The crystallogenesis of bacteriophage P22 tail-fiber gp26 is described. To study possible pH-induced conformational changes in gp26 structure, native trimeric gp26 has been crystallized at acidic pH (4.6) and a chimera of gp26 fused to maltose-binding protein (MBP-gp26) has been crystallized at neutral and alkaline pH (7-10).
Keywords: bacteriophage P22, membrane-fusion proteins, coiled coil, cryo-annealing
Abstract
Gp26 is one of three phage P22-encoded tail accessory factors essential for stabilization of viral DNA within the mature capsid. In solution, gp26 exists as an extended triple-stranded coiled-coil protein which shares profound structural similarities with class I viral membrane-fusion protein. In the cryo-EM reconstruction of P22 tail extracted from mature virions, gp26 forms an ∼220 Å extended needle structure emanating from the neck of the tail, which is likely to be brought into contact with the cell’s outer membrane when the viral DNA-injection process is initiated. To shed light on the potential role of gp26 in cell-wall penetration and DNA injection, gp26 has been crystallized at acidic, neutral and alkaline pH. Crystals of native gp26 grown at pH 4.6 diffract X-rays to 2.0 Å resolution and belong to space group P21, with a dimer of trimeric gp26 molecules in the asymmetric unit. To study potential pH-induced conformational changes in the gp26 structure, a chimera of gp26 fused to maltose-binding protein (MBP-gp26) was generated. Hexagonal crystals of MBP-gp26 were obtained at neutral and alkaline pH using the high-throughput crystallization robot at the Hauptman–Woodward Medical Research Institute, Buffalo, NY, USA. These crystals diffract X-rays to beyond 2.0 Å resolution. Structural analysis of gp26 crystallized at acidic, neutral and alkaline pH is in progress.
1. Introduction
P22 is a well characterized double-stranded DNA (dsDNA) bacteriophage that infects the Gram-negative bacterium Salmonella enterica serovar typhimurium. The mature virion is an icosahedral T = 7 capsid ∼650 Å in diameter (Casjens, 1979 ▶; Prasad et al., 1993 ▶; Thuman-Commike et al., 1996 ▶, 1999 ▶, 2000 ▶), which has a 2.8 MDa tail apparatus at a single vertex (Tang et al., 2005 ▶). The tail complex, also known as the ‘portal vertex structure’, protrudes ∼320 Å outside the coat protein shell (Tang et al., 2005 ▶) and is formed by five proteins: the dodecameric portal protein gp1 (Bazinet et al., 1988 ▶), six copies of a trimer of the tail-spike protein gp9 (Goldenberg & King, 1982 ▶) and the three tail accessory proteins gp4, gp10 and gp26 (Casjens & Huang, 1982 ▶; Casjens & King, 1974 ▶; Hartweig et al., 1986 ▶; Strauss & King, 1984 ▶).
The P22 virus cycle is characterized by the formation of a spherical metastable procapsid of ∼600 Å in diameter that expands to form an infectious particle of ∼650 Å in diameter via a series of exothermic structural rearrangements (Thuman-Commike et al., 1996 ▶; Zhang et al., 2000 ▶). Packaging of the P22 genome (∼43 kbps) into the procapsid takes place through the portal protein in an ATP-dependent process (Casjens & Huang, 1982 ▶; Jackson et al., 1982 ▶; Poteete et al., 1979 ▶, 1983 ▶) that requires a virus-encoded terminase complex. After encapsidation of a single copy of the P22 genome, the DNA is cleaved and the portal channel is closed by the three tail accessory factors gp4, gp10 and gp26 (Strauss & King, 1984 ▶), also known as ‘head-completion proteins’. Six trimeric tail spikes (gp9) then attach to the nascent tail (Israel, 1978 ▶), yielding the mature infectious particle. P22 phages bearing mutations in each of the three genes 4, 10 and 26 (gp4−, gp10− and gp26−) assemble procapsids normally and package DNA, but some particles lose their DNA in the infected cell (Lenk et al., 1975 ▶) and all particles lose their DNA within minutes of being released from cells (Strauss & King, 1984 ▶). This indicates that gp4, gp10 and gp26 are not directly involved in DNA packaging, but are critical to retention of the encapsidated dsDNA inside the capsid.
We recently cloned the gene encoding gp26 protein and showed that gp26 forms an extended triple-stranded coiled coil in solution which is likely to be held together by five conserved trimerization heptads (Andrews et al., 2005 ▶). Similarly to class I viral fusion proteins, gp26 is characterized by an exceedingly high structural stability and high resistance to both thermal and chemical denaturation. Likewise, the N-terminal region of gp26 (residues 1–37) presents a highly hydrophobic region (Olia, personal communication) which closely resembles a class I viral fusion peptide (Eckert & Kim, 2001 ▶). In the cryo-EM reconstruction of P22 tail extracted from the mature virus (Tang et al., 2005 ▶), gp26 forms a 218 Å long needle-like density emanating for the virus tail, which could form a membrane-puncturing device. These preliminary data led us to hypothesize that gp26 functions by penetrating the first lipid bilayer of the Salmonella cell wall and then, perhaps with the help of several other proteins that are injected with the DNA (Israel, 1977 ▶), fusing outer and inner bilayers to generate a transient pore through which P22 DNA is injected. By analogy to viral class I fusion proteins such as influenza haemagglutinin, gp26 may undergo large conformational changes during membrane fusion in response to local variation in the periplasmic pH. In Gram-negative bacteria, a proton motive force is maintained across the cytoplasmic membrane. In a typical Escherichia coli cell, for instance, the periplasmic pH is about 1.7 pH units lower than the cytoplasmic compartment (Kashket, 1982 ▶). In turn, lower pH values may exist in the local proximity of proton pumps (e.g. ATPase). Therefore, it is fascinating to speculate that gp26 may undergo conformational changes in response to the lower pH found in the periplasm compartment. To test this structural hypothesis and shed light on the molecular structure of gp26, we have initiated structural studies on native gp26 as well as gp26 fused to maltose-binding protein (MBP-gp26) at different pH values.
2. Material and methods
2.1. Expression, purification and crystallization of native gp26
The gene encoding the tail accessory factor gp26 was cloned in expression vector pET-21b (Novagen) and purified as described previously (Andrews et al., 2005 ▶). Gp26 concentrated to 7–10 mg ml−1 was used to set up Hampton Research Crystal Screen I using the hanging-drop vapor-diffusion method. The best crystals were obtained at pH 4.6 using sodium acetate as the crystallization buffer and either 8–10% PEG 8000 or 10–12% PEG 4000 in the presence of 0.1 M ammonium sulfate. In both cases, gp26 crystals grew within one week at 293 K to reach maximum dimensions of 50 × 100 × 500 µm. The quality, size and reproducibility of the gp26 crystals were dramatically improved by setting up large crystallization droplets (3.5 + 3.5 µl) on thick ‘unsiliconized’ glass cover slips, which allow crystallization droplets to spread, generating a large drop–air interface.
2.2. Generating a chimera of maltose-binding protein fused to gp26 (MBP-gp26)
The gene encoding the gp26 protein was PCR amplified and ligated into the BamHI/PstI sites of the maltose-binding protein expression vector pMAL c-2e (New England Biolabs). The MBP fused to the N-terminus of gp26 (MBP-gp26) was expressed in E. coli strain BL21 (DE3/pLysS) cells at room temperature for 8 h. The MBP-gp26 chimera was affinity purified from soluble E. coli extracts using amylose–agarose beads (New England Biolabs) and eluted with 10 mM maltose. Further analysis by Superdex 200 gel-filtration chromatography revealed that the MBP-gp26 chimera elutes as a single species at a position corresponding to 250 kDa. The MBP-gp26 chimera was crystallized using the hanging-drop vapor-diffusion method. Crystals were obtained at pH 7 and 10 using 40% PEG 1K and 4K, respectively.
2.3. Data collection and analysis
Single crystals of native gp26 and the MBP-gp26 chimera were flash-frozen in 27.5% ethylene glycol. Several data sets were collected at SSRL (Stanford Synchrotron Radiation Laboratory) beamline 9-2, CHESS (Cornell High Energy Synchrotron Source) beamlines F-1 and A-1 National Synchrotron Light Source (NSLS) beamline X6A. Diffraction data were reduced to intensities using the programs DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶) and further analyzed with programs from CCP4 (Collaborative Computational Project, Number 4, 1994 ▶). Native gp26 crystals belong to space group P21, with unit-cell parameters a = 40.5, b = 116.4, c = 170.4 Å, β ≃ 90.5°. MBP-gp26 crystals obtained between pH 7 and 10 diffracted X-rays to beyond 1.8 Å resolution and belong to space group P32, with average unit-cell parameters a = 43, b = 43, c = 272 Å. A complete summary of unit-cell data and processing statistics is given in Table 1 ▶.
Table 1. Summary of diffraction data statistics for native gp26 crystallized at low pH and MBP-gp26 chimera crystallized at neutral and alkaline pH.
Values in parentheses are for the highest resolution shell.
| Data set | Native-gp26, pH 4.6 | MBP-gp26, pH 7.0 | MBP-gp26, pH 10.0 |
|---|---|---|---|
| Crystallization conditions | 10% PEG 8K, 0.1 M sodium acetate pH 4.6 | 40% PEG 1K, 0.1 M MOPS pH 7.0 | 40% PEG 4K, 0.1 M CAPS pH 10.0 |
| Beamline | SSRL 9-2 | BNLS X6A | BNLS X6A |
| Space group | P21 | P32 | P32 |
| Reflections (total/unique) | 870135/71223 | 338926/32511 | 310479/19166 |
| Unit-cell parameters | |||
| a (Å) | 9.97 | 43.85 | 43.20 |
| b (Å) | 14.52 | 43.85 | 43.20 |
| c (Å) | 71.17 | 272.31 | 271.23 |
| α (°) | 90.0 | 90.0 | 90.0 |
| β (°) | 90.5 | 90.0 | 120.0 |
| γ (°) | 90.0 | 90.0 | 120.0 |
| Resolution (Å) | 40–2.2 | 30–2.0 | 40–2.5 |
| Completeness (%) | 90.8 (65.4) | 95.2 (70.4) | 96.9 (84.6) |
| Rmerge (%) | 9.8 (18.4) | 8.4 (20.6) | 9.9 (24.2) |
| 〈I〉/〈σ(I)〉 | 17.2 (5.6) | 163 (4.3) | 13.6 (4.4) |
3. Results and discussion
3.1. Crystallographic characterization of gp26 crystals at acidic pH
Crystals of phage P22 gp26 protein diffracted anisotropically to beyond 2.2 Å resolution (Fig. 1 ▶) and displayed a high degree of internal disorder along the longer crystal dimension. This resulted in smeared diffraction spots (Fig. 2 ▶ a) and high mosaicity spread (0.75–1.0°). To improve the diffraction quality, we subjected the crystals to cryo-annealing treatments. This was performed by simply blocking the cryostream for ∼10 s, often repeating the procedure two or three times consecutively. In almost all cases (over 30 crystals), cryo-annealing dramatically improved the quality of diffraction spots as well as decreasing the mosaicity spread to a typical value of 0.3–0.5° (Figs. 2 ▶ a and 2 ▶ b). Crystallographic analysis indicates that gp26 crystallizes in a primitive monoclinic unit cell that has a volume of ∼792 903.4 Å3. Assuming one gp26 trimer to be present in the asymmetric unit, the Matthews coefficient V M is ∼5.1 Å3 Da−1 and the solvent content is ∼74%. Both values are significantly higher that those observed for most globular proteins (1.62 < V M < 3.53 Å3 Da−1), but not uncommon for exceptionally elongated trimeric coiled-coil proteins. For instance, in several crystal forms of influenza hemagglutinin (Rosenthal et al., 1998 ▶; Zhang et al., 1999 ▶), which resemble gp26 in topology and size, the solvent content observed in the unit cell is as high as 75%. Alternatively, the asymmetric unit of gp26 may contain a dimer of gp26 trimers, which results in a V M of ∼2.55 Å3 Da−1 and a solvent content of ∼52%. To solve this ambiguity, we investigated the local symmetry of gp26 crystals by self-rotation functions using polar angles (Rossmann & Blow, 1962 ▶). Self-rotation functions were computed with GLRF (Tong & Rossmann, 1997 ▶) using approximately 800 reflections with an I/σ(I) higher than 5 in the resolution range 8.0–4.0 Å and a radius of integration of 25 Å. To investigate the existence of local threefold non-crystallographic symmetry (NCS) axes, we computed a self-rotation function in space group P21 for κ = 120°. As shown in Fig. 2 ▶(c), the stereographic projections at κ = 120° revealed a distinct peak of ∼5.5σ on the self-rotation equator at polar angles ψ = 90° and ϕ = −65°. This maximum of the self-rotation function corresponds to the threefold NCS axis running along the trimeric coiled coil. In a search for sixfold NCS axes, a stereographic projection computed at κ = 60° also gave an identical peak at polar angles ψ = 90° and ϕ = −65° (data not shown), which supports the idea that two parallel gp26 trimers exist in the asymmetric unit. The stereographic plot allowed us to determine the orientation of the gp26 non-crystallographic threefold axis with respect to the crystallographic twofold screw axis along the b axis. The gp26 oligomer is orthogonal to the crystallographic b axis and rotated ∼65° from the a axis. In a search for twofold NCS axes, a stereographic projection at κ = 180° (Fig. 2 ▶ d) gave six strong peaks (∼4.0 height/σ), with a periodicity of 30°. These peaks were observed at all resolutions using different data shells and are likely to indicate the symmetry axis relating the six gp26 monomers in the hexameric oligomer. In summary, self-rotation function indicates the low-pH crystal form of gp26 is likely to contain a dimer of gp26 trimers in the asymmetric unit, which results in a V M of ∼2.55 Å3 Da−1 and a solvent content of ∼52%.
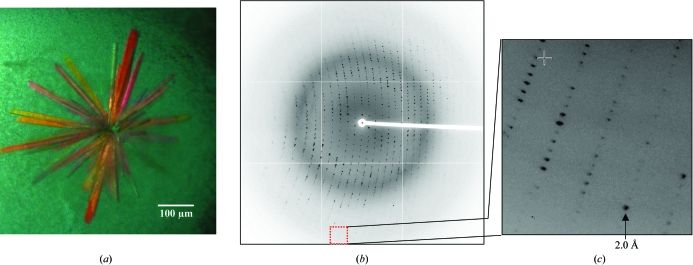
Figure 1.
Crystallization of gp26 at acidic pH. (a) Crystals of P22 tail accessory factor gp26 grew in clusters of shafts joined at the center. Isolated individual crystals have average dimensions of 80 × 100 × 350 µm. (b) Diffraction image recorded from a gp26 crystal oscillated 0.7° with an exposure time of 30 s. The image was measured at SSRL beamline 9-2 using an X-ray wavelength of 0.98 Å. (c) Enlargement of gp26 diffraction pattern showing distinct diffraction spots to 2 Å resolution. Owing to the anisotropic diffraction, X-ray data were only complete to 2.2 Å resolution (Table 1 ▶).
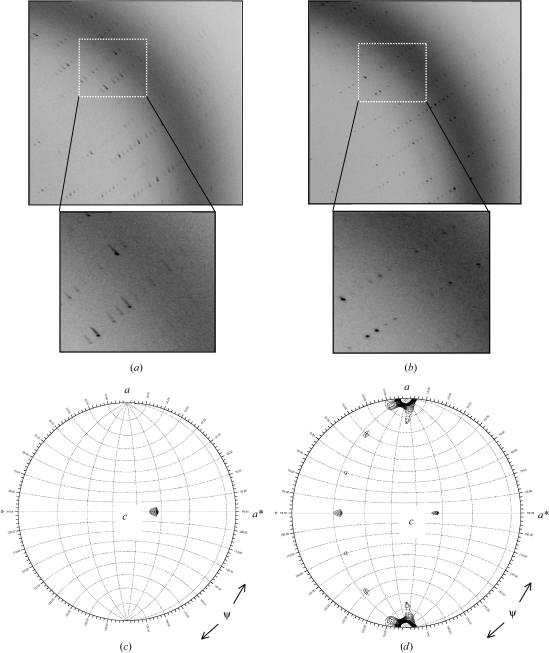
Figure 2.
Cryo-annealing dramatically improves the diffraction quality of gp26 crystals. (a) Diffraction image recorded from a gp26 crystal prior to cryo-annealing. Bragg spots are elongated and smeared, with mosaicity spread often higher than 1°. (b) The same diffraction image was recorded after three 10 s rounds of cryo-annealing. In (b), diffraction spots are more circular and the overall quality of the image has improved. (c) and (d), stereographic projections of the κ = 120 and κ = 180° sections of the P22 tail accessory factor gp26 self-rotation function showing the direction of threefold and twofold non-crystallographic symmetry axes, respectively. The maps were contoured at 4σ in steps of 0.25σ. The functions were computed with program GLRF (Tong & Rossmann, 1997 ▶). The polar angles ψ, ϕ and κ follow the Rossmann convention (Rossmann & Blow, 1962 ▶), with the orthogonal y axis aligned with the crystal b axis, the x axis lying in the crystal ab plane and the z axis coinciding with the c axis. The polar angle ψ denotes the inclination of the rotation axis from the y axis, which in space group P21 corresponds to the crystallographic screw axis. The polar angle ϕ denotes the inclination of the rotation axis from the x axis.
3.2. Crystallization of maltose-binding protein fused to gp26 (MBP-gp26)
All crystallization hits for native gp26 were obtained at acidic pH. In the attempt to obtain crystals of gp26 at neutral pH, we generated a chimera of maltose-binding protein (MBP) fused to gp26 (MBP-gp26). As observed for native gp26, unboiled MBP-gp26 chimera migrated on SDS–PAGE in its native trimeric state (Fig. 3 ▶, lane 1), suggesting that the trimer is SDS-resistant at the SDS concentration present in the SDS–PAGE running buffer (0.1%). In contrast, a sample of MBP-gp26 boiled for at least 5 min migrated on SDS–PAGE as an ∼75–80 kDa band (Fig. 3 ▶, lane 2), which is consistent with a chimera of MBP (∼44 kDa) fused to gp26 (∼26 kDa). This simple assay confirmed that MBP does not interfere with the trimerization of gp26 and that, similarly to native gp26, MBP-gp26 fusion protein almost certainly adopts a triple-stranded trimeric coiled-coil quaternary structure. To obtain crystals of MBP-gp26 at neutral pH, we used the high-throughput crystallization laboratory at the Hauptman–Woodward Medical Research Institute, Buffalo, NY, USA. A screen of 1536 crystallization conditions conducted by microbatch under paraffin oil gave around ten crystallization hits at pH values between 7 and 10. Both at neutral and alkaline pH, MBP-gp26 crystallized under highly concentrated solutions of various PEGs to form elongated crystals of average dimensions 80 × 80 × 500 µm (Fig. 3 ▶ b). These hits were repeated using conventional hanging-drop vapor diffusion and yielded a hexagonal crystal form of MBP-gp26 that diffracts X-rays to beyond 2.0 Å resolution (Table 1 ▶). The hexagonal asymmetric unit contains a single MBP-gp26 monomer and has a Matthews coefficient V M of ∼2.1 Å3 Da−1 and a solvent content of ∼40.5%. The elongated shape the unit cell and the small unit-cell volume (437 332.9 Å3) are incompatible with either a dimer or a trimer of MBP-gp26 in the asymmetric unit, as they would result in a negative solvent content. This implies that the MBP-gp26 trimeric coiled coil is assembled in the hexagonal unit cell using the crystallographic threefold axis running along the c axis.
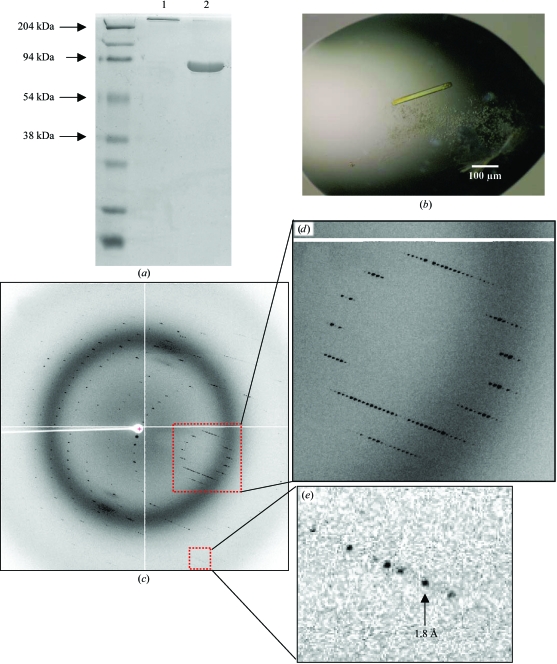
Figure 3.
Crystallization of gp26 fused to maltose-binding protein at neutral and alkaline pH. (a) A chimera of gp26 fused to maltose-binding protein (MBP-gp26) retains full oligomerization on SDS–PAGE. Unboiled MBP-gp26 in lane 1 runs on SDS–PAGE in its native trimeric state (3 × 70 kDa = ∼210 kDa), while an aliquot of MBP-gp26 fusion protein boiled for 5 min migrated as an ∼75 kDa band (lane 2). (b) Light micrograph of a typical MBP-gp26 crystal. The average dimensions of these crystals are 80 × 80 × 500 µm. The morphology of the crystal represents a macroscopic description of the hexagonal unit cell, which has averaged unit-cell parameters a = 43, b = 43, c = 272 Å. (c) Diffraction pattern of MBP-gp26 crystal oscillated by 0.5° with an exposure time of 10 s. The diffraction image was collected at BNLS beamline X6A on an ADSC Quantum-210 CCD detector. (d) and (e), enlargements of MBP-gp26 diffraction pattern showing intense diffraction spots to 1.8 Å resolution.
3.3. Conclusions
We have described a preliminary crystallographic analysis of P22 tail accessory factor gp26 expressed using a recombinant bacterial system. Native gp26 crystallizes at acidic pH (∼4.6) as a dimer of trimeric coiled coils. At neutral and alkaline pH, gp26 fused to maltose-binding protein crystallizes in an elongated hexagonal unit cell in which the trimeric coiled coil is generated by crystallographic threefold symmetry. In both cases, high-resolution diffraction data were carefully measured using synchrotron radiation. The work presented this paper provides the necessary prerequisites to solve the first three-dimensional structure of tail factor gp26. Independent analysis of the two crystal forms of gp26 will shed light on the possible pH-induced conformational changes and allow visualization of the conformation and position of the hydrophobic fusion peptide-like region (residues 1–37) of gp26 in the context of the tertiary and quaternary structure of the protein.
Acknowledgments
We thank Nancy Walker and Adam Olia at SUNY Upstate Medical University for excellent technical support and help in the X-ray data collection, respectively. We are grateful to the staff of Stanford Synchrotron Radiation Laboratory (SSRL), National Synchrotron Light Source (NSLS) beamline X6a as well as Cornell High Energy Synchrotron Source (CHESS) for beam-time availability and support during the data collection.
References
- Andrews, D., Butler, J. S., Al-Bassam, J., Joss, L., Winn-Stapley, D. A., Casjens, S. & Cingolani, G. (2005). J. Biol. Chem.280, 5929–5933. [DOI] [PubMed] [Google Scholar]
- Bazinet, C., Benbasat, J., King, J., Carazo, J. M. & Carrascosa, J. L. (1988). Biochemistry, 27, 1849–1856. [DOI] [PubMed] [Google Scholar]
- Casjens, S. (1979). J. Mol. Biol.131, 1–14. [DOI] [PubMed] [Google Scholar]
- Casjens, S. & Huang, W. M. (1982). J. Mol. Biol.157, 287–298. [DOI] [PubMed] [Google Scholar]
- Casjens, S. & King, J. (1974). J. Supramol. Struct.2, 202–224. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Eckert, D. M. & Kim, P. S. (2001). Annu. Rev. Biochem.70, 777–810. [DOI] [PubMed] [Google Scholar]
- Goldenberg, D. & King, J. (1982). Proc. Natl Acad. Sci. USA, 79, 3403–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartweig, E., Bazinet, C. & King, J. (1986). Biophys. J.49, 24–26. [DOI] [PMC free article] [PubMed]
- Israel, V. (1977). J. Virol.23, 91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel, V. (1978). J. Gen. Virol.40, 669–673. [DOI] [PubMed] [Google Scholar]
- Jackson, E. N., Laski, F. & Andres, C. (1982). J. Mol. Biol.154, 551–563. [DOI] [PubMed] [Google Scholar]
- Kashket, E. R. (1982). Biochemistry, 21, 5534–5538. [DOI] [PubMed] [Google Scholar]
- Lenk, E., Casjens, S., Weeks, J. & King, J. (1975). Virology, 68, 182–199. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Poteete, A. R., Jarvik, V. & Botstein, D. (1979). Virology, 95, 550–564. [DOI] [PubMed] [Google Scholar]
- Poteete, A. R., Sauer, R. T. & Hendrix, R. W. (1983). J. Mol. Biol.171, 401–418. [DOI] [PubMed] [Google Scholar]
- Prasad, B. V., Prevelige, P. E., Marietta, E., Chen, R. O., Thomas, D., King, J. & Chiu, W. (1993). J. Mol. Biol.231, 65–74. [DOI] [PubMed] [Google Scholar]
- Rosenthal, P. B., Zhang, X., Formanowski, F., Fitz, W., Wong, C. H., Meier-Ewert, H., Skehel, J. J. & Wiley, D. C. (1998). Nature (London), 396, 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann, M. G. & Blow, D. M. (1962). Acta Cryst.15, 24–31. [Google Scholar]
- Strauss, H. & King, J. (1984). J. Mol. Biol.172, 523–543. [DOI] [PubMed] [Google Scholar]
- Tang, L., Marion, W. R., Cingolani, G., Prevelige, P. E. & Johnson, J. E. (2005). EMBO J.24, 2087–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuman-Commike, P. A., Greene, B., Jakana, J., McGough, A., Prevelige, P. E. & Chiu, W. (2000). J. Virol.74, 3871–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuman-Commike, P. A., Greene, B., Jakana, J., Prasad, B. V., King, J., Prevelige, P. E. Jr & Chiu, W. (1996). J. Mol. Biol.260, 85–98. [DOI] [PubMed] [Google Scholar]
- Thuman-Commike, P. A., Greene, B., Malinski, J. A., Burbea, M., McGough, A., Chiu, W. & Prevelige, P. E. Jr (1999). Biophys. J.76, 3267–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, L. & Rossmann, M. G. (1997). Methods Enzymol.276, 594–611. [PubMed] [Google Scholar]
- Zhang, X., Rosenthal, P. B., Formanowski, F., Fitz, W., Wong, C. H., Meier-Ewert, H., Skehel, J. J. & Wiley, D. C. (1999). Acta Cryst. D55, 945–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Greene, B., Thuman-Commike, P. A., Jakana, J., Prevelige, P. E. Jr, King, J. & Chiu, W. (2000). J. Mol. Biol.297, 615–626. [DOI] [PubMed] [Google Scholar]