Abstract
Embryonic morphogenesis is driven by a suite of cell behaviours, including coordinated shape changes, cellular rearrangements and individual cell migrations, whose molecular determinants are largely unknown. In the zebrafish, Dani rerio, trilobite mutant embryos have defects in gastrulation movements1–4 and posterior migration of hindbrain neurons5. Here, we have used positional cloning to demonstrate that trilobite mutations disrupt the transmembrane protein Strabismus (Stbm)/Van Gogh (Vang), previously associated with planar cell polarity (PCP) in Drosophila melanogaster6,7, and PCP and canonical Wnt/β-catenin signalling in vertebrates8,9. Our genetic and molecular analyses argue that during gastrulation, trilobite interacts with the PCP pathway without affecting canonical Wnt signalling. Furthermore, trilobite may regulate neuronal migration independently of PCP molecules. We show that trilobite mediates polarization of distinct movement behaviours. During gastrulation convergence and extension movements, trilobite regulates mediolateral cell polarity underlying effective intercalation and directed dorsal migration at increasing velocities. In the hindbrain, trilobite controls effective migration of branchiomotor neurons towards posterior rhombomeres. Mosaic analyses show trilobite functions cell-autonomously and non-autonomously in gastrulae and the hindbrain. We propose Trilobite/Stbm mediates cellular interactions that confer directionality on distinct movements during vertebrate embryogenesis.
To determine the molecular underpinnings of various cell movements in vertebrate embryos, we undertook a molecular characterization of the trilobite (tri) locus1,4. Through positional cloning, we identified a stbm/vang homologue9, a gene that is broadly expressed during early development9, as a candidate for tri (see Supplementary Information, Fig. S1). Analysis of the trim209, trim747, tritc240a and tritk50f chemically-induced alleles10,11 demonstrated that each contains a disruption in stbm (Fig. 1a). In trim747, a nonsense mutation at Ser 427 is predicted to shorten Stbm by 93 amino acids. The trim209 allele harbours a 13 base pair insertion of intronic sequence, resulting in a frameshift at Ala 441 and premature termination of translation. The tritc240a allele carries a 39 base pair insertion, resulting in the in-frame addition of 13 amino acids at Arg 21. Transcripts for stbm were not detected in tritk50f homozygous mutant embryos and genomic PCR indicated that at least part of the coding sequence is deleted (data not shown). To confirm that stbm represents the tri gene, we injected stbm RNA into embryos produced by tri heterozygous parents and scored its effect on the mutant phenotype. Partial suppression of the convergence and extension defect was observed in all mutant embryos (n = 75) at 1 day post-fertilization (Fig. 1b–d). In contrast, mutated stbm RNA encoded by the trim747 allele did not affect wild-type or tri phenotypes (data not shown), suggesting this is a loss-of-function and likely a null mutation. A hallmark of genes controlling tissue or planar cell polarity (PCP) is that their increased activity also impairs gastrulation8,12–14. Overexpression of stbm RNA in wild-type embryos strongly inhibited convergence and extension, as indicated by shorter embryonic axes, synopthalmia, and dorsal flexure (Fig. 1e), effects resembling interference with Dishevelled (Dsh) function12 and tri mutant phenotypes1,4. In loss-of-function experiments, injection of stbm antisense morpholino (MO) oligonucleotides15 caused a modest convergence and extension defect in 95% of wild-type embryos (n = 374; Fig. 1f), consistent with previous experiments in zebrafish9 and Xenopus laevis8. Injection of stbm MO into tri embryos enhanced the convergence and extension defect, suggesting that maternally-derived tri (ref. 9) influences gastrulation movements (Fig. 1g). Moreover, the tri motor neuron migration defect was also phenocopied by injection of stbm MO into wild-type embryos (62%, n = 262; Fig. 1h–j). Conversely, injection of stbm RNA into tri embryos frequently restored wild-type motor neuron migration (33%, n = 43; Fig. 1k–m). Together, these results establish that genetic lesions in a zebrafish stbm homologue cause the tri phenotype and hereafter we will refer to this gene as tri.
Figure 1. tri encodes a Stbm homologue.
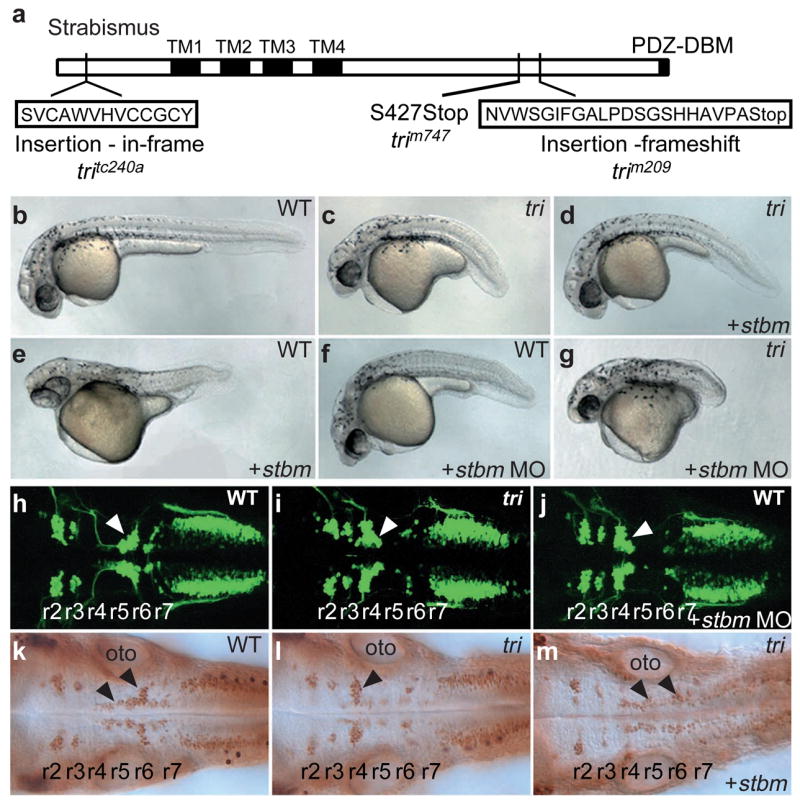
a, A schematic representation of Stbm, showing mutations in three tri alleles (TM, putative transmembrane domains; PDZ-DBM, putative PDZ-domain binding motif). b–g, Stbm controls convergence and extension cell movements. Wild-type (b) and trim209 (c) embryos at 1 day post-fertilization. Injection of stbm RNA partially suppresses the tri convergence and extension defect (d). Wild-type embryos injected with stbm RNA (e) or stbm MO (f) show inhibited convergence and extension. g, stbm MO enhances the convergence and extension defect in tri mutant embryos. h–j, stbm MO phenocopies tri neuronal migration phenotype. Embryos at 36 h post-fertilization with gfp expression in branchiomotor neurons under control of the islet1 promoter (dorsal views). Motor neurons (arrowheads) induced in rhombomere 4 (r4) migrate into r6 and r7 in wild-type (h) but remain in r4 in trim209 embryos (i) and wild-type embryos injected with stbm MO (j). k–m, Stbm suppresses tri neuronal migration defect. Embryos at 36 h post-fertilization with anti-Islet antibody-labelled branchiomotor neurons. Motor neurons migrated into r6 in wild-type (k) and trim209 embryos injected with stbm RNA (m), but not in trim209 (l) embryos. oto, otocyst.
In Drosophila, stbm/vang is instructive during the establishment of cell fates within ommatidia6,7 and genetically interacts with the PCP pathway to establish wing tissue polarity6. In vertebrates, Stbm/Vang was proposed to function downstream of Wnt and Frizzled to activate the PCP pathway while antagonizing canonical Wnt/β-catenin signalling, and thus anteroposterior neural patterning16. However, there are conflicting reports that argue for and against an involvement of mouse, fish and frog stbm in neural patterning8,9,17,18. Previous analyses of tri phenotypes did not identify anteroposterior neural patterning defects1–5, a conclusion we have extended here for simultaneous interference with maternal and zygotic tri functions (Supplementary Information, Fig. S2). Given the evidence implicating canonical Wnt signalling as the key regulator of anteroposterior neural patterning in vertebrates19–21, we argue that Tri/Stbm is neither required nor capable of significantly modulating canonical Wnt signalling in zebrafish gastrulae9,18.
With regard to the PCP pathway, genetic epistasis experiments identified functional interactions between tri and silberblick (slb/wnt11)22 and knypek (glypican 4/6)3 PCP signalling mutants, suggesting they function in the same or parallel pathways during convergence and extension. However, we were unable to suppress the tri convergence and extension defect by injecting either dsh-ΔN or rho kinase 2 RNAs (Supplementary Information, Fig. S3), downstream mediators of PCP signalling that are capable of suppressing slb gastrulation defects13,23. Moreover, neither excess tri function, nor its reduction with MOs could suppress the slbtz216 gastrulation mutant phenotype (Supplementary Information, Fig. S3). We next showed that although overexpression of tri did not alter the intra-cellular distribution of Rho kinase 2 (ref. 23), it did interfere with the ability of excess Wnt11 to do so (Supplementary Information, Fig. S3). Together, these results suggest Tri/Stbm can modulate PCP signalling, but that it is not simply a positive or negative component of a linear PCP pathway.
Although interference with Tri/Stbm function inhibits convergence and extension in fish and frogs2,8,9, the cellular basis of this defect is unknown. We asked whether tri mediates mediolateral cell elongation, a behaviour critical for cell intercalations underlying convergence and extension movements in the dorsal regions of late zebrafish gastrulae24,25 and dependent on PCP pathway components23,26,27. Using confocal microscopy, we demonstrated that the length-to-width ratio (LWR) of paraxial ectodermal cells is reduced in tri gastrulae, compared to wild type (Fig. 2a–d and Table 1). The mediolateral alignment (MLA) of paraxial ectodermal cells is also reduced in tri embryos (Fig. 2a,e and Table 1). To test the cell-autonomy of tri function in convergence and extension, we performed transplantations at the blastula stage and assessed the morphology of donor-derived paraxial ectodermal cells at late gastrulation. Wild-type cells transplanted into wild-type hosts were elongated and mediolaterally aligned (Fig. 2d,e and Table 1). In tri hosts, wild-type cells were rounded and showed no mediolateral bias (Fig. 2d–f and Table 1). Labelled tri cells also did not elongate or mediolaterally align in wild-type hosts (Fig. 2d,e,g and Table 1). The lack of elongation and alignment of wild-type and tri donor-derived ectodermal cells was similar to that observed for tri mutant ectodermal cells (Table 1). The inability of tri cells to behave normally in a wild-type environment indicates that tri functions autonomously in cell polarization. Moreover, failure of wild-type cells to elongate and mediolaterally align in tri embryos demonstrates that tri function also has a non-autonomous component. Therefore, as proposed for Drosophila Stbm/Vang14, Tri/Stbm seems to influence intercellular communication that is essential for the establishment of tissue polarity during gastrulation.
Figure 2. tri functions autonomously and non-autonomously to control cell polarity.
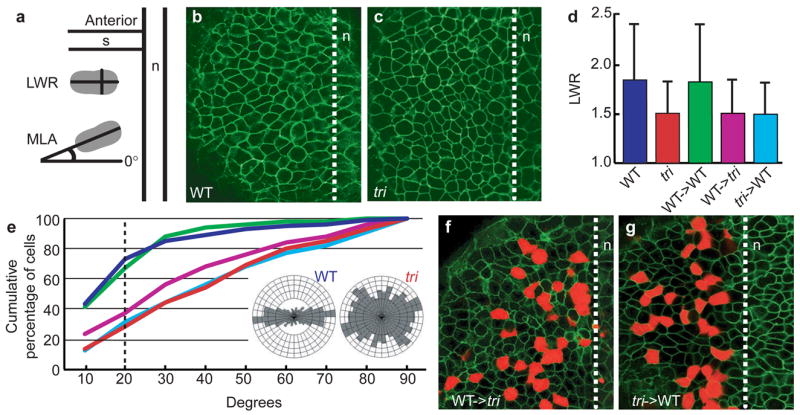
a, A schematic representation of the methods used to measure cell elongation (LWR) and mediolateral alignment (MLA) relative to the notochord (n). Wild-type (b) and tri (c) ectodermal cells labelled with membrane-localized GFP (green). d, LWRs of wild-type and tri paraxial ectodermal cells and donor cells (wild-type->wild-type; wild-type->tri; tri->wild-type) with their standard deviations. e, Cumulative percentage of mediolaterally aligned paraxial ectodermal cells as a function of their angle, relative to a line perpendicular to the notochord. Graph line colours correspond to bar graph colours in d. Dashed line indicates sector ± 20° perpendicular to the notochord27. Rose diagrams (insets) depict cell orientations in wild-type and tri paraxial ectoderm of late gastrulae. The mediolateral axis corresponds to the horizontal plane (0°), perpendicular to the notochord, and the antero-posterior axis is aligned vertically (90°). Transplanted wild-type (f) and tri (g) donor-derived ectodermal cells (red) surrounded by host ectoderm (green) are shown. The dashed lines in b,c,f and g indicate notochord boundary. s, somite.
Table 1.
Mediolateral elongation of ectodermal cells requires autonomous and non-autonomous tri function
| Genotype | LWR | MLA (% ± 20°) | cells/embryos (n) | |
|---|---|---|---|---|
| Donor | Host | |||
| WT* | 1.85 ± 0.56 | 73 | 274/4 | |
| tri* | 1.51 ± 0.31a | 27e | 142/4 | |
| WT | WT | 1.83 ± 0.58b | 66f | 47/3 |
| WT | tri | 1.51 ± 0.33c | 37g | 91/7 |
| Tri | WT | 1.50 ± 0.31d | 30h | 116/4 |
control embryos
vs. WT, significantly different at P <0.0001
vs. WT, not significantly different at P > 0.5
vs. WT, P < 0.0001; vs. tri, P > 0.5
vs. WT, P < 0.0001; vs. tri, P > 0.5
vs. WT, significantly different at P < 0.0001
vs. WT, P > 0.5
vs. WT, P < 0.0001; vs. tri, not significantly different at P > 0.1
vs. WT, P < 0.0001; vs. tri, P > 0.5
In zebrafish gastrulae, lateral mesodermal and ectodermal cell populations move dorsally at increasing speed2,24 (Fig. 3a). The acceleration of convergence movements is compromised in tri mutant embryos2. We used time-lapse Nomarski imaging to identify specific cell behaviours underlying the increased rates of dorsal convergence and asked whether these behaviours require tri function. At mid-gastrulation, we observed that wild-type lateral mesodermal cells are not elongated and migrate as individuals with a slow net dorsal speed along indirect paths (Fig. 3b–d and Table 2). At this stage, the morphology and behaviour of tri cells does not differ from wild type (Fig. 3b,c,e and Table 2). By late gastrulation, wild-type mesodermal cells are significantly more elongated and migrate dorsally at increased net speeds with more direct trajectories (Fig. 3b,c,f and Table 2; see also Supplementary Information, Movie S1). By contrast, late gastrulation tri cells are more rounded and move with a significantly reduced net dorsal speed along less direct trajectories when compared with their wild-type counterparts (Fig. 3b,c,g and Table 2; see also Supplementary Information, Movie S2). Although increasing rates of convergence have been observed in gastrulae of the teleost fish, Fundulus heteroclitus28, the mediolateral lengthening of zebrafish lateral mesodermal cells associated with their increased net dorsal velocities represents a new behaviour. We propose that tri-dependent mediolateral cell polarization is required for persistent dorsal migration of cells along straight paths and consequently for the increased velocities of their convergence movements.
Figure 3. tri is required for the increased net speed of directed dorsal migration.
a, Domains of convergence and extension cell movements in zebrafish gastrulae24. Yellow arrows in dorsal region indicate strong extension movements with little convergence. Light and dark blue arrows indicate domains of slow and fast convergence and extension, respectively. b, LWR of lateral mesodermal cells in wild-type and tri embryos, and tri embryos injected with tri MO. c, Total and net dorsal migration speeds of lateral mesodermal cells at 80% epiboly, yolk-plug closure (YPC)-tailbud (TB), and TB-1 somite stages. d–g, Shape changes and dorsal migration trajectories of lateral mesodermal cells. Colours match domains of slow and fast convergence and extension movement in a. D, dorsal. V, ventral. Scale bars represent 10 μm.
Table 2.
The net dorsal migration speed of wild-type lateral mesodermal cells increases as cells become mediolaterally elongated.
| Genotype | Stage | LWR | Cells/embryos (n) | Total speed (μm h−1) | Net dorsal speed (μm h−1) | Cells/embryos (n) |
|---|---|---|---|---|---|---|
| WT | 80% | 1.40 ± 0.28 | 182/4 | 168 ± 41 | 81 ± 21 | 209/6 |
| tri | 80% | 1.45 ± 0.26a | 103/2 | – | – | – |
| tri+MO | 80% | 1.34 ± 0.25b | 100/2 | 168 ± 30 | 89 ± 30f | 62/2 |
| WT | TB-1s | 1.74 ± 0.45c | 402/13 | 133 ± 25 | 105 ± 22g | 32/2 |
| tri | TB-1s | 1.50 ± 0.33d | 88/2 | 76 ± 8 | 53 ± 6h | 22/1 |
| tri+MO | TB-1s | 1.40 ± 0.26e | 28/1 | 159 ± 12 | 5 ± 7 | 21/1 |
vs. WT at 80%, not significantly different at P > 0.2
vs. WT at 80%, not significantly different at P > 0.07
vs. WT at 80%, significantly different at p < 0.0001
vs. WT at TB-1s, P < 0.0001
vs. WT at TB-1s, P < 0.0001
vs. WT at 80%, not significantly different at P > 0.3
vs. WT at 80%, P < 0.0001
vs. WT at TB-1s, P < 0.0001
Similar to the movements of lateral gastrula cells, facial (nVII) hindbrain branchiomotor neurons undergo directed migration from rhombomere 4 (r4) into r6 and r7 (refs 29,30). In tri mutant embryos, motor neurons induced in r4 fail to migrate tangentially into more posterior rhombomeres (see Supplementary Information, Movies S3 and S4), whereas radial migration of the motor neurons is unaffected5. We used mosaic analysis31 to ask whether tri functions cell-autonomously in this process. About 75% of wild-type motor neurons migrated normally out of r4 into r6 and r7 after transplantation into wild-type hosts (n = 175 cells, 22 embryos; Fig. 4a,d). In contrast, wild-type motor neurons failed to migrate out of r4 in tri hosts (n = 66 cells, 11 embryos; Fig. 4b,d), identifying a non-cell-autonomous role for tri in the surrounding hindbrain tissue. In the reciprocal experiment, 33% of tri motor neurons migrated normally into r6 and r7 in wild-type hosts (n = 90 cells, 12 embryos; Fig. 4c,d) indicating that the tri phenotype can be rescued by the wild-type environment. However, as a high percentage of tri motor neurons failed to migrate in the wild-type environment, we suggest that tri function also has a cell-autonomous component during tangential migration of hindbrain motor neurons. Cumulatively, these experiments indicate that Tri/Stbm is required for cellular interactions that mediate migratory events in the gastrula and hindbrain.
Figure 4. tri functions autonomously and non-autonomously during tangential neuronal migration.
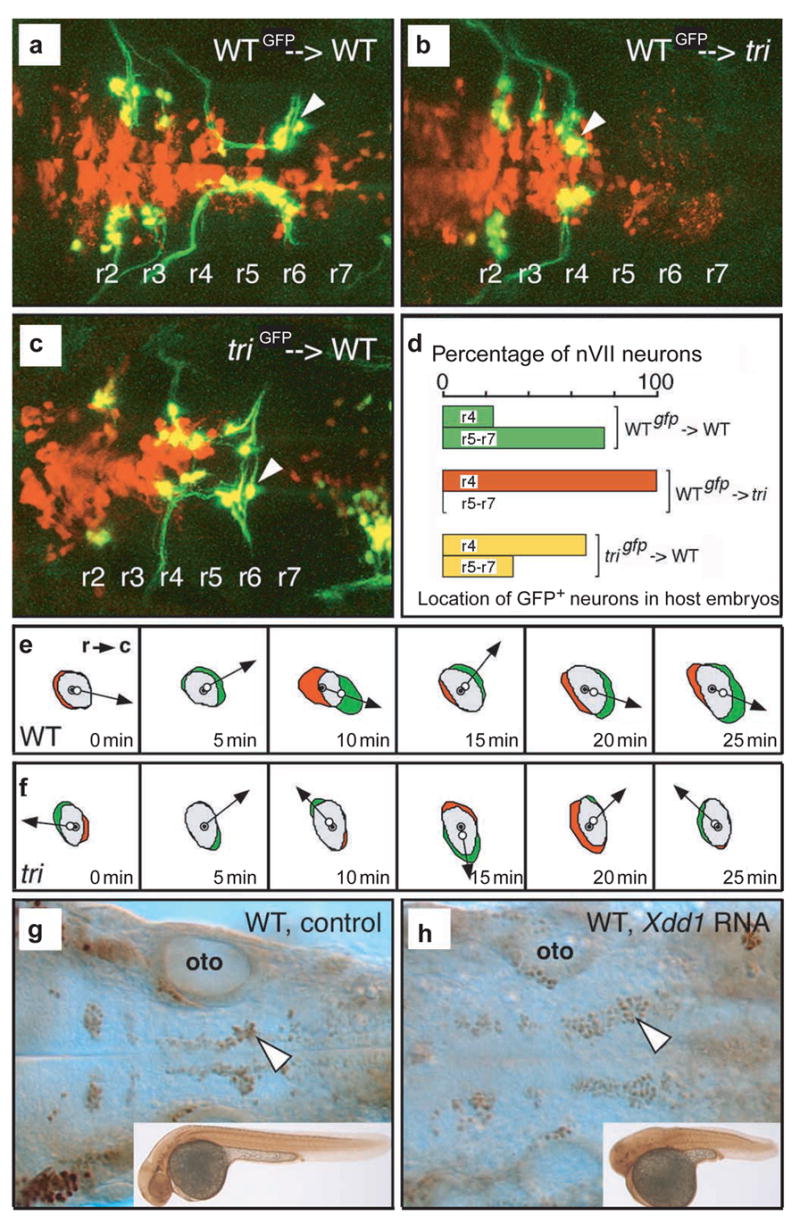
a–c, Dorsal views of hindbrain in unlabelled host embryos containing transplanted rhodamine–dextran-labelled donor cells (red), some of which differentiated into GFP-expressing motor neurons (green/yellow). Wild-type motor neurons (arrowhead) migrated out of r4 and into r6 in a wild-type host (a), but not in a tritc240a host (b). tritc240a motor neurons (arrowhead) migrated into r6 in a wild-type host (c). d, A quantitative summary of the transplantation data. e,f, Representative behaviours of wild-type and tritc240a motor neurons during a 25-min time-lapse recording. Red indicates area of retraction from previous cell position (closed circle marks cell centre) and green indicates area of expansion in current cell position (open circle marks cell centre). Arrows point in direction of cell movement. r, rostral; c, caudal. e, Wild-type motor neurons show biased caudal migration. f, tritc240a motor neurons meander showing no directionality. g,h, Dorsal views of wild-type hindbrains labelled with an Islet antibody. Motor neurons migrated into r6, posterior to the otocyst (oto), in control wild-type embryos (inset; g) and in Xdd1-injected wild-type embryos (h) showing a strong convergence and extension defect (inset).
We used time-lapse analysis of green fluorescent protein (GFP)-expressing nVII motor neurons to identify the cellular basis of the neuronal migration defect5. Whereas wild-type and tri neurons have similar LWRs (wild-type = 1.5 ± 0.2, n = 9; tri = 1.6 ± 0.3, n = 10; p = 0.32), tri neurons move slower (wild-type = 19.4 ± 6.7 μm h−1, n = 9; tri = 11.9 ± 2.5 μm h−1, n = 10; p = 0.01) and in random directions. Further analyses demonstrated that wild-type cells have caudally-biased expansion and rostrally-biased retraction zones (Fig. 4e), resulting in effective caudal translocation of cell bodies. In contrast, expansion and retraction zones form randomly around the periphery of tri neurons (Fig. 4f), producing insignificant caudal movement (wild-type = 11.3 ± 9.3 μm h−1, n = 9; tri = 1.6 ± 3.2 μm h−1, n = 10; p = 0.02). As dynamic changes in cell area are similar between wild-type and tri neurons (wild-type = 3.5 ± 0.8 μm2min−1, n = 9; tri = 3.1 ± 0.9 μm2min−1, n = 10; p = 0.32), we propose that tri neurons either cannot stabilize their protrusions or cannot polarize their orientation. Interestingly there is strong expression of Ltap, the mouse stbm homologue most similar to tri, in hindbrain r4 (ref. 32), suggesting a possible role for the mouse protein in tangential motor neuron migration. Taken together, our studies suggest that Tri/Stbm confers directionality on both gastrula and tangential neuronal cell migration.
Previously, it was shown that disruption of knypek (glypican 4/6), a positive modulator of PCP signalling that is broadly expressed in zebrafish embryos at the time of motor neuron migration27, had no effect on tangential migration of motor neurons5. This suggested that these movements may occur independently of PCP signalling. Consistently, facial (nVII) motor neurons migrate normally in slb (wnt11; n = 19) and pipetail (wnt5a; n = 53) mutant embryos (data not shown). As Tri/Stbm can bind to Dsh in vitro9, we examined motor neuron migration in wild-type embryos expressing Xdd1, a PCP-specific dominant-negative Dsh12. Ubiquitous overexpression of Myc-tagged xdd1 in zebrafish embryos through 1-day post-fertilization caused severe convergence and extension defects that are consistent with previous results13, but did not impair neuronal migration (n = 77; Fig. 4g,h and data not shown). Similarly, although tri expression can rescue neuronal migration in tri mutant embryos, tri overexpression in wild-type embryos caused severe convergence and extension defects without affecting motor neuron migration (n = 54; data not shown). These results suggest that Tri/Stbm may control tangential migration of hindbrain neurons through a Dsh-independent pathway distinct from non-canonical Wnt signalling.
It is noteworthy that Tri/Stbm has been employed by vertebrate embryos to control mediolateral intercalation during gastrulation and such distinct movement behaviours as directed migration of gastrula cells and hindbrain motor neurons. One question is whether there is a common role for Tri/Stbm in the regulation of these distinct cell behaviours. During gastrulation, tri function is required for cell polarization associated with intercalation and directed migration at increasing velocities. By contrast, migrating motor neurons do not exhibit an elongated morphology. However, in the absence of tri function, gastrula cells are unable to converge dorsally along straight trajectories and motor neurons move their cell bodies in random directions, rather then exclusively towards posterior rhombomeres. We propose that Tri/Stbm confers directionality on cell intercalation and migration of gastrula cells and hindbrain motor neurons, affording effective movement towards their targets. Disruption of the tri homologue Ltap in Loop-tail mice results in severe spina bifida17,32. It will be important to determine whether neural tube closure defects are caused by disrupted gastrulation movements, or whether they reflect later Ltap functions in neurulation-specific cell behaviours.
There is considerable evidence that several Wnt signalling pathways, including canonical Wnt/β-catenin and non-canonical Wnt/calcium and PCP, regulate morphogenesis during vertebrate development33. Among these, the PCP pathway mediates medio-lateral cell elongation that underlies convergence and extension movements23,26,27. Inhibition of convergence and extension movements in frog and fish embryos overexpressing stbm or injected with stbm MOs8,9 suggests that Tri/Stbm is a mediator of tissue polarity. Our finding that inactivation of tri in zebrafish gastrulae impairs mediolateral cell elongation provides the first genetic evidence that homologous molecules underlie tissue polarity in flies and vertebrates. Genetic epistasis studies in Drosophila suggest that Stbm/Vang functions within, or parallel to, the Frizzled/PCP pathway14. In vertebrates, Tri/Stbm was shown to associate with Dsh in vitro, to recruit Dsh to the membrane in vivo and to promote c-Jun phosphorylation, suggesting that it functions as a positive component of PCP signalling9. Our results support the notion that the relationship between Tri/Stbm and the PCP pathway is not simply quantitative, but rather that Tri/Stbm modulates PCP signalling, perhaps functioning in a parallel pathway to control cell polarity underlying cell movements during gastrulation. It is possible that Tri/Stbm cooperates with a vertebrate homologue of the Drosophila Prickle protein in a feedback amplification mechanism similar to that proposed for wing tissue polarity34. However, the interaction between stbm/vang and prickle is complex and other data from Drosophila suggests these genes may be antagonists6. Moreover, our data suggests Tri/Stbm may mediate motor neuron migration independently of PCP signalling, thus providing an entry point to elucidate the molecular basis of this class of neuronal migration. This study underscores the complexity of genetic interactions Tri/Stbm engages in to establish tissue polarity and implicates this protein in the regulation of distinct movement behaviours during vertebrate development.
Methods
Zebrafish maintenance and staging
Zebrafish embryos were produced by natural matings and adults maintained as described1. Embryo staging was performed according to morphology as described35.
Genetic mapping and positional cloning
Detailed methods are described in Supplementary Information. The stbm coding sequence was amplified by reverse transcription (RT)-PCR from trim209, trim747 and tritc240a homozygous mutant alleles and then directly sequenced.
Morpholino and RNA injections
Injections were performed at the one-cell stage as described3,15. The stbm/tri MO was obtained from Gene Tools, LLC (Philomath, OR), and was previously described9. The tri/stbm and mutated trim747 open reading frames were subcloned into an expression vector and verified by sequencing. Sense-capped RNA was synthesized using mMessage mMachine (Ambion, Austin, TX) after template linearization. For phenotype rescue and phenocopy experiments, 5–50 pg tri RNA and 1–4 ng tri MO were used per embryo. Injection of greater than 8 ng of MO caused increasing central nervous system degeneration commonly observed with MO injections15. Embryos scored for suppression of the tri phenotype were genotyped by PCR using the simple sequence length polymorphism marker Z17411.
Immunostaining
Islet antibody staining and GFP imaging were performed as described5 and phenylthiourea (0.003% w/v) was used to inhibit pigmentation. Expression of Myc-tagged Xdd1 was detected using an anti-Myc antibody (Calbiochem, San Diego, CA) as described12, and imaged using a Zeiss LSM 510 laser scanning inverted confocal microscope (Zeiss, Thornwood, NY).
Transplantation experiments
To determine the autonomy of tri function during gastrulation, host embryos were injected with RNA encoding membrane-localized GFP26 and donor embryos with rhodamine–dextran (Molecular Probes, Eugene, OR) and RNA encoding membrane GFP at the one-cell stage. Cells were extracted from donor embryos at the 1000-cell stage and injected close to the margin of dome stage hosts using standard methods36. At tailbud-2 somite stages, embryos were mounted in 1.5% methyl cellulose and imaged using a Zeiss LSM 510 confocal microscope. z-series images of donor-derived ectodermal cells were acquired at 2-μm intervals. Determination of cell orientation, LWR and statistical analyses were performed as described27. The cell-autonomy of tri function during motor neuron migration was determined as described31.
Gastrula and neuronal cell movement analyses
Time-lapse analyses of lateral gastrula mesodermal cells were performed as described24 and movie images were collected at 30-s intervals. LWRs were measured from single-frame Nomarski images using Object-Image (Norbert Vischer) and calculated using Microsoft Excel software. Statistical analyses were performed using Student’s t-test. Our observations indicate that cell behaviours driving convergence and extension movements change continuously. Thus, our methods do not provide a comprehensive description of cell behaviours observed between 80% epiboly and the end of gastrulation. GFP-expressing motor neurons in wild-type and tri embryos were imaged at 200× magnification on an Olympus BX60 microscope (Melville, NY). Time-lapse recordings were made using Cytos software (Applied Scientific Instrumentation, Eugene, OR). Image processing and analysis was performed using DIAS software (Solltech, Iowa City, IA).
Supplementary Material
Acknowledgments
We thank B. Appel, R. Blakely, A. Schier, and C.V.E. Wright for critical comments. We thank J. Clanton and C. Baccam for excellent fish care, M. Halpern for γ-ray-mutagenized fish, M. Westerfield, H. Takeda, P. Ingham, M. Ekker, Y. Grinblat, B. Thisse, C. Thisse, E. Weinberg and T. Jowett for probes, R. Harland, S. Sokol and M. Tada for constructs, and C.-P. Heisenberg and H. Okamoto for fish. S.B. and A.C. are indebted to K. Cooper and C. Moens for invaluable guidance in transplantation procedures. The Zeiss confocal microscope is supported by National Insitutes of Health (NIH) grant 1S10RR015682. J.R.J. and D.S.S. are supported by a National Institutes of Health Vascular Biology Training Grant (T32HL07751). S.B. is supported by a NSF-Missouri’s Alliance for Graduate Education and the Professoriate (MAGEP) fellowship and A.C. by NIH grant NS40449. L.S.K. is supported by NIH grant GM55101 and Pew Scholars Program in the Biomedical Sciences.
Footnotes
Supplementary Information is available on Nature Cell Biology’s website (http://cellbio.nature.com).
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
References
- 1.Solnica-Krezel L, et al. Development. 1996;123:67–80. doi: 10.1242/dev.123.1.67. [DOI] [PubMed] [Google Scholar]
- 2.Sepich DS, et al. Genesis. 2000;27:159–173. doi: 10.1002/1526-968x(200008)27:4<159::aid-gene50>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 3.Marlow F, et al. Dev Biol. 1998;203:382–399. doi: 10.1006/dbio.1998.9032. [DOI] [PubMed] [Google Scholar]
- 4.Hammerschmidt M, et al. Development. 1996;123:143–151. doi: 10.1242/dev.123.1.143. [DOI] [PubMed] [Google Scholar]
- 5.Bingham S, Higashijima S, Okamoto H, Chandrasekhar A. Dev Biol. 2002;242:149–160. doi: 10.1006/dbio.2001.0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor J, Abramova N, Charlton J, Adler PN. Genetics. 1998;150:199–210. doi: 10.1093/genetics/150.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolff T, Rubin GM. Development. 1998;125:1149–1159. doi: 10.1242/dev.125.6.1149. [DOI] [PubMed] [Google Scholar]
- 8.Darken RS, et al. EMBO J. 2002;21:976–985. doi: 10.1093/emboj/21.5.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park M, Moon RT. Nature Cell Biol. 2002;4:20–25. doi: 10.1038/ncb716. [DOI] [PubMed] [Google Scholar]
- 10.Haffter P, et al. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 11.Driever W, et al. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- 12.Sokol SY. Curr Biol. 1996;6:1456–1467. doi: 10.1016/s0960-9822(96)00750-6. [DOI] [PubMed] [Google Scholar]
- 13.Heisenberg CP, et al. Nature. 2000;405:76–81. doi: 10.1038/35011068. [DOI] [PubMed] [Google Scholar]
- 14.Adler PN, Lee H. Curr Opin Cell Biol. 2001;13:635–640. doi: 10.1016/s0955-0674(00)00263-5. [DOI] [PubMed] [Google Scholar]
- 15.Nasevicius A, Ekker SC. Nature Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 16.Heisenberg CP, Tada M. Curr Biol. 2002;12:R126–R128. doi: 10.1016/s0960-9822(02)00704-2. [DOI] [PubMed] [Google Scholar]
- 17.Murdoch JN, Doudney K, Paternotte C, Copp AJ, Stanier P. Hum Mol Genet. 2001;10:2593–2601. doi: 10.1093/hmg/10.22.2593. [DOI] [PubMed] [Google Scholar]
- 18.Park M, Moon RT. Nature Cell Biol. 2002;4:467. doi: 10.1038/ncb716. [DOI] [PubMed] [Google Scholar]
- 19.Lekven AC, Thorpe CJ, Waxman JS, Moon RT. Dev Cell. 2001;1:103–114. doi: 10.1016/s1534-5807(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 20.Erter CE, Wilm TP, Basler N, Wright CV, Solnica-Krezel L. Development. 2001;128:3571–3583. doi: 10.1242/dev.128.18.3571. [DOI] [PubMed] [Google Scholar]
- 21.Wilson SW, Rubenstein JL. Neuron. 2000;28:641–651. doi: 10.1016/s0896-6273(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 22.Heisenberg CP, Nusslein-Volhard C. Dev Biol. 1997;184:85–94. doi: 10.1006/dbio.1997.8511. [DOI] [PubMed] [Google Scholar]
- 23.Marlow F, Topczewski J, Sepich DS, Solnica-Krezel L. Curr Biol. 2002;12:876–884. doi: 10.1016/s0960-9822(02)00864-3. [DOI] [PubMed] [Google Scholar]
- 24.Myers DC, Sepich DS, Solnica-Krezel L. Dev Biol. 2002;243:81–98. doi: 10.1006/dbio.2001.0523. [DOI] [PubMed] [Google Scholar]
- 25.Concha ML, Adams RJ. Development. 1998;125:983–994. doi: 10.1242/dev.125.6.983. [DOI] [PubMed] [Google Scholar]
- 26.Wallingford JB, et al. Nature. 2000;405:81–85. doi: 10.1038/35011077. [DOI] [PubMed] [Google Scholar]
- 27.Topczewski J, et al. Dev Cell. 2001;1:251–264. doi: 10.1016/s1534-5807(01)00005-3. [DOI] [PubMed] [Google Scholar]
- 28.Trinkaus JP. J Exp Zool. 1998;281:328–335. doi: 10.1002/(sici)1097-010x(19980701)281:4<328::aid-jez7>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 29.Chandrasekhar A, Moens CB, Warren JT, Jr, Kimmel CB, Kuwada JY. Development. 1997;124:2633–2644. doi: 10.1242/dev.124.13.2633. [DOI] [PubMed] [Google Scholar]
- 30.Higashijima S, Hotta Y, Okamoto H. J Neurosci. 2000;20:206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moens CB, Fritz A. Methods Cell Biol. 1999;59:253–272. doi: 10.1016/s0091-679x(08)61829-4. [DOI] [PubMed] [Google Scholar]
- 32.Kibar Z, et al. Nature Genet. 2001;28:251–255. doi: 10.1038/90081. [DOI] [PubMed] [Google Scholar]
- 33.Peifer M, McEwen DG. Cell. 2002;109:271–274. doi: 10.1016/s0092-8674(02)00739-0. [DOI] [PubMed] [Google Scholar]
- 34.Tree DRP, et al. Cell. 2002;109:371–381. [Google Scholar]
- 35.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 36.Westerfield M. The zebrafish book. University Oregon Press; Eugene: 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



