Abstract
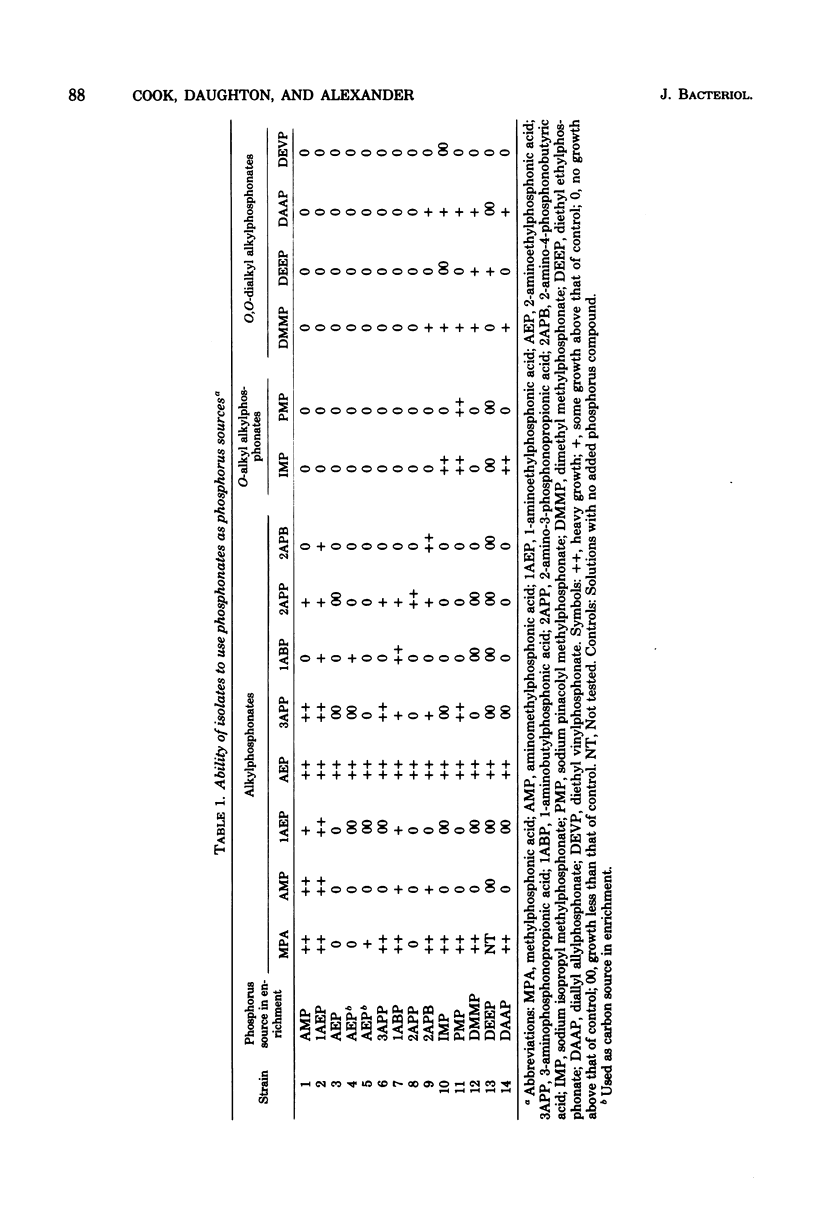
Bacteria able to use at least one of 13 ionic alkylphosphonates of O-alkyl or O,O-dialkyl alkylphosphonates as phosphorus sources were isolated from sewage and soil. Four of these isolates used 2-aminoethylphosphonic acid (AEP) as a sole carbon, nitrogen, and phosphorus source. None of the other phosphonates served as a carbon source for the organisms. One isolate, identified as Pseudomonas putida, grew with AEP as its sole carbon, nitrogen, and phosphorus source and released nearly all of the organic phosphorus as orthophosphate and 72% of the AEP nitrogen as ammonium. This is the first demonstration of utilization of a phosphonoalkyl moiety as a sole carbon source. Cell-free extracts of P. putida contained an inducible enzyme system that required pyruvate and pyridoxal phosphate to release orthophosphate from AEP; acetaldehyde was tentatively identified as a second product. Phosphite inhibited the enzyme system.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cassaigne A., Lacoste A. M., Neuzil E. Recherches sur le catabolisme des acides phosphoniques: biodégradation de la liaison C--P par Pseudomonas aeruginosa. C R Acad Sci Hebd Seances Acad Sci D. 1976 May 3;282(17):1637–1639. [PubMed] [Google Scholar]
- Fourche J., Jensen H., Neuzil E. Fluorescence reactions of aminophosphonic acids. Anal Chem. 1976 Jan;48(1):155–159. doi: 10.1021/ac60365a046. [DOI] [PubMed] [Google Scholar]
- HOSKIN F. C. Some observations concerning the biochemical inertness of methylphosphonic and isopropyl methylphosphonic acids. Can J Biochem Physiol. 1956 Jul;34(4):743–746. [PubMed] [Google Scholar]
- Harvey N. L., Fewson C. A., Holms W. H. Apparatus for batch culture of micro-organisms. Lab Pract. 1968 Oct;17(10):1134–1136. [PubMed] [Google Scholar]
- Hilderbrand R. L., Henderson T. O., Glonek T., Myers T. C. Isolation and characterization of a phosphonic acid rich glycoprotein preparation from Metridium dianthus. Biochemistry. 1973 Nov 6;12(23):4756–4762. doi: 10.1021/bi00747a601. [DOI] [PubMed] [Google Scholar]
- Kennedy S. I., Fewson C. A. Enzymes of the mandelate pathway in Bacterium N.C.I.B. 8250. Biochem J. 1968 Apr;107(4):497–506. doi: 10.1042/bj1070497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn E. D., Dearborn D. G., Fales H. M., Sokoloski E. A. Phosphonoglycan. A major polysaccharide constituent of the amoeba plasma membrane contains 2-aminoethylphosphonic acid and 1-hydroxy-2-aminoethylphosphonic acid. J Biol Chem. 1973 Mar 25;248(6):2257–2259. [PubMed] [Google Scholar]
- La Nauze J. M., Rosenberg H., Shaw D. C. The enzymic cleavage of the carbon-phosphorus bond: purification and properties of phosphonatase. Biochim Biophys Acta. 1970 Aug 15;212(2):332–350. doi: 10.1016/0005-2744(70)90214-7. [DOI] [PubMed] [Google Scholar]
- Lacoste A. M., Neuzil E. Transamination de l'acide amino-2-éthylphosphonique par Pseudomonas aeruginosa. C R Acad Sci Hebd Seances Acad Sci D. 1969 Jul 16;269(2):254–257. [PubMed] [Google Scholar]
- Murai T., Tomizawa C. Chemical transformation of S-benzyl O-ethyl phenylphosphonothiolate (Inezin) by ultraviolet light. J Environ Sci Health B. 1976;11(2):185–197. doi: 10.1080/03601237609372034. [DOI] [PubMed] [Google Scholar]
- Rosenberg H., La Nauze J. M. The metabolism of phosphonates by microorganisms. The transport of aminoethylphosphonic acid in Bacillus cereus. Biochim Biophys Acta. 1967 Jun 13;141(1):79–90. doi: 10.1016/0304-4165(67)90247-4. [DOI] [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966 May;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]



