XAC1151, a small heat-shock protein from X. axonopodis pv. citri belonging to the α-crystallin family, was crystallized using the sitting-drop vapour-diffusion method in the presence of ammonium phosphate. X-ray diffraction data were collected to 1.65 Å resolution using a synchrotron-radiation source.
Keywords: XAC1151, small heat-shock proteins, α-crystallins, Xanthomonas axonopodis
Abstract
The hspA gene (XAC1151) from Xanthomonas axonopodis pv. citri encodes a protein of 158 amino acids that belongs to the small heat-shock protein (sHSP) family of proteins. These proteins function as molecular chaperones by preventing protein aggregation. The protein was crystallized using the sitting-drop vapour-diffusion method in the presence of ammonium phosphate. X-ray diffraction data were collected to 1.65 Å resolution using a synchrotron-radiation source. The crystal belongs to the rhombohedral space group R3, with unit-cell parameters a = b = 128.7, c = 55.3 Å. The crystal structure was solved by molecular-replacement methods. Structure refinement is in progress.
1. Introduction
Xanthomonas axonopodis pv. citri (Xac) is a phytopathogenic bacterium that causes citrus canker disease in most commercial citrus cultivars, resulting in significant losses worldwide. Symptoms of infected plants include canker lesions on fruits and leaves associated with progressive tree decline. Sequencing and annotation of the Xac genome (da Silva et al., 2002 ▶) allowed us to initiate structural and functional studies of genes and proteins in order to elucidate the mechanisms involved in plant–pathogen interaction.
In X. axonopodis pv. citri, the hspA gene (XAC1151) encodes a small heat-shock protein (sHSP) with 158 amino-acid residues. This protein shows high identity to sHSPs from various species from the genus Xanthomonas and also to another citrus pathogen Xylella fastidiosa (strain 9a5c).
Heat-shock proteins (HSPs), also called stress proteins or molecular chaperones, are a superfamily of proteins that are present in various organisms. They are essential for normal cell function and confer stability on the cell proteome by protecting a diverse group of proteins engaged in signal transduction, metabolism, translation and other activities, improving the resistance of the cell under stress conditions. However, HSPs are also present under normal cell conditions (Sun & MacRae, 2005 ▶). HSP synthesis is stimulated in response to environmental challenges when organisms are exposed to abnormal or extreme environmental stress conditions such as heat shock, desiccation, humidity, light, starvation, chemical stress caused by organic compounds or strong oxidants, oxygen deprivation, exposure to heavy metals or oxidative stress (Laksanalamai & Robb, 2004 ▶).
Protein folding and refolding are mediated by ATP-dependent molecular-chaperones, including HSP60 (chaperonins), HSP70, HSP90, HSP104/C1pb and HSP110. The sHSPs appear to prevent irreversible protein aggregation and insolubilization of unfolded proteins, without chaperone activity, in an ATP-independent process. They have a high capacity to bind unfolded proteins and deliver them to the ATP-dependent chaperone systems (Sun & MacRae, 2005 ▶). Members of this protein family have a central conserved domain of approximately 90 amino-acid residues called the α-crystallin domain (Kappe et al., 2002 ▶). Furthermore, the sHSP family members share common features such as small molecular weight (12–30 kDa) and the formation of large oligomers (9–30 subunits), resulting in a dynamic quaternary structure (Stromer et al., 2003 ▶).
The amino-acid sequences of the N- and C-termini are not conserved and contribute to the structural diversity among sHSPs. Both ends are essential for multimerization and molecular-chaperone activity (Sun & MacRae, 2005 ▶; Ganea, 2001 ▶). αA-Crystallin, a classical member of the sHSP protein family, is a large protein composed of 30–40 identical subunits. Deletion of the first 19 N-terminal amino-acid residues showed little effect on the quaternary structure, while removal of the first 56 or more N-terminal residues converts αA-crystallin into a predominantly small multimeric complex with 3–4 subunits, suggesting an essential function for the N-terminus in oligomerization (Bova et al., 2000 ▶). The C-terminal extension is flexible and is enriched in polar and charged amino-acid residues that contribute to the solubility of the sHSP (Ganea, 2001 ▶). Loss of chaperone activity was observed when the last 11–12 residues from the rat α-crystallin C-terminus were removed with calpain II (Kelley et al., 1993 ▶). The importance of the C-terminal end to the chaperone activity was confirmed by using Escherichia coli IbpB, another member of the sHSP protein family. A truncated protein lacking the last 11 residues was produced in E. coli, which showed loss of chaperone activity, although the protein was able to dimerize (Jiao et al., 2005 ▶).
In this report, we describe the cloning, purification, crystallization, data collection and preliminary X-ray diffraction analysis of a recombinant sHSP encoded by the gene hspA from X. axonopodis pv. citri.
2. Cloning, expression and purification
The full-length hspA gene (XAC1151) was amplified by PCR from the genomic DNA of X. axonopodis pv. citri strain 306 using the primers XAC1151F (5′-CGGAATTCATGAACATCGTTCGTTAT-3′) and XAC1151R (5′-CCCAAGCTTTTACTGCACCGTGCTGCC-3′), which were designed based on the genome sequence (da Silva et al., 2002 ▶). The sequences in bold correspond to EcoRI and HindIII sites, respectively. The PCR product was subcloned into the pMOS-Blue PCR cloning kit (Amersham Bioscience) and the whole sequence was confirmed by DNA sequencing. The insert was then transferred to the pET28a expression vector (Novagen) using EcoRI/HindIII restriction sites and the plasmid was used to transform E. coli strain BL21 (DE3) competent cells. A single clone of BL21 (DE3) cells harbouring the hspA gene was grown overnight in 2YT medium containing 0.2% glucose and 30 µg ml−1 kanamycin at 303 K and 250 rev min−1. Cells were transferred to 2 l fresh medium and cultivated under the same conditions until the OD600 reached 0.6. Protein production was induced with 0.1 mM IPTG and cells were harvested after 4 h induction. Cells were resuspended in 50 ml lysis buffer containing 50 mM Tris–HCl pH 8.0, 500 mM NaCl, 30 mM imidazole and 1 mM benzamidine and lysed by sonication (ten cycles of 30 s sonication followed by 30 s on ice). Cell debris and nucleic acids was separated by centrifugation at 30 000g for 30 min and the supernatant was applied onto a nickel-affinity column in an Äkta Prime Purification System (Amersham Bioscience). Recombinant protein was eluted in a linear elution gradient of 0–500 mM imidazole and the fractions were analyzed by SDS–PAGE. Fractions containing high amounts of the pure protein were pooled and dialyzed against 5 mM Tris–HCl buffer pH 8.0. Recombinant sHSP from X. axonopodis pv. citri was overexpressed in E. coli in a soluble form with a yield of ∼15 mg purified protein per litre of culture. Single-step purification by immobilized metal-affinity chromatography was sufficient to produce crystallization-quality protein.
3. Crystallization
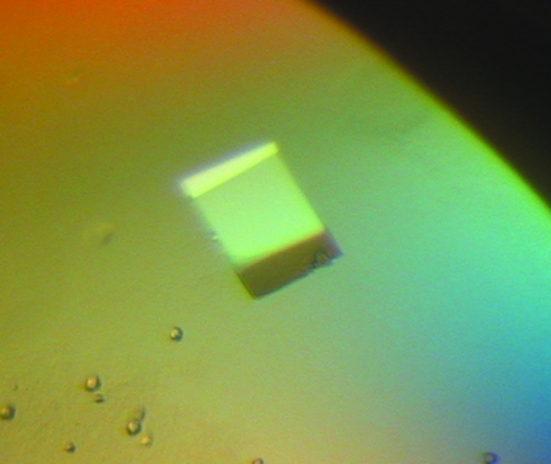
Initial crystallization experiments were carried out at a constant temperature of 293 K by the sitting-drop vapour-diffusion method (McPherson, 1982 ▶) using the sparse-matrix crystallization screening protocols described by Jancarik & Kim (1991 ▶). All crystallization trials were performed in Chryschem multi-well plates (sitting drop) with 300 µl reservoir solution using Crystal Screen and Crystal Screen II from Hampton Research, Wizard I and II from Emerald and JB Screens 1–10 from Jena Biosciences. Drops consisted of 2 µl protein solution (7 mg ml−1 in 5 mM Tris–HCl buffer pH 8.0) and 2 µl reservoir solution. Small crystals were observed in solution Nos. 3, 12, 15, 21, 29 and 33 from Crystal Screen, solution Nos. 2, 7, 23 and 43 from Crystal Screen II, solution Nos. 32 and 43 from Wizard I and solution Nos. 25, 37 and 48 from Wizard II. New screenings varying the pH of the buffer and the concentration of the precipitant were performed and we obtained crystals suitable for X-ray data collection (Fig. 1 ▶) in 0.1 M Tris–HCl buffer pH 7.7 containing 1.2 M (NH4)2HPO4. The crystals grew in 4–5 weeks.
Figure 1.
Crystal of the small heat-shock protein Xac1151 from X. axonopodis pv. citri. The approximate dimensions of the crystal are 0.40 × 0.35 × 0.20 mm.
4. Data collection
Crystals were mounted in nylon loops and flash-frozen in a nitrogen stream at 100 K in mother liquor containing 25% glycerol as a cryoprotectant. X-ray diffraction intensities were collected at the D03B-MX1 beamline, Laboratório Nacional de Luz Síncrotron (Campinas, Brazil) using a wavelength of 1.43 Å and a MAR CCD 165 detector (MAR Research) with 35 s exposures. Diffraction data were collected from 260 images using the oscillation method; individual frames consisted of 0.7° oscillation steps over a range of 182°. Diffraction data were indexed, integrated, scaled and merged using the HKL2000 package (Otwinowski & Minor, 1997 ▶).
5. Results
The crystals of XAC1151 belong to the rhombohedral space group R3 (No. 146), with unit-cell parameters a = b = 128.7, c = 55.3 Å. The calculated packing parameter, based on a molecular weight of 21.7 kDa, indicates the presence of two monomers in the asymmetric unit. This corresponds to a typical Matthews coefficient (V M) of 2.2 Å3 Da−1, which is within the expected range (Matthews, 1968 ▶). This V M corresponds to a solvent content of approximately 43.6%. The data-collection statistics are shown in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the last resolution shell.
| Space group | R3 |
| Unit-cell parameters | a = b = 128.7, c = 55.3 |
| Resolution range | 50.0–1.65 (1.71–1.65) |
| Total No. of reflections | 207262 |
| No. of unique reflections | 40597 |
| Rsym (%) | 4.3 (47.6) |
| Completeness (%) | 98.9 (89.2) |
| 〈I/σ(I)〉 | 24.5 (2.2) |
| Multiplicity | 5.1 (3.5) |
| Solvent content (%) | 43.6 |
| Subunits per ASU | 2 |
A molecular-replacement solution was found using the crystallographic structure of the small heat-shock protein sHSP from Triticum aestivum (PDB code 1gme; van Monfort et al., 2001 ▶) as a search model. Molecular-replacement procedures were performed with the program AMoRe (Navaza, 1994 ▶) implemented in the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶). The best solution was obtained using the dimer with 45 residues from the amino-terminus and 15 residues from the carboxy-terminus removed. The solution has a correlation coefficient of 36.5% and an R cryst of 48.6% after rigid-body refinement. Structure refinement is in progress.
In summary, we have obtained well diffracting crystals of a small heat-shock protein from the phytopathogenic bacterium X. axonopodis pv. citri. There are three structures of small heat-shock proteins deposited in the Protein Data Bank: two eukaryotic (Tenia saginata and T. aestivum) and one from a hyperthermophilic bacteria (Methanococcus jannashii). These structures have been solved at relatively poor resolution (2.5, 2.7 and 2.9 Å, respectively). We have collected data to a resolution of 1.65 Å; this higher resolution will help to better define the structure of this important family of proteins involved in the folding process of proteins.
Acknowledgments
This work was supported by Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP) through grant 01/07536-6 (SMolBNet) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). EH and ECT were supported by FAPESP fellowships Proc. 03/01646-0 and 02/07218-7, respectively. GAP is an undergraduate student supported by a PIBIC/CNPq fellowship.
References
- Bova, M. P., McHaourab, H. S., Han, Y. & Fung, B. K. (2000). J. Biol. Chem.275, 1035–1042. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- da Silva, A. C. R. et al. (2002). Nature (London), 417, 459–463. [Google Scholar]
- Ganea, E. (2001). Curr. Protein Pept. Sci.2, 205–225. [DOI] [PubMed] [Google Scholar]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411. [Google Scholar]
- Jiao, W., Qian, M., Li, P., Zhao, L. & Chang, Z. (2005). J. Mol. Biol.347, 871–884. [DOI] [PubMed] [Google Scholar]
- Kappe, G., Leunissen, J. A. & de Jong, W. W. (2002). Prog. Mol. Subcell. Biol.28, 1–17. [DOI] [PubMed] [Google Scholar]
- Kelley, M. J., David, L. L., Iwasaki, N., Wright, J. & Shearer, T. R. (1993). J. Biol. Chem.268, 18844–18849. [PubMed] [Google Scholar]
- Laksanalamai, P. & Robb, F. T. (2004). Extremophiles, 8, 1–11. [DOI] [PubMed] [Google Scholar]
- McPherson, A. (1982). Preparation and Analysis of Protein Crystals. New York: Wiley.
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Monfort, R. L. M. van, Basha, E., Friedrich, K. L., Slingsby, C. & Vierling, E. (2001). Nature Struct. Biol.8, 1025–1030. [DOI] [PubMed] [Google Scholar]
- Navaza, J. (1994). Acta Cryst. A50, 157–163. [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Stromer, T., Ehrnsperger, M., Gaestel, M. & Buchner, J. (2003). J. Biol. Chem.278, 18015–18021. [DOI] [PubMed] [Google Scholar]
- Sun, Y. & MacRae, T. H. (2005). Cell. Mol. Life Sci.62, 2460–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]