression
| Gene | Description | GenBank accession no. |
|---|---|---|
| 4CL3 | 4-Coumarate:CoA ligase 3 | AF283553 |
| ADH | Alcohol dehydrogenase | DT504809 |
| ADK | Adenylate kinase | DT496348 |
| C3HC4 | C3HC4-type RING finger | CN550424 |
| CHI | Chalcone flavanone isomerase | DT517112 |
| CaMBP | Calmodulin-binding proteins | CX177282 |
| Cel9B | Family 9 glycoside hydrolase | DT510114 |
| DXPS | Deoxyxylulose-5-phosphate synthase | AARH01010919 (6042–6581) |
| F5H | Ferulate 5-hydroxylase | CV252951 |
| GT47C | Glycosyltransferase GT47C | DQ899955 |
| HAT22 | Homeodomain-leucine zipper protein 22, adjacent to KNAT1 | AARH01001104 (48991–49477) |
| ISP | Signal peptidase I | DT520192 |
| KNAT1 | Knotted 1-like | DT509858 |
| LFY | Leafy | AARH01006471 (326067–326315) |
| MATE | Multi-antimicrobial extrusion protein, adjacent to DXPS | DT515805 |
| MP | Monopteros | AARH01001041 (107896–108549) |
| NAK | Serine/threonine kinase | AARH01010612 (3382–3763) |
| NBS-LRR | Nucleotide-binding site–leucine rich repeat | AARH01009055 (9726–10186) |
| NPR1 | Nonexpresser of PR genes | AARH01003035 (164386–164911) |
| P4H | Prolyl 4-hydroxylase | AARH01001302 (23930–24300) |
| PPO3 | Polyphenol oxidase | AY665682 |
| PPR | Pentatricopeptide repeat-containing protein | DN491158 |
| PREG1-like | PREG1-like negative regulator | CX174618 |
| RAR1 | RAR1 disease resistance gene | DT517811 |
| Rps19 | Mitochondrial ribosomal protein S19 | DT487761 |
| SKOR | Stelar K+ outward rectifying channel | AARH01005031 (50653–51295) |
| SPB | Squamosa promoter-binding protein like | CV243662 |
| SUS | Sucrose synthase | DT497251 |
| TI5 | Kunitz trypsin inhibitor 5 | AY378090 |
| Unknown1 | Adjacent to ISP | CV131225 |
| Unknown2a | Adjacent to ADK | CV230181 |
Gene surveyed for the cis- and trans-regulatory variation analysis but not for assays of the four hybrids from central British Columbia.
To identify SNPs in the selected genes in each of the four P. trichocarpa × P. deltoides hybrids, we PCR amplified and sequenced both alleles simultaneously. A total of 38 genes were sequenced from each of the hybrids and 31 genes with SNPs between the alleles were identified for expression analysis (Table 1).
Allele-specific gene expression analysis:
We studied the allelic expression variation for 30 genes in four Populus hybrids using a single base primer extension assay (Figure 1). This method has been shown to be effective in distinguishing between and quantifying sequence variants by a single SNP site (Cowles et al. 2002; Norton et al. 2002; Yan et al. 2002; Bray et al. 2003; Wang et al. 2005). Relative expression of the P. trichocarpa derived allele (Pt) for the four hybrids is shown in Table 2. We used 1.5-fold (that is a 60:40 ratio) as a minimum threshold ratio for allelic differential expression because it encompasses the standard error for all genes and represents a conservative estimation of unequal expression. To test the consistency of the expression data among replicates, eight leaf replicates (separate RNA extractions from different leaves) from one of the hybrids were tested for two genes, GT47C and TI5, and the resulting standard errors were 3 and 2%, respectively.
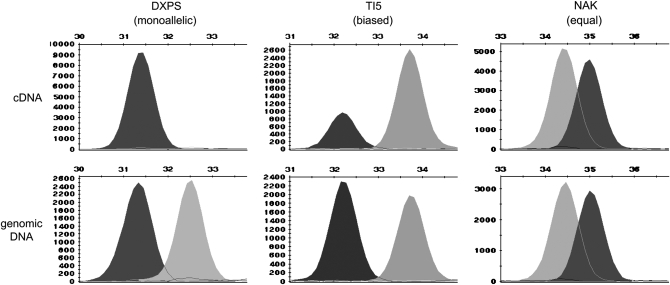
Figure 1.—
Example outputs from single base primer extension assays in the GeneMapper 3.0 software. The marker SNPs between two homologous alleles result in detectable fluorescent peaks. Relative expression levels of the species-specific alleles were read from the heights of the fluorescent peaks. Readings for genomic DNAs were used to normalize those of cDNAs.
TABLE 2.
Allelic expression ratios in leaves and stems of four Populus hybrids
| Leaf
|
Stem
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | HY 1 | SE (%) | HY 2 | SE (%) | HY 3 | SE (%) | HY 4 | SE (%) | HY 1 | SE (%) | HY 2 | SE (%) | HY 3 | SE (%) | HY 4 | SE (%) |
| Differential allelic expression of 60:40 or greater in the majority of the four hybrids in both leaves and stems | ||||||||||||||||
| 4CL3 | 40 | 2 | 40 | 0 | 39 | 2 | 30 | 2 | ||||||||
| Cel9Ba | 72 | 1 | 77 | 2 | 69 | 0 | 84 | 1 | 61 | 1 | 64 | 6 | 55 | 1 | 65 | 1 |
| DXPSa | 25 | 1 | 18 | 0 | 10 | 3 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NBS-LRR | 43 | 5 | 33 | 4 | 30 | 0 | 36 | 0 | 51 | 7 | 35 | 2 | 31 | 1 | 22 | 2 |
| PREG1-like | 16 | 1 | 17 | 0 | 30 | 6 | 26 | 1 | 28 | 2 | 26 | 1 | 30 | 4 | 27 | 1 |
| RPS19 | 79 | 0 | 70 | 1 | 74 | 1 | 66 | 0 | 75 | 1 | 67 | 1 | 73 | 2 | 80 | 1 |
| SPB | 66 | 6 | 66 | 2 | 65 | 4 | 63 | 5 | 64 | 0 | 63 | 6 | 62 | 3 | 69 | 0 |
| TI5a | 84 | 0 | 73 | 1 | 76 | 0 | 66 | 2 | 83 | 0 | 82 | 3 | 89 | 3 | 80 | 2 |
| Differential allelic expression of 60:40 or greater only in leaves of the majority of the four hybrids | ||||||||||||||||
| MATEa | 60 | 0 | 63 | 0 | 55 | 2 | 60 | 1 | 47 | 4 | 43 | 4 | 40 | 1 | 40 | 1 |
| SKORa | 37 | 2 | 32 | 0 | 30 | 5 | 28 | 1 | 51 | 6 | 40 | 1 | 48 | 4 | 43 | 6 |
| Differential allelic expression of 60:40 or greater only in stems of the majority of the four hybrids | ||||||||||||||||
| ADKa | 41 | 0 | 35 | 1 | 44 | 1 | 48 | 1 | 34 | 1 | 33 | 2 | 40 | 2 | 40 | 1 |
| CaMBPa | 51 | 3 | 46 | 2 | 41 | 4 | 51 | 4 | 43 | 1 | 40 | 3 | 39 | 0 | 29 | 3 |
| F5H | 74 | 6 | 66 | 2 | 61 | 3 | ||||||||||
| HAT22 | 75 | 1 | 80 | 4 | 70 | 1 | 79 | 6 | ||||||||
| ISPa | 41 | 4 | 43 | 0 | 41 | 2 | 39 | 3 | 27 | 3 | 33 | 1 | 23 | 3 | 26 | 3 |
| PPO3a | 37 | 0 | 44 | 2 | 49 | 1 | 59 | 3 | 23 | 3 | 26 | 3 | 36 | 4 | 27 | 1 |
| PPRa | 58 | 0 | 55 | 4 | 37 | 2 | 51 | 5 | 72 | 3 | 63 | 2 | 54 | 2 | 72 | 2 |
| Less than 60:40 allelic expression bias in the majority of the four hybrids | ||||||||||||||||
| C3HC4 | 52 | 1 | 46 | 3 | 50 | 0 | 55 | 2 | 42 | 0 | 44 | 1 | 56 | 2 | 55 | 1 |
| CHI | 46 | 2 | 55 | 3 | 53 | 2 | 59 | 3 | ||||||||
| KNAT1 | 56 | 2 | 54 | 1 | 58 | 0 | ||||||||||
| LFY | 56 | 0 | 51 | 5 | 47 | 4 | ||||||||||
| NAK | 54 | 6 | 33 | 5 | 46 | 3 | 43 | 0 | 45 | 3 | 44 | 1 | 47 | 1 | 42 | 3 |
| P4H | 59 | 0 | 62 | 0 | 46 | 0 | 50 | 1 | 54 | 0 | 56 | 7 | 46 | 2 | 51 | 2 |
| RAR1 | 51 | 3 | 40 | 3 | 45 | 1 | 43 | 3 | 43 | 2 | 42 | 1 | 46 | 3 | 36 | 1 |
| Unknown1 | 54 | 1 | 55 | 6 | 70 | 2 | 56 | 0 | 56 | 1 | 51 | 2 | 65 | 0 | 53 | 1 |
| Varied expression among the four hybrids | ||||||||||||||||
| ADH | 67 | 0 | 49 | 0 | 56 | 3 | 49 | 1 | 62 | 3 | 52 | 0 | 40 | 2 | 50 | 2 |
| GT47C | 63 | 2 | 62 | 0 | 57 | 2 | 58 | 1 | 61 | 2 | 58 | 2 | 57 | 1 | 57 | 3 |
| MP | 32 | 1 | 47 | 2 | 25 | 2 | 43 | 3 | ||||||||
| NPR1 | 40 | 5 | 37 | 5 | 43 | 4 | 45 | 0 | ||||||||
| SUS | 59 | 0 | 49 | 7 | 62 | 4 | 61 | 3 | 50 | 4 | 53 | 1 | 60 | 4 | 59 | 4 |
The percentage of transcripts derived from the P. trichocarpa allele in each hybrid is shown. HY 1, 2, 3, and 4 refer to hybrids 11-BULH-4-1, 9-KTWD-4, 7-IRVC.3-1, and 7-IRVD-5-1, respectively. “L” and “S” represent leaf and stem. SE represents standard error of two biological replicates.
Genes showing significantly different allelic expression ratios between leaves and stems as determined by two-tailed homoscedastic variance t-tests (P = 0.05).
Among 30 genes examined, 17 genes showed allelic expression bias in either leaves or stems, or both, in the majority of the hybrids. Notably, the DXPS gene (for deoxyxylulose-5-phosphate synthase) showed monoallelic expression of the P. deltoides allele in stems (Figure 1) of all four hybrids, and this was confirmed by direct sequencing of the RT–PCR products. For the other 13 genes, 8 showed equal allelic expression in the majority or all of the examined hybrids, and 5 genes displayed varied expression among different hybrids without a common pattern in the majority of the hybrids. Comparisons of expression ratios in different hybrids for the same gene showed that the allelic expression biases were usually, but not always, in the same direction and relatively similar in each of the four hybrids. Examples of opposite allelic expression biases in one hybrid compared to the other three hybrids include NBS-LRR in stems of hybrid 1, PPO3 in leaves of hybrid 1, and ADH in stems of hybrid 3. When comparing three genes, DXPS, KNAT1, and ISP with their chromosomally adjacent genes MATE, HAT22, and Unknown1, no correlated expression was detected among adjacent genes.
Organ-specific allelic expression patterns:
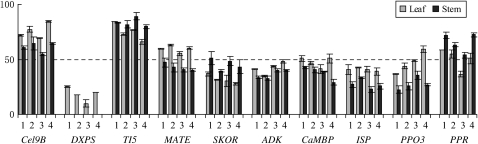
We examined leaves and stems to detect organ-specific allelic expression patterns. Expression of 24 genes was assayed in both leaves and stems. Eight genes showed biased allelic expression in the majority of the four hybrids in both leaves and stems, 2 genes only in leaves, and 5 genes only in stems (Table 2). Significant expression differences between leaves and stems were detected for 10 genes when all hybrids and replicates were analyzed together, with a sample size of 8 for each gene in each organ. DXPS showed biased expression in leaves but monoallelic expression in stems; ADK, CaMBP, ISP, PPO3, PPR, and TI5 showed greater allelic expression bias in stems than in leaves; Cel9B and SKOR showed higher bias in leaves than in stems; and MATE showed different parental alleles being preferably expressed between leaves and stems (Figure 2).
Figure 2.—
Graphical display of organ-specific allelic expression. Shown are data for 10 genes in leaves (shaded bars) and stems (solid bars) of four F1 hybrids. The bars indicate the percentage of transcripts from the P. trichocarpa allele in the F1 hybrid. Numbers 1, 2, 3, and 4 are used to indicate hybrid 11-BULH-4-1, 9-KTWD-4, 7-IRVC.3-1, and 7-IRVD-5-1, respectively. Error bars show standard errors of two replicates. Data are from Table 2.
Comparisons of allelic expression ratios in a hybrid vs. its parents:
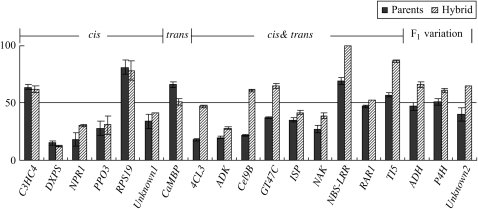
Allelic biases in gene expression in F1 hybrids could be caused by hybridization, or they could reflect unequal expression levels in the two parents that are inherited in the hybrid. To distinguish between these possibilities the expression ratio of the P. deltoides allele to the P. trichocarpa allele (Pd:Pt) was compared between an equal mix of parental RNAs and RNA from the F1 hybrid. Because the parental plants were not available for the four hybrids whose expression was assayed above we used another P. trichocarpa × P. deltoides F1 hybrid (see materials and methods) for comparisons to the parents. Nineteen genes were selected, most of which had SNPs at the same positions as in the other hybrids. There were 6 of 19 genes that showed the same ratio in the hybrids as in the parental RNA mixture, and the remaining 13 genes showed different ratios between the parents and the hybrids. (Table 3, Figure 3). An NBS-LRR gene displayed monoallelic expression in the hybrid, but expression of both alleles in the parents, suggesting hybridization-induced silencing of the P. deltoides allele. Direct sequencing of the RT–PCR products confirmed the monoallelic expression of NBS-LRR in the hybrid. Four genes (ADH, P4H, TI5, and Unknown2) showed a higher allelic bias in the hybrid than between the two parents. In contrast, 5 of the other 12 genes including 4CL3, ADK, ISP, CaMBP, and NAK demonstrated a higher allelic expression bias between the two parents than in the hybrid, but in the same direction. Three genes, Cel9B, GT47C, and Unknown2, displayed opposite ratios of allelic expression levels in the hybrid vs. its parents; that is, there was a bias toward one allele in the hybrid and toward the other allele when comparing the parents.
TABLE 3.
Classification of regulation mode using the allele-specific transcript ratios (Pd:Pt) in mixed parental RNA and in the F1 hybrid
| Parents
|
Hybrid
|
Pd:Pt (parent) ≠ 50:50 | Pd:Pt (hybrid) ≠ 50:50 | Pd:Pt (parent) ≠ Pd:Pt (hybrid) | Regulation classification | |||
|---|---|---|---|---|---|---|---|---|
| Gene | Pd:Pt | SE (%) | Pd:Pt | SE (%) | ||||
| C3HC4 | 36:64 | 2 | 38:62 | 3 | Yes | Yes | No | cis |
| DXPS | 85:15 | 2 | 88:12 | 1 | Yes | Yes | No | cis |
| NPR1 | 82:18 | 6 | 70:30 | 1 | Yes | Yes | No | cis |
| PPO3 | 72:28 | 6 | 69:31 | 8 | Yes | Yes | No | cis |
| RPS19 | 19:81 | 6 | 22:78 | 8 | Yes | Yes | No | cis |
| Unknown1 | 66:34 | 6 | 58:42 | 0 | Yes | Yesa | No | cis |
| CaMBP | 34:66 | 2 | 49:51 | 3 | Yes | No | Yes | trans |
| 4CL3 | 82:18 | 1 | 53:47 | 1 | Yes | Yesa | Yes | cis and trans |
| ADK | 80:20 | 1 | 72:28 | 1 | Yes | Yes | Yes | cis and trans |
| Cel9B | 78:22 | 1 | 39:61 | 1 | Yes | Yes | Yes | cis and trans |
| GT47C | 63:37 | 1 | 35:65 | 2 | Yes | Yes | Yes | cis and trans |
| ISP | 65:35 | 2 | 58:42 | 2 | Yes | Yesa | Yes | cis and trans |
| NAK | 73:27 | 3 | 61:39 | 3 | Yes | Yes | Yes | cis and trans |
| NBS-LRR | 31:69 | 3 | 0:100 | 0 | Yes | Yes | Yes | cis and trans |
| RAR1 | 53:47 | 1 | 48:52 | 0 | Yesa | Yesa | Yes | cis and trans |
| TI5 | 43:57 | 2 | 14:86 | 1 | Yesa | Yes | Yes | cis and trans |
| ADH | 53:47 | 3 | 34:66 | 2 | No | Yes | Yes | F1 variation |
| P4H | 49:51 | 3 | 39:61 | 2 | No | Yes | Yes | F1 variation |
| Unknown2 | 60:40 | 6 | 35:65 | 0 | No | Yes | Yes | F1 variation |
SE refers to standard error of three replicates. Two-tailed homoscedastic variance t-tests (P = 0.05) were performed to test the deviation in allelic expression ratio of the mixed parental RNA from equal expression, the F1 hybrid allelic ratios from equal expression, and mixed parental RNA vs. the F1 hybrid.
Ratios that were statistically different from 50:50 but were less than the 60:40 ratio used to define biased expression in Table 2.
Figure 3.—
Comparisons of the percentages of P. trichocarpa (Pt) transcripts in equal mixes of parental RNAs and in the F1 hybrid. Solid columns and hatched columns represent percentages of Pt transcripts in the mix of parental RNAs and those in the F1 hybrid, respectively. Error bars indicate standard errors of three replicates. Genes are grouped by the regulation patterns. Data are from Table 3.
Cis- and trans-regulatory variation analysis:
On the basis of different hypotheses for cis- and trans-regulation (see Introduction), three different expression patterns in the hybrid compared with its parents can be classified as to mode of gene regulation (Wittkopp et al. 2004):
Cis-regulation: the same allelic expression bias in both parents and hybrid. Pd:Pt (parent) = Pd:Pt (hybrid) ≠ 50:50.
Trans-regulation: biased expression in the parents but equal expression in the hybrid. Pd:Pt (parent) ≠ 50:50, Pd:Pt (hybrid) = 50:50.
Cis- and trans-regulation: biased expression in the hybrid that is different from the expression bias in the parents. Pd:Pt (parent) ≠ Pd:Pt (hybrid), Pd:Pt (hybrid) ≠ 50:50, Pd:Pt (parent) ≠ 50:50.
For the 19 genes examined, 6 were classified as cis-regulated, 1 gene as trans-regulated (CaMBP for a calmodulin binding protein), and 9 were considered to be adjusted by combined cis- and trans-regulation (Figure 3, Table 3). Among these 9 genes, 5 showed allelic biases in the same direction and 2 (GT47C and Cel9B) had allelic expression biases in the opposite direction. Three genes (ADH, P4H, and Unknown2) showed equal expression in the parents but biased allelic expression in the hybrid and these genes do not show evidence of either cis- or trans-regulation. Instead those genes show allelic variation in the F1 hybrid (Table 3).
DISCUSSION
Prevalence of unequal allelic expression:
We surveyed 30 genes for their allelic expression in four P. trichocarpa × P. deltoides F1 hybrids, and the results show that a considerable percentage of genes show variation in allelic expression levels in Populus interspecific F1 hybrids. Using a threshold cutoff of 1.5-fold (60:40), 17 of the 30 (57%) genes showed differential allelic expression in at least one organ in the majority of the four hybrids. Considering the 3 and 2% standard error results from genes GT47C and TI5 with eight tested leaf replicates, 60:40 should be a conservative threshold for classifying differential allelic expression. Indeed 57% might be an underestimate of the true percentage of genes with unequal allelic expression. A previous study in maize intraspecific hybrids, using less stringent criteria for differential expression, identified 11 of 15 (73%) genes that showed <0.85- or >1.18-fold differences in allelic expression ratios (Guo et al. 2004). Research done concurrently to our study examined allelic expression in 316 genes in maize intraspecific hybrids and found that 43–53% of the genes (depending on the cross) showed unequal allelic expression (Springer and Stupar 2007b). In contrast, a study of mouse hybrids found only ∼10% of genes with >1.5-fold allelic expression difference (Cowles et al. 2002). There appears to be a higher degree of allelic expression variation in the Populus hybrids and the maize hybrids than in the mouse hybrids. It has been suggested that the highly polymorphic maize genome could account for its relatively high degree of allelic expression variation (Guo et al. 2004). Similarly, the genetic divergence between P. trichocarpa and P. deltoides probably contributes to a higher allelic expression variation compared to the mouse intraspecific hybrids.
Organ-specific differences in allelic expression:
We observed allelic expression differences between leaves and stems for 10 of 15 genes (67%) that were expressed in both organs and showed allelic expression variation. Although the sample size is relatively small, the results still suggest a surprisingly high degree of organ-specific differences in allelic expression. It was previously shown in a diploid F1 cotton hybrid that the AdhA gene showed organ-specific allelic silencing (Adams and Wendel 2005). A study done concurrently to the research reported here examined allelic expression in three organs of intraspecific maize hybrids and found that about half of the genes that were expressed in all three organs showed different allelic ratios in at least one of the three organs (Springer and Stupar 2007b). Similarly, a study of mouse intraspecific hybrids identified two genes with diverged allelic expression patterns in different tissues (Cowles et al. 2002). The above studies have established that allelic expression in hybrids can be highly tissue- and organ-specific. In future studies of individual cell types, it would be interesting to characterize allelic expression at a finer scale.
Gene regulatory variation in hybrids:
The comparison of expression between a mix of parental RNAs and F1 hybrid RNA revealed 6 of 19 (32%) genes under mainly cis-regulation, 1 of 19 (5%) under primarily trans-regulation, and 9 genes (47%) controlled coordinately by cis- and trans-regulation. Therefore cis-regulation, sometimes in combination with trans-regulation, appears to be largely responsible for the regulation of the genes in our study. Studies of interspecific Drosophila hybrids reported 12 of 28 (43%) genes to be completely explained by cis-regulation, and the remaining 16 all explained by cis- and trans-regulation (Wittkopp et al. 2004). This contrasts to the findings in maize intraspecific hybrids that showed pure cis-regulation accounting for allelic expression in 18 of 35 (51%), a majority of the sampled genes (Stupar and Springer 2006). Although variable proportions of complete cis-regulation are found in the various studies of different organisms, cis-effects were consistently involved in most if not all of the assayed genes, and pure trans-regulation is rare, affecting only 1 of 19 genes in the Populus hybrids, 1 of 35 in the maize hybrids (Stupar and Springer 2006), and none of 28 in the Drosophila hybrids (Wittkopp et al. 2004). Cis- and trans-regulation were explored at a much larger scale in a recently published study of maize hybrids that examined 316 genes and found that pure cis-regulation predominates. As cis-elements function in an allele-specific manner, allelic expression following cis-regulation reflects an inheritance of the regulatory pattern from the two parents to the hybrid.
Hybridization induced changes in allelic expression:
After hybridization both alleles are exposed to common trans-regulators in the same cellular environment, and so trans-regulation and combined cis- and trans-regulation could be induced by hybridization to harmonize the two heterozygous genomes (Landry et al. 2005). There is a hypothesis that cis- and trans-compensatory evolution is important in leading to novel gene expression and performance in the hybrids (Landry et al. 2005). Compensatory cis- and trans-regulation is inferred when the allelic expression difference in an F1 hybrid is more extreme than, or in the opposite direction from, that in the parents, suggesting changes in trans- compensate for the already existing cis-divergence. In this study 4 of 19 genes display allelic expression biases to a larger extent in the hybrid than in the parents, and 3 other genes, Cel9B, GT47C, and Unknown2 clearly show opposite allelic divergence in the hybrid compared with the parental divergence. It has been proposed that reuniting diverged regulatory factors and hierarchies in hybrids can lead to altered gene expression patterns (Riddle and Birchler 2003). The cases involving trans-regulation in Populus hybrids suggest a modification of the regulatory network upon interspecific hybridization that affects expression of some genes.
Three genes in this study (ADH, P4H, and Unknown2) showed equal expression in the parents but biased allelic expression in the F1 hybrid indicating expression variation in the hybrid that could not necessarily be classified as trans-regulation according to the test we used. Other factors, such as epigenetic variation, might account for the expression changes in those genes.
The Populus F1 hybrids used in this study show strong heterosis, particularly in regards to growth rate, trunk diameter, stem diameter, and leaf size. Might altered gene regulation in F1 hybrids observed in this study be involved in generating the heterotic phenotypes observed in these hybrids? Although no data from this study provide evidence for that possibility, it has been proposed that another type of altered gene regulation in F1 hybrids, deviations from mid-parent expression levels, may contribute to heterosis (Birchler et al. 2003; Swanson-Wagner et al. 2006). It is tempting to speculate that altered gene regulation in the Populus hybrids, especially monoallelic expression, may play a role in the heterosis seen in this system, although future studies will be needed to test this hypothesis.
Expression changes upon hybridization in diploid hybrids compared with allopolyploid hybrids:
Allopolyploid hybrids can be formed by hybridization between two diploid species followed by spontaneous or induced chromosome doubling. Altered gene expression levels and patterns in an F1 hybrid could be directly passed on to the allopolyploid if there is chromosome doubling in the F1 hybrid. Indeed studies of newly synthesized allopolyploids have revealed considerable alterations in gene expression compared with their parents (Comai et al. 2000; Kashkush et al. 2002; Adams et al. 2004; Wang et al. 2004, 2006a; Hegarty et al. 2005, 2006), much of which have been shown to be caused by interspecific hybridization instead of chromosome doubling. An important distinction between expression changes caused by hybridization in diploid hybrids vs. allopolyploid hybrids is that expression patterns of alleles in diploid hybrids are more likely to experience homogenization in subsequent generations, if there is recombination between the alleles, than homeologous genes in allopolyploids that may maintain distinct expression patterns over evolutionary time if there is no intergenomic recombination.
Acknowledgments
We thank Dan Carson from Kruger Products (formerly Scott Paper) in New Westminster, British Columbia, for letting us make cuttings from his P. trichocarpa x P. deltoides F1 hybrids, Carl Douglas for providing a cutting of P. deltoides, and the University of British Columbia Botanical Garden for growing four of the hybrids and P. trichocarpa on Totem Field. We also thank Erin Gilchrist for providing SNP data for five genes used in this study, Quentin Cronk for helpful discussions, and Jeannette Whitton and Sean Graham for comments on an earlier version of the manuscript. This study was funded by a grant from the National Science and Engineering Research Council of Canada, by start-up funds from the Faculty of Land and Food Systems at the University of British Columbia, and by infrastructure funds from the Canadian Foundation for Innovation.
References
- Adams, K. L., 2007. Evolution of duplicate gene expression in polyploid and hybrid plants. J. Hered. 98: 136–141. [DOI] [PubMed] [Google Scholar]
- Adams, K. L., R. Cronn, R. Percifield and J. F. Wendel, 2003. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100: 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, K. L., R. Percifield and J. F. Wendel, 2004. Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics 168: 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, K. L., and J. F. Wendel, 2005. Allele-specific, bidirectional silencing of an alcohol dehydrogenase gene in different organs of interspecific diploid cotton hybrids. Genetics 171: 2139–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, M. L., 2004. Natural hybridization and the evolution of domesticated, pest and disease organisms. Mol. Ecol. 13: 997–1007. [DOI] [PubMed] [Google Scholar]
- Auger, D. L., A. D. Gray, T. S. Ream, A. Kato, E. H. Coe, Jr. et al., 2005. Nonadditive gene expression in diploid and triploid hybrids of maize. Genetics 169: 389–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, J., S. Lee, C. Chen, X. Zhang, Y. Zhang et al., 2005. Serial analysis of gene expression study of a hybrid rice strain (LYP9) and its parental cultivars. Plant Physiol. 138: 1216–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler, J. A., D. L. Auger and N. C. Riddle, 2003. In search of the molecular basis of heterosis. Plant Cell 15: 2236–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler, J. A., H. Yao and S. Chudalayandi, 2006. Unraveling the genetic basis of hybrid vigor. Proc. Natl. Acad. Sci. USA 103: 12957–12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw, Jr., H. D., and R. F. Stettler, 1995. Molecular genetics of growth and development in populus. IV. Mapping QTLs with large effects on growth, form, and phenology traits in a forest tree. Genetics 139: 963–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, N. J., P. R. Buckland, M. J. Owen and M. C. O'Donovan, 2003. Cis-acting variation in the expression of a high proportion of genes in human brain. Hum. Genet. 113: 149–153. [DOI] [PubMed] [Google Scholar]
- Comai, L., A. P. Tyagi, K. Winter, R. Holmes-Davis, S. H. Reynolds et al., 2000. Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12: 1551–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles, C. R., J. N. Hirschhorn, D. Altshuler and E. S. Lander, 2002. Detection of regulatory variation in mouse genes. Nat. Genet. 32: 432–437. [DOI] [PubMed] [Google Scholar]
- Gilchrist, E. J., G. W. Haughn, C. C. Ying, S. P. Otto, J. Zhuang et al., 2006. Use of ecotilling as an efficient SNP discovery tool to survey genetic variation in wild populations of Populus trichocarpa. Mol. Ecol. 15: 1367–1378. [DOI] [PubMed] [Google Scholar]
- Guo, M., M. A. Rupe, O. N. Danilevskaya, X. Yang and Z. Hu, 2003. Genome-wide mRNA profiling reveals heterochronic allelic variation and a new imprinted gene in hybrid maize endosperm. Plant J. 36: 30–44. [DOI] [PubMed] [Google Scholar]
- Guo, M., M. A. Rupe, X. Yang, O. Crasta, C. Zinselmeier et al., 2006. Genome-wide transcript analysis of maize hybrids: allelic additive gene expression and yield heterosis. Theor. Appl. Genet. 113: 831–845. [DOI] [PubMed] [Google Scholar]
- Guo, M., M. A. Rupe, C. Zinselmeier, J. Habben, B. A. Bowen et al., 2004. Allelic variation of gene expression in maize hybrids. Plant Cell 16: 1707–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty, M. J., G. L. Barker, I. D. Wilson, R. J. Abbott, K. J. Edwards et al., 2006. Transcriptome shock after interspecific hybridization in senecio is ameliorated by genome duplication. Curr. Biol. 16: 1652–1659. [DOI] [PubMed] [Google Scholar]
- Hegarty, M. J., and S. J. Hiscock, 2005. Hybrid speciation in plants: new insights from molecular studies. New Phytol. 165: 411–423. [DOI] [PubMed] [Google Scholar]
- Hegarty, M. J., J. M. Jones, I. D. Wilson, G. L. Barker, J. A. Coghill et al., 2005. Development of anonymous cDNA microarrays to study changes to the Senecio floral transcriptome during hybrid speciation. Mol. Ecol. 14: 2493–2510. [DOI] [PubMed] [Google Scholar]
- Kashkush, K., M. Feldman and A. A. Levy, 2002. Gene loss, silencing, and activation in a newly synthesized wheat allotetraploid. Genetics 160: 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry, C. R., P. J. Wittkopp, C. H. Taubes, J. M. Ranz, A. G. Clark et al., 2005. Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics 171: 1813–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., G. T. Howe and R. Wu, 1998. Developmental factors responsible for heterosis in aspen hybrids (Populus tremuloides × P. tremula). Tree Physiol. 18: 29–36. [DOI] [PubMed] [Google Scholar]
- Liu, B., and J. F. Wendel, 2000. Retrotransposon activation followed by rapid repression in introgressed rice plants. Genome 43: 874–880. [PubMed] [Google Scholar]
- Meyer, S., H. Pospisil and S. Scholten, 2007. Heterosis associated gene expression in maize embryos 6 days after fertilization exhibits additive, dominant and overdominant pattern. Plant Mol. Biol. 63: 381–391. [DOI] [PubMed] [Google Scholar]
- Norton, N., N. M. Williams, H. J. Williams, G. Spurlock, G. Kirov et al., 2002. Universal, robust, highly quantitative SNP allele frequency measurement in DNA pools. Hum. Genet. 110: 471–478. [DOI] [PubMed] [Google Scholar]
- Pinsonneault, J., C. U. Nielsen and W. Sadee, 2004. Genetic variants of the human H+/dipeptide transporter PEPT2: analysis of haplotype functions. J. Pharmacol. Exp. Ther. 311: 1088–1096. [DOI] [PubMed] [Google Scholar]
- Riddle, N. C., and J. A. Birchler, 2003. Effects of reunited diverged regulatory hierarchies in allopolyploids and species hybrids. Trends Genet. 19: 597–600. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., 1997. Hybrid origins of plant species. Annu. Rev. Ecol. Syst. 28: 359–389. [Google Scholar]
- Rieseberg, L. H., M. A. Archer and R. K. Wayne, 1999. Transgressive segregation, adaptation and speciation. Heredity 83(Pt 4): 363–372. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., O. Raymond, D. M. Rosenthal, Z. Lai, K. Livingstone et al., 2003. a Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301: 1211–1216. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., B. Sinervo, C. R. Linder, M. C. Ungerer and D. M. Arias, 1996. Role of gene interactions in hybrid speciation: evidence from ancient and experimental hybrids. Science 272: 741–745. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., A. Widmer, A. M. Arntz and J. M. Burke, 2003. b The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 1141–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald, J., J. M. Akey, J. Whittle, E. N. Smith, G. Yvert et al., 2005. Simultaneous genotyping, gene-expression measurement, and detection of allele-specific expression with oligonucleotide arrays. Genome Res. 15: 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon, A., M. L. Ainouche and J. F. Wendel, 2005. Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae). Mol. Ecol. 14: 1163–1175. [DOI] [PubMed] [Google Scholar]
- Shaked, H., K. Kashkush, H. Ozkan, M. Feldman and A. A. Levy, 2001. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13: 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, X., Z. Liu, Z. Dong, Y. Wang, Y. Chen et al., 2005. Mobilization of the active MITE transposons mPing and Pong in rice by introgression from wild rice (Zizania latifolia Griseb.). Mol. Biol. Evol. 22: 976–990. [DOI] [PubMed] [Google Scholar]
- Song, R., G. Segal and J. Messing, 2004. Expression of the sorghum 10-member kafirin gene cluster in maize endosperm. Nucleic Acids Res. 32: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, N. M., and R. M. Stupar, 2007. a Allelic variation and heterosis in maize: How do two halves make more than a whole? Genome Res. 17: 264–275. [DOI] [PubMed] [Google Scholar]
- Springer, N. M., and R. M. Stupar, 2007. b Allele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. Plant Cell 19: 2391–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar, R. M., P. J. Hermanson and N. M. Springer, 2007. Non-additive expression and parent-of-origin effects identified by microarray and allele-specific expression profiling of maize endosperm. Plant Physiol. (in press). [DOI] [PMC free article] [PubMed]
- Stupar, R. M., and N. M. Springer, 2006. Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics 173: 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Wagner, R. A., Y. Jia, R. DeCook, L. A. Borsuk, D. Nettleton et al., 2006. All possible modes of gene action are observed in a global comparison of gene expression in a maize F1 hybrid and its inbred parents. Proc. Natl. Acad. Sci. USA 103: 6805–6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan, G. A., S. Difazio, S. Jansson, J. Bohlmann, I. Grigoriev et al., 2006. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604. [DOI] [PubMed] [Google Scholar]
- Uzarowska, A., B. Keller, H. P. Piepho, G. Schwarz, C. Ingvardsen et al., 2007. Comparative expression profiling in meristems of inbred-hybrid triplets of maize based on morphological investigations of heterosis for plant height. Plant Mol. Biol. 63: 21–34. [DOI] [PubMed] [Google Scholar]
- Vuylsteke, M., F. van Eeuwijk, P. Van Hummelen, M. Kuiper and M. Zabeau, 2005. Genetic analysis of variation in gene expression in Arabidopsis thaliana. Genetics 171: 1267–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., A. D. Johnson, A. C. Papp, D. L. Kroetz and W. Sadee, 2005. Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenet. Genomics 15: 693–704. [PubMed] [Google Scholar]
- Wang, Z., Z. Ni, H. Wu, X. Nie and Q. Sun, 2006. Heterosis in root development and differential gene expression between hybrids and their parental inbreds in wheat (Triticum aestivum L.). Theor. Appl. Genet. 113: 1283–1294. [DOI] [PubMed] [Google Scholar]
- Wittkopp, P. J., B. K. Haerum and A. G. Clark, 2004. Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88. [DOI] [PubMed] [Google Scholar]
- Wu, L. M., Z. F. Ni, F. R. Meng, Z. Lin and Q. X. Sun, 2003. Cloning and characterization of leaf cDNAs that are differentially expressed between wheat hybrids and their parents. Mol. Genet. Genomics 270: 281–286. [DOI] [PubMed] [Google Scholar]
- Yan, H., W. Yuan, V. E. Velculescu, B. Vogelstein and K. W. Kinzler, 2002. Allelic variation in human gene expression. Science 297: 1143. [DOI] [PubMed] [Google Scholar]