A complex of the essential splicing factor U2AF65 and a deoxyuridine oligonucleotide has been crystallized by modification of an interdomain linker.
Keywords: U2AF65, polypyrimidine tract, protein engineering
Abstract
The large subunit of the essential pre-mRNA splicing factor U2 auxiliary factor (U2AF65) binds the polypyrimidine tract near the 3′ splice site of pre-mRNA introns and directs the association of the U2 small nuclear ribonucleoprotein particle (U2 snRNP) of the spliceosome with the pre-mRNA. Protein engineering, in which the flexible linker region connecting tandem RNA-recognition motifs (RRMs) within the U2AF65 RNA-binding domain was partially deleted, allowed successful crystallization of the protein–nucleic acid complex. Cocrystals of a U2AF65 variant with a deoxyuridine dodecamer diffract X-rays to 2.9 Å resolution and contain one complex per asymmetric unit.
1. Introduction
U2AF is an essential heterodimeric pre-mRNA splicing factor that is required during the earliest steps of 3′ splice-site recognition (Ruskin et al., 1988 ▶; Zamore & Green, 1989 ▶). The large subunit, U2AF65, recognizes a pre-mRNA consensus sequence near the 3′ splice site termed the polypyrimidine tract (Py-tract; Zamore & Green, 1989 ▶; Singh et al., 1995 ▶). Subsequently, the U2AF65–Py-tract complex facilitates the stable association of the U2 snRNP with the pre-mRNA in the first ATP-dependent step of pre-mRNA splicing (Zamore & Green, 1989 ▶). U2AF65 recognizes the Py-tract through a pair of RNA-recognition motifs (RRM1 and RRM2) that are arranged in tandem (Banerjee et al., 2004 ▶). The RNA-recognition motif (RRM) is one of the most common RNA-binding folds in eukaryotes and a single polypeptide chain often contains several RRM domains that are involved in recognition of the target RNA (Maris et al., 2005 ▶). Two other examples of tandem RRM-containing proteins that recognize Py-tracts are Py-tract-binding protein (PTB) and Sex-lethal (SXL) (Oberstrass et al., 2005 ▶; Handa et al., 1999 ▶). Despite their similar domain structure, PTB, SXL and U2AF65 differ in target sequence and specificity. Whereas PTB prefers 5′-CUUC-3′ sequences (Singh et al., 1995 ▶), SXL specifically associates with Py-tracts containing 5′-GUUG-3′ sites (Valcarcel et al., 1993 ▶). U2AF65, in contrast, displays a preference for polyuridine sequences, but recognizes a wide range of Py-tract sequences consistent with its role as an essential splicing factor (Senapathy et al., 1990 ▶; Zamore et al., 1992 ▶). U2AF65, unlike SXL, shows little preference for binding ribose versus deoxyribose Py-tracts (Singh et al., 2000 ▶), allowing DNA to be substituted for RNA in structural studies.
Tandem RRM-containing proteins vary in the length and structure of the amino-acid linker connecting the RRM domains. In PTB, RRM1 and RRM2 are connected by a linker 51 amino acids in length that allows the two RRMs to tumble independently (Oberstrass et al., 2005 ▶). However, SXL RRM1 and RRM2 are connected by a shorter ten-amino-acid-long linker which becomes structured upon RNA binding (Handa et al., 1999 ▶). U2AF65 resembles the former example, with a 30-amino-acid-long linker that is predicted to lack RNA interactions based on RNA affinity analysis (Shamoo et al., 1995 ▶).
Without detailed structural information on the complex of U2AF65 and pre-mRNA, it remains unclear how the common RRM scaffold of U2AF65 prefers polyuridine sequences yet adapts to recognize a wide variety of naturally occurring Py-tract sequences. The structures of isolated U2AF65 RRM1 and RRM2 domains have been determined using nuclear magnetic resonance spectroscopy (Ito et al., 1999 ▶) and we have recently further detailed the U2AF65 RRM1 structure using X-ray crystallography (PDB code 2fzr). Although structural information is critical to understand the role of U2AF65 in splicing, the RNA-binding domain of U2AF65 (U2AF65 RRM1,2) in complex with nucleic acid has resisted crystallization. To investigate the key interactions with the Py-tract that guide 3′ splice-site selection by U2AF, we have used protein engineering to carry out cocrystallization and preliminary X-ray analysis of a human U2AF65 fragment with a modified linker in complex with a deoxyuridine dodecamer (dU12).
2. Methods and results
2.1. Overexpression and purification
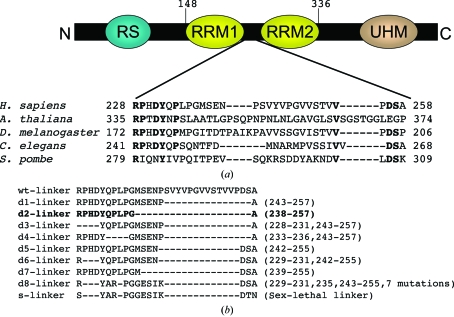
The RNA-binding domain of Homo sapiens U2AF65 contains RRM1 and RRM2 between residues 148 and 336 and these 189 residues have a molecular weight of 20.7 kDa. The linker region between RRM1 and RRM2 is poorly conserved (Fig. 1 ▶ a) and has an amino-acid sequence that is predicted to be intrinsically unstructured (Dosztanyi et al., 2005 ▶). Shortening the poorly conserved loop regions has been shown to promote crystallization without interfering with the activity of a protein (Mazza et al., 2002 ▶; Nolen et al., 2001 ▶). Several U2AF65 RRM1,2 variants (d1–d8-s-U2AF65 RRM1,2; Fig. 1 ▶ b) with shortened or modified linker regions were constructed for cocrystallization using the overlap extension polymerase chain reaction (PCR; Horton et al., 1993 ▶). In brief, an initial round of PCR individually amplified the coding sequences upstream and downstream of the desired deletion. In a second round of PCR, the upstream and downstream fragments were joined by virtue of the complementary ends engineered into the first-round primers, thereby eliminating the linker region. BamHI and EcoRI sites were used to insert the overlap PCR products as N-terminal glutathione-S-transferase (GST) fusions in the pGEX-6P-1 vector (Amersham Biosciences).
Figure 1.
(a) Domain map of human U2AF65 showing the limits of RRM1 and RRM2. UHM (U2AF homology motif) is a protein-interaction domain (Gozani et al., 1998 ▶) and RS denotes an arginine/serine-rich domain. The RRM1–RRM2 linker is expanded to show a phylogenetic sequence comparison. Residues that are conserved in at least four of the five species shown are indicated in bold. (b) Variants of the linker region (residues 228–258) between human U2AF65 RRM1 and RRM2 used for cocrystallization with oligonucleotides. Dashes and numbers in parentheses indicate deleted residues. The ‘d8-linker’ variant is derived from the linker sequence of splicing factor Sex-lethal (‘s-linker’). The successfully cocrystallized variant is shown in bold.
The recombinant U2AF65 RRM1,2 proteins with wild-type or modified linkers were overexpressed in Escherichia coli BL21 Rosetta cells (Novagen). Cells were grown to an OD of 0.6 in Luria–Bertani (LB) broth and induced with 0.2 mM IPTG for 16 h at 192 K. Pelleted cells were resuspended in 1 M NaCl, 15%(v/v) glycerol and 25 mM HEPES pH 7.4 and lysed using a French press. The lysate was pelleted at 16 000 rev min−1 and soluble protein was recovered in the supernatant. Supernatant was bound to a GS-Trap column (Amersham Biosciences) in 1 M NaCl and 25 mM HEPES pH 7.4 and eluted using 0.15 M NaCl, 0.1 M Tris pH 8 with 10 mM glutathione. The GST tag was cleaved from the U2AF65 RRM1,2 variants by treatment with PreScission Protease (Amersham Biosciences) during dialysis against a buffer containing 150 mM NaCl, 25 mM HEPES pH 7.4 and 5%(v/v) glycerol. Cleaved GST was separated from the U2AF65 RRM1,2 fragments by subtractive glutathione affinity chromatography (Amersham Biosciences) in 50 mM NaCl and 25 mM Tris pH 8. Subtractive anion-exchange chromatography with a HiTrap Q HP column (Amersham Biosciences) was used to remove the remaining GST and protease, which bound to the column while the U2AF65 RRM1,2 fragments flowed through in 50 mM NaCl and 25 mM Tris pH 8. The final purification step was gel filtration on a Superdex-75 prep-grade column (Amersham Biosciences), which was previously equilibrated with a buffer containing 100 mM NaCl and 10 mM HEPES pH 6.8. The purified U2AF65 RRM1,2 variants were concentrated to a final concentration of 45 mg ml−1 using an Amicon Ultra-15 ultrafiltration device (Millipore). The protein concentration was estimated by measuring the absorbance at 280 nm using an extinction coefficient of 6400 M −1 cm−1 calculated using the ProtParam tool of SWISS-PROT (http://www.expasy.ch). The final crystallized d2-U2AF65 RRM1,2 fragment includes residues 148–336 excluding residues 238–257 of the full-length human protein (accession code X64044) and five additional N-terminal residues from the expression vector (Gly-Pro-Leu-Gly-Ser).
2.2. Crystallization
Complexes of the U2AF65 RRM1,2 variants with deoxyuridine dodecamer (dU12) were formed by mixing the components in a 1:1.2 molar ratio of protein to DNA to give a final protein concentration of 1 mM. DNAs were synthesized by the Oligonucleotide Synthesis Core Facility of the Bloomberg School of Public Health, resuspended in 0.1 M NaCl and 15 mM HEPES pH 6.8 and then used without further purification. Following incubation of the U2AF65 RRM1,2–dU12 mixture on ice for 30 min, screens for crystallization were conducted using the hanging-drop vapor-diffusion method. In a typical crystallization experiment and the final optimized crystallizations, a mixture of the U2AF65 RRM1,2–dU12 complex and reservoir solution (1 µl each) was equilibrated against 700 µl reservoir solution. Sparse-matrix screens (Crystal Screens 1 and 2, Hampton Research) identified initial crystallization conditions, which were optimized using Additive Screen 1. Crystallization screens with 12- and 14-nucleotide polyuridine DNAs gave similar results. Only one of the nine U2AF65 RRM1,2 variants cocrystallized with deoxyuridine sequences (d2-U2AF65 RRM1,2; Fig. 1 ▶ b).
The optimal reservoir solution contained 1.6 M ammonium sulfate, 100 mM MES pH 6.5, 10% dioxane, 200 mM non-detergent sulfobetaine (NDSB) 195 at 277 K and was mixed into drops in a 5:4(v:v) ratio of U2AF65 RRM1,2–dU12 to reservoir. Needles with approximate dimensions of 0.05 × 0.05 × 0.25 mM appeared from precipitate after 1–2 weeks. Several previously determined cocrystal structures of RRM-containing proteins bound to nucleic acid have been obtained using ammonium sulfate as the precipitant (Deo et al., 1999 ▶; Oubridge et al., 1994 ▶), illustrating the ability of high-salt conditions to promote crystallization of this type of complex.
2.3. X-ray data collection and structure solution
To cryoprotect the crystals for data collection, the d2-U2AF65 RRM1,2 crystals were dipped in a solution of a 1:1(v:v) ratio of mineral oil and Paratone-N and flash-frozen in liquid nitrogen. A native data set was collected with an oscillation range of 1° per 10 s exposure at 100 K using beamline 8.2.2 of the Advanced Light Source (ALS; Berkeley, CA, USA). Data in the 20.0–2.9 Å resolution range were indexed, integrated and scaled using HKL2000 (Otwinowski & Minor, 1997 ▶) and the data-collection statistics are summarized in Table 1 ▶. The crystals belong to the hexagonal space group P6522. The Matthews coefficient (V M) is calculated to be 2.5 Å3 Da−1, with 50% solvent content if one d2-U2AF65 RRM1,2–dU12 complex (molecular weight 22 478 Da) is present per asymmetric unit (Matthews, 1968 ▶). Completion and analysis of the structure will be published elsewhere.
Table 1. Crystallographic data.
Values in parentheses are for the highest resolution shell.
| X-ray source | ALS beamline 8.2.2 |
| Wavelength (Å) | 1.0 |
| Space group | P6522 (or P6122) |
| Unit-cell parameters (Å, °) | a = b = 58.3, c = 229.3, α = β = 90, γ = 120 |
| Diffraction limit | 20.0–2.9 (3.00–2.90) |
| Redundancy | 15.4 (15.9) |
| Completeness (%) | 99.9 (100.0) |
| Rsym† (%) | 8.6 (49.5) |
| 〈I/σ(I)〉 | 25.7 (4.95) |
R
sym = 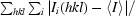
 , where Ii(hkl) is the intensity I for the ith measurement of a reflection with indices hkl and 〈I〉 is the weighted mean of all measurements of I.
, where Ii(hkl) is the intensity I for the ith measurement of a reflection with indices hkl and 〈I〉 is the weighted mean of all measurements of I.
Acknowledgments
We thank Dr C. Wolberger and the Johns Hopkins School of Medicine Department of Biophysics and Biophysical Chemistry for generously providing temporary access to their X-ray facility, the staff at ALS for assistance with data collection and Dr S. Morrow for oligonucleotide synthesis. KEF was supported in part by a training grant (T32 GM08403) from the NIH and EAS was a Lang Fellow. This work was supported by a grant from the National Institutes of Health (NIH; GM070503-01).
References
- Banerjee, H., Rahn, A., Gawande, B., Guth, S., Valcarcel, J. & Singh, R. (2004). RNA, 10, 240–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo, R. C., Bonanno, J. B., Sonenberg, N. & Burley, S. K. (1999). Cell, 98, 835–845. [DOI] [PubMed] [Google Scholar]
- Dosztanyi, Z., Csizmok, V., Tompa, P. & Simon, I. (2005). J. Mol. Biol.347, 827–839. [DOI] [PubMed] [Google Scholar]
- Gozani, O., Potashkin, J. & Reed, R. (1998). Mol. Cell Biol.18, 4752–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa, N., Nureki, O., Kurimoto, K., Kim, I., Sakamoto, H., Shimura, Y., Muto, Y. & Yokoyama, S. (1999). Nature (London), 398, 579–585. [DOI] [PubMed] [Google Scholar]
- Horton, R. M., Ho, S. N., Pullen, J. K., Hunt, H. D., Cai, Z. & Pease, L. R. (1993). Methods Enzymol.217, 270–279. [DOI] [PubMed] [Google Scholar]
- Ito, T., Muto, Y., Green, M. R. & Yokoyama, S. (1999). EMBO J.18, 4523–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris, C., Dominguez, C. & Allain, F. H. (2005). FEBS J.272, 2118–2131. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Mazza, C., Segref, A., Mattaj, I. W. & Cusack, S. (2002). Acta Cryst. D58, 2194–2197. [DOI] [PubMed] [Google Scholar]
- Nolen, B., Yun, C. Y., Wong, C. F., McCammon, J. A., Fu, X. D. & Ghosh, G. (2001). Nature Struct. Biol.8, 176–183. [DOI] [PubMed] [Google Scholar]
- Oberstrass, F. C., Auweter, S. D., Erat, M., Hargous, Y., Henning, A., Wenter, P., Reymond, L., Amir-Ahmady, B., Pitsch, S., Black, D. L. & Allain, F. H. (2005). Science, 309, 2054–2057. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Oubridge, C., Ito, N., Evans, P. R., Teo, C. H. & Nagai, K. (1994). Nature (London), 372, 432–438. [DOI] [PubMed] [Google Scholar]
- Ruskin, B., Zamore, P. D. & Green, M. R. (1988). Cell, 52, 207–219. [DOI] [PubMed] [Google Scholar]
- Senapathy, P., Shapiro, M. B. & Harris, N. L. (1990). Methods Enzymol.183, 252–278. [DOI] [PubMed] [Google Scholar]
- Shamoo, Y., Abdul-Manan, N. & Williams, K. R. (1995). Nucleic Acids Res.23, 725–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R., Banerjee, H. & Green, M. R. (2000). RNA, 6, 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R., Valcarcel, J. & Green, M. R. (1995). Science, 268, 1173–1176. [DOI] [PubMed] [Google Scholar]
- Valcarcel, J., Singh, R., Zamore, P. D. & Green, M. R. (1993). Nature (London), 362, 171–175. [DOI] [PubMed] [Google Scholar]
- Zamore, P. D. & Green, M. R. (1989). Proc. Natl Acad. Sci. USA, 86, 9243–9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore, P. D., Patton, J. G. & Green, M. R. (1992). Nature (London), 355, 609–614. [DOI] [PubMed] [Google Scholar]