Abstract
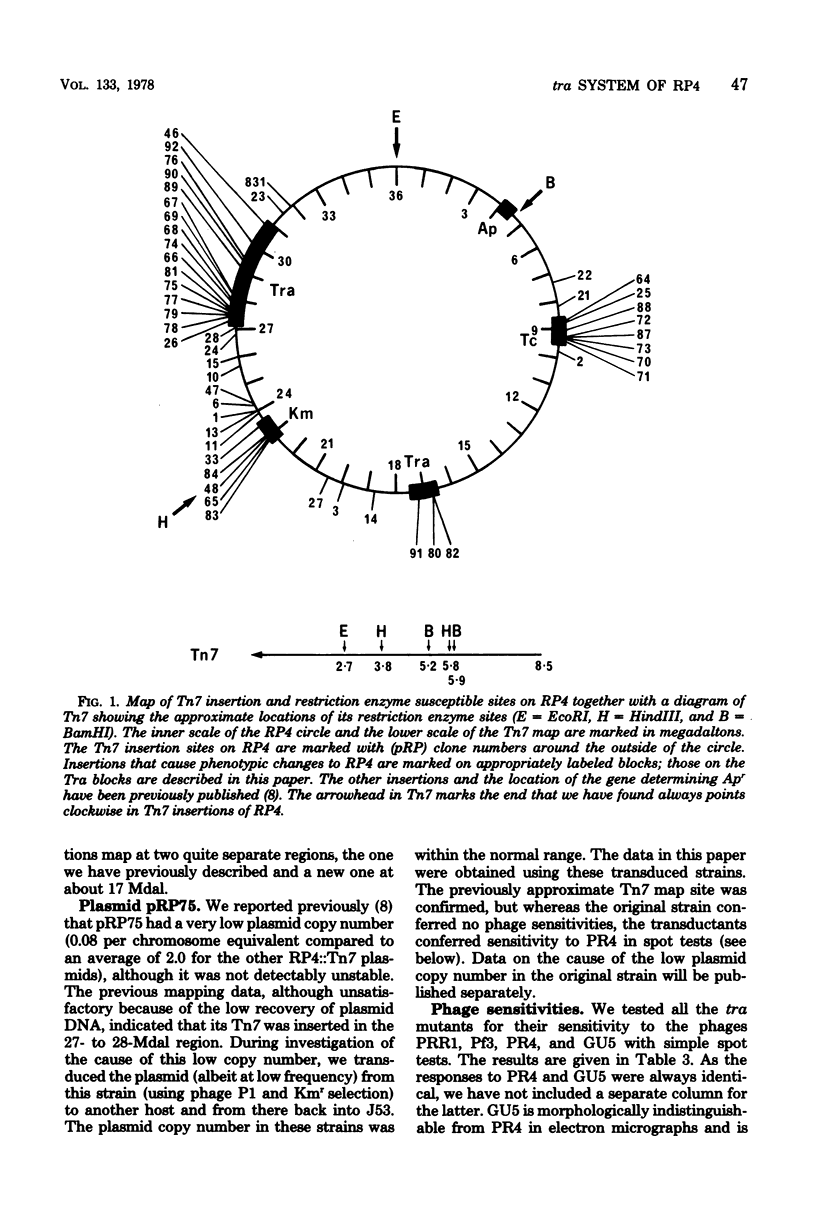
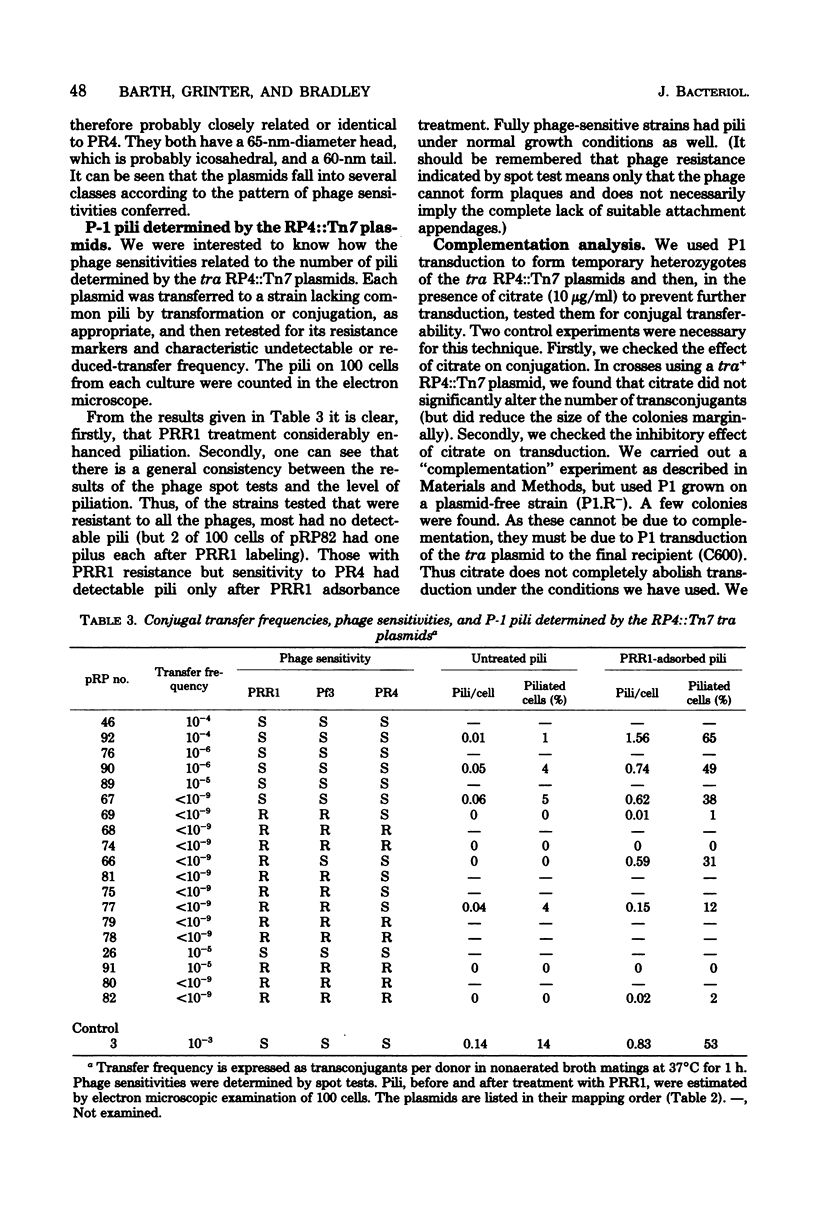
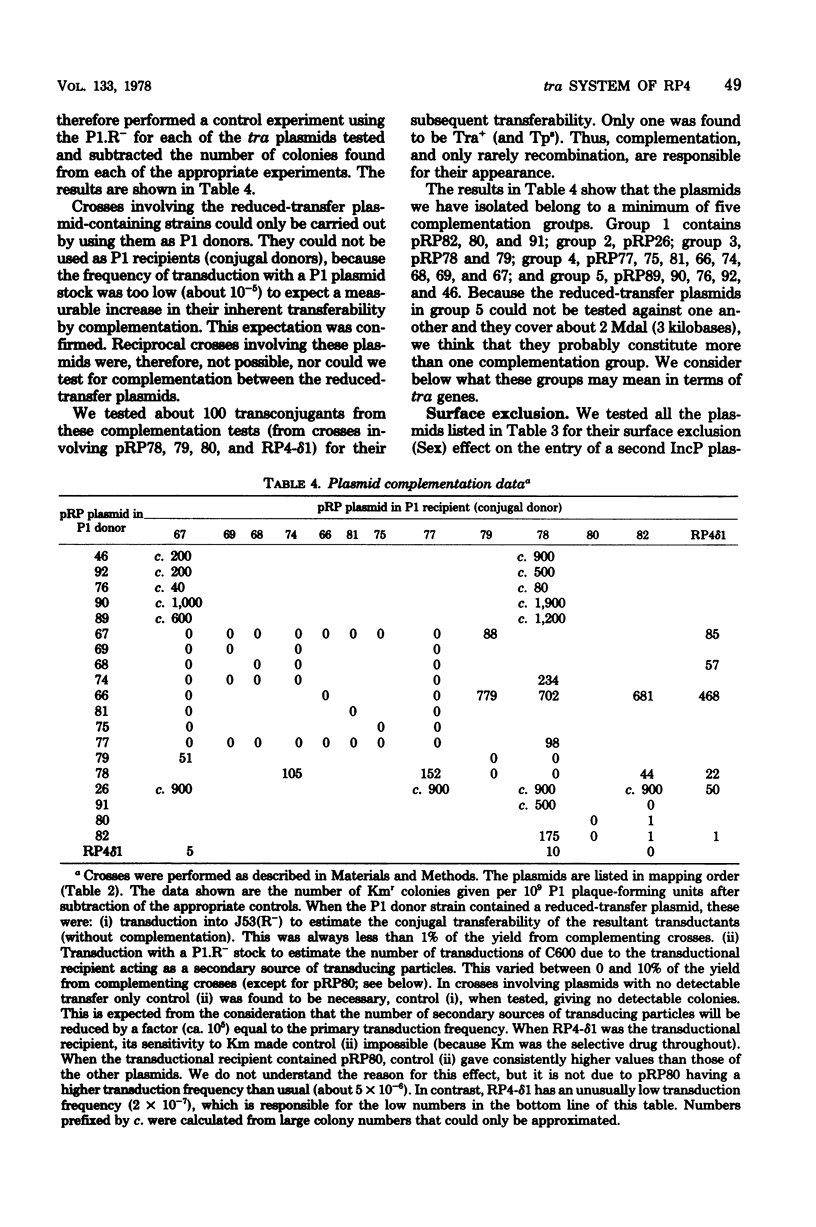
We have begun an analysis in Escherichia coli of the conjugal transfer functions of the broad-host-range plasmid RP4. We have isolated 19 tra mutants of RP4, generated by insertion of transposon 7, and mapped their insertion sites by restriction endonuclease analysis. These sites fall into two separate regions on either side of the kanamycin resistance determinant. The transfer rates of the mutants range from 10% of that of RP4 to an undetectable level. Spot tests with the P-1 pilus-specific phages PRR1, Pf3, and PR4 and electron microscopic examination for pili have classified the mutants into several phenotypes consistent with their having normal, retracted, or no pili. Analysis of transient plasmid heterozygotes, created by P1 transduction, divided the tra mutants into a minimum of five complementation groups. Some of these groups contain more than one phenotypic class and may represent more than one gene because of the possible polar and deletion effects of Tn7 insertion.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achtman M. Genetics of the F sex factor in enterobacteriaceae. Curr Top Microbiol Immunol. 1973;60:79–123. doi: 10.1007/978-3-642-65502-9_3. [DOI] [PubMed] [Google Scholar]
- Achtman M., Willetts N., Clark A. J. Beginning a genetic analysis of conjugational transfer determined by the F factor in Escherichia coli by isolation and characterization of transfer-deficient mutants. J Bacteriol. 1971 May;106(2):529–538. doi: 10.1128/jb.106.2.529-538.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achtman M., Willetts N., Clark A. J. Conjugational complementation analysis of transfer-deficient mutants of Flac in Escherichia coli. J Bacteriol. 1972 Jun;110(3):831–842. doi: 10.1128/jb.110.3.831-842.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P. T., Datta N., Hedges R. W., Grinter N. J. Transposition of a deoxyribonucleic acid sequence encoding trimethoprim and streptomycin resistances from R483 to other replicons. J Bacteriol. 1976 Mar;125(3):800–810. doi: 10.1128/jb.125.3.800-810.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P. T., Grinter N. J. Comparison of the deoxyribonucleic acid molecular weights and homologies of plasmids conferring linked resistance to streptomycin and sulfonamides. J Bacteriol. 1974 Nov;120(2):618–630. doi: 10.1128/jb.120.2.618-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P. T., Grinter N. J. Map of plasmid RP4 derived by insertion of transposon C. J Mol Biol. 1977 Jul 5;113(3):455–474. doi: 10.1016/0022-2836(77)90233-9. [DOI] [PubMed] [Google Scholar]
- Bradley D. E. Adsorption of bacteriophages specific for Pseudomonas aeruginosa R factors RP1 and R1822. Biochem Biophys Res Commun. 1974 Apr 8;57(3):893–900. doi: 10.1016/0006-291x(74)90630-5. [DOI] [PubMed] [Google Scholar]
- Bradley D. E. Adsorption of the R-specific bacteriophage PR4 to pili determined by a drug resistance plasmid of the W compatibility group. J Gen Microbiol. 1976 Jul;95(1):181–185. doi: 10.1099/00221287-95-1-181. [DOI] [PubMed] [Google Scholar]
- Bradley D. E., Cohen D. R. Basic characterization of W-pili. J Gen Microbiol. 1976 Nov;97(1):91–103. doi: 10.1099/00221287-97-1-91. [DOI] [PubMed] [Google Scholar]
- Bradley D. E., Rutherford E. L. Basic characterization of a lipid-containing bacteriophage specific for plasmids of the P, N, and W compatibility groups. Can J Microbiol. 1975 Feb;21(2):152–163. doi: 10.1139/m75-023. [DOI] [PubMed] [Google Scholar]
- Cohen S. N. Transposable genetic elements and plasmid evolution. Nature. 1976 Oct 28;263(5580):731–738. doi: 10.1038/263731a0. [DOI] [PubMed] [Google Scholar]
- Datta N., Hedges R. W., Shaw E. J., Sykes R. B., Richmond M. H. Properties of an R factor from Pseudomonas aeruginosa. J Bacteriol. 1971 Dec;108(3):1244–1249. doi: 10.1128/jb.108.3.1244-1249.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski D., Meyer R., Miller D. S., Helinski D. R. Generation in vitro of deletions in the broad host range plasmid RK2 using phage Mu insertions and a restriction endonuclease. Gene. 1976;1(1):107–119. doi: 10.1016/0378-1119(76)90010-x. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Cresswell J. M., Jacob A. E. A non-transmissible variant of RP4 suitable as cloning vehicle for genetic engineering. FEBS Lett. 1976 Jan 15;61(2):186–188. doi: 10.1016/0014-5793(76)81033-2. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Jacob A. E. Transposition of ampicillin resistance from RP4 to other replicons. Mol Gen Genet. 1974;132(1):31–40. doi: 10.1007/BF00268228. [DOI] [PubMed] [Google Scholar]
- Heffron F., Rubens C., Falkow S. Translocation of a plasmid DNA sequence which mediates ampicillin resistance: molecular nature and specificity of insertion. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3623–3627. doi: 10.1073/pnas.72.9.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Chan R. K., Tye B. K., Botstein D. Mutagenesis by insertion of a drug-resistance element carrying an inverted repetition. J Mol Biol. 1975 Oct 5;97(4):561–575. doi: 10.1016/s0022-2836(75)80059-3. [DOI] [PubMed] [Google Scholar]
- Newton W. A., Beckwith J. R., Zipser D., Brenner S. Nonsense mutants and polarity in the lac operon of Escherichia coli. J Mol Biol. 1965 Nov;14(1):290–296. doi: 10.1016/s0022-2836(65)80250-9. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Clowes R. C., Cohen S. N., Curtiss R., 3rd, Datta N., Falkow S. Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol Rev. 1976 Mar;40(1):168–189. doi: 10.1128/br.40.1.168-189.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., Shipley P. Host range and properties of the Pseudomonas aeruginosa R factor R1822. J Bacteriol. 1973 Feb;113(2):772–780. doi: 10.1128/jb.113.2.772-780.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., Siak J. S., Gray R. H. Characteristics of PRD1, a plasmid-dependent broad host range DNA bacteriophage. J Virol. 1974 Sep;14(3):689–699. doi: 10.1128/jvi.14.3.689-699.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., Thomas D. D. Characteristics and purification of PRR1, an RNA phage specific for the broad host range Pseudomonas R1822 drug resistance plasmid. J Virol. 1973 Dec;12(6):1560–1567. doi: 10.1128/jvi.12.6.1560-1567.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsubo E., Nishimura Y., Hirota Y. Transfer-defective mutants of sex factors in Escherichia coli. I. Defective mutants and complementation analysis. Genetics. 1970 Feb;64(2):173–188. doi: 10.1093/genetics/64.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanisich V. A., Bennett P. M. Isolation and characterisation of deletion mutants involving the transfer genes of P-group plasmids in Pseudomonas aeruginosa. Mol Gen Genet. 1976 Dec 8;149(2):211–216. doi: 10.1007/BF00332891. [DOI] [PubMed] [Google Scholar]
- Stanisich V. A. The properties and host range of male-specific bacteriophages of Pseudomonas aeruginosa. J Gen Microbiol. 1974 Oct;84(2):332–342. doi: 10.1099/00221287-84-2-332. [DOI] [PubMed] [Google Scholar]
- Walker D. H., Jr, Anderson T. F. Morphological variants of coliphage P1. J Virol. 1970 Jun;5(6):765–782. doi: 10.1128/jvi.5.6.765-782.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]


