Lipoprotein e (P4) is a class C acid phosphatase and a potential vaccine candidate for nontypeable H. influenzae infections. This paper reports the crystallization of recombinant e (P4) and the acquisition of a 1.7 Å resolution native X-ray diffraction data set.
Keywords: Haemophilus influenzae, lipoprotein e (P4), class C acid phosphatases
Abstract
Haemophilus influenzae infects the upper respiratory tract of humans and can cause infections of the middle ear, sinuses and bronchi. The virulence of the pathogen is thought to involve a group of surface-localized macromolecular components that mediate interactions at the host–pathogen interface. One of these components is lipoprotein e (P4), which is a class C acid phosphatase and a potential vaccine candidate for nontypeable H. influenzae infections. This paper reports the crystallization of recombinant e (P4) and the acquisition of a 1.7 Å resolution native X-ray diffraction data set. The space group is P42212, with unit-cell parameters a = 65.6, c = 101.4 Å, one protein molecule per asymmetric unit and 37% solvent content. This is the first report of the crystallization of a class C acid phosphatase.
1. Introduction
The bacterium Haemophilus influenzae is a Gram-negative facultative anaerobic coccobacillus and a common commensal of the human upper respiratory tract (Rao et al., 1999 ▶). The organism is the etiologic agent of a variety of local and invasive infections in humans. Isolates of H. influenzae are separated into two groups based on the presence or absence of capsular carbohydrate. Encapsulated organisms are distinguished serologically into seven different serotypes designated a–f, whereas nonencapsulated strains are designated as nontypeable. These latter strains account for the majority of mucosal diseases, including infections of the middle ear, sinuses and bronchi.
A group of surface-localized macromolecules of H. influenzae, including six abundant proteins designated P1–P6 in order of decreasing molecular weight, are thought to be involved in pathogenesis (Loeb & Smith, 1980 ▶; Vachon et al., 1985 ▶). Among these proteins, the 28 kDa cationic lipoprotein e (P4) is one of the best characterized. This protein is an acid phosphatase encoded by the hel gene and is conserved in size and antigenicity within and between strains (Granoff & Munson, 1986 ▶; Green et al., 1991 ▶). Its optimum phosphomonoesterase activity is achieved at pH 5.0 in the presence of divalent copper with arylphosphate substrates (Reilly et al., 1999 ▶). The enzyme is resistant to tartrate, inorganic phosphate, fluoride and p-hydroxymercuriphenylsulfonate, but is inhibited by orthovanadate, molybdate and EDTA (Reilly et al., 1999 ▶). Sequence alignments suggest that e (P4) contains the DDDD motif characteristic of class C bacterial nonspecific acid phosphatases (Rossolini et al., 1998 ▶).
The recombinant form of e (P4), designated rP4, has been expressed previously in Escherichia coli, purified and characterized (Reilly & Smith, 1999 ▶). Physicochemical characterization showed that rP4 maintained similar features as the wild-type enzyme (Reilly & Smith, 1999 ▶). Enzymatically inactive mutants of rP4 have recently been shown to be potential vaccine candidates for H. influenzae (Green et al., 2005 ▶).
Crystal structure determination of rP4 is an important step in elucidating the molecular basis for the biochemical roles of e (P4). The structure will also assist in the design of rP4 mutants for use in a H. influenzae vaccine. Here, we report the crystallization and preliminary X-ray crystallographic analysis of rP4. To our knowledge, this is the first report of the crystallization of a bacterial class C acid phosphatase.
2. Methods and results
2.1. Protein purification
Recombinant e (P4) was expressed in E. coli strain BL21(DE3) as described previously (Reilly & Smith, 1999 ▶). The harvested bacteria were pelleted at 5000g, resuspended in 10 mM Na2HPO4 pH 7.2 and frozen at 253 K in preparation for protein purification. All purification procedures were conducted at 277 K. All chromatography steps were performed with an AKTA FPLC using columns purchased from Amersham Biosciences (GE Healthcare).
Frozen cells were thawed, supplemented with the protease inhibitor PMSF (1 mM final concentration) and disrupted with two passes through a French pressure cell at 110 MPa. Cell debris was removed by centrifugation at 5000g for 10 min. NaCl was added to the supernatant to a final concentration of 1 M and the mixture was stirred for 1 h. This step was necessary to release cationic rP4 from negatively charged phospholipid head groups of membranes. The sample was then subjected to a low-speed centrifugation step (5000g, 10 min), followed by ultracentrifugation at 184 000g for 1 h in order to pellet bacterial membranes. The resulting supernatant contained the majority of the acid phosphatase activity. The supernatant from the ultracentrifugation step was dialyzed against 10 mM Na2HPO4 pH 7.2, 300 mM NaCl (buffer A) and filtered through a 0.45 µm filter. The sample was loaded onto a 5 ml HiTrap Chelating HP column that had been charged with 2.5 ml 0.1 M CuSO4 and equilibrated with 25 ml buffer A. Bound proteins were eluted with a 150 ml linear gradient of 0–50 mM imidazole in buffer A. We note that rP4 does not have a polyhistidine affinity tag and thus the protein presumably binds to the Cu-affinity column via an endogenous metal-binding site. The phosphomonoesterase activities of the eluted fractions were measured using a discontinuous colorimetric assay with p-nitrophenylphosphate as the substrate (Reilly et al., 1999 ▶). Highly active fractions were pooled and dialyzed into 50 mM sodium acetate pH 6.0 (buffer B) containing 50 mM NaCl and loaded onto a 5 ml HiTrap SP Sepharose cation-exchange column that had been equilibrated with 25 ml buffer B. Elution of rP4 was achieved with a 150 ml linear gradient of 0.05–2 M NaCl in buffer B.
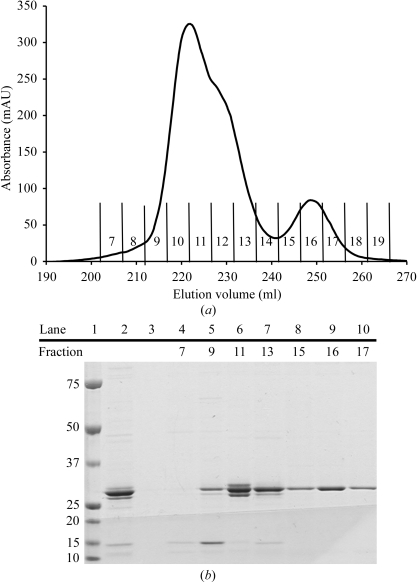
Results from SDS–PAGE indicated that eluted rP4 fractions exhibited minor but unacceptable levels of contaminating proteins, so the active fractions were pooled and dialyzed back into buffer A for further purification. The metal-affinity and cation-exchange steps were repeated using the procedures described above, except that a shallower linear gradient consisting of 0.05–2 M NaCl in 500 ml buffer B was used in the cation-exchange step. The shallower gradient allowed the resolution of two main protein peaks, which eluted in the range 140–380 mM NaCl (Fig. 1 ▶ a). Both protein peaks displayed phosphomonoesterase activity, but differed markedly in the level of protein purity. The larger peak (fractions 8–14) clearly had a shoulder visible on the high ionic strength side (Fig. 1 ▶ a) and SDS–PAGE confirmed the presence of multiple protein species in these fractions (Fig. 1 ▶ b). In contrast, protein samples taken from the smaller peak (fractions 15–18) showed a single band in SDS–PAGE (Fig. 1 ▶ b). We note that the results shown in Fig. 1 ▶ were reproducible, but the relative areas under the two chromatogram peaks varied from preparation to preparation. Fractions 8–14 (pool A) and 15–18 (pool B) were pooled separately for crystallization trials. The two pools of protein were dialyzed into buffer B and concentrated to 13 mg ml−1 using an Amicon ultrafiltration cell with a 10 kDa molecular-weight cutoff. The protein concentration was determined by the Coomassie Plus assay (Pierce).
Figure 1.
Purification of rP4 by cation-exchange chromatography. (a) The chromatogram is a plot of absorbance (λ = 280 nm) as a function of elution volume obtained with a HiTrap SP Sepharose cation-exchange column and AKTA FPLC protein-purification system. Fractions eluted with a linear NaCl gradient are labelled 7–19, with fractions 7 and 18 corresponding to NaCl concentrations of 140 and 380 mM, respectively. (b) SDS–PAGE analysis (Coomassie stain) of the chromatogram in (a). Lanes 1, 2 and 3 correspond to molecular-weight markers (kDa), protein prior to the cation-exchange chromatography step and flowthrough, respectively. Lanes 4–10 correspond to elution fractions as indicated above the gel.
Mass-spectrometric analysis revealed two components with apparent molecular masses of 28 378 and 28 509, which differ by the mass equivalent of one methionine. The 28 378 Da protein was the major component of pool A and the 28 509 Da protein was the major component of pool B. The species with apparent mass 28 509 is likely to correspond to full-length rp4, which has a theoretical molecular weight of 28 569 Da, while the 28 378 Da protein possibly represents rP4 devoid of the N-terminal Met. These results suggest the possibility of proteolytic degradation of the N-terminal Met during expression and purification.
2.2. Crystallization
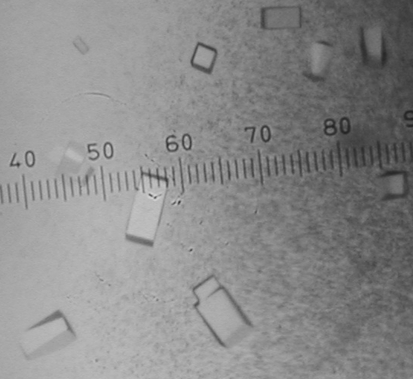
All crystallization experiments were performed at 295 K using Cryschem 24-well sitting-drop plates (Hampton Research) with a reservoir volume of 0.75 ml. Drops were formed by mixing equal volumes of the reservoir (2 µl) and protein solutions (2 µl). Commercially available crystal screens were used to identify initial crystallization conditions. Both protein samples yielded small crystals in these initial screens, but the most promising crystals were obtained with pool B protein and Hampton Research Crystal Screen reagent No. 6 [0.2 M MgCl2, 0.1 M Tris–HCl pH 8.5, 30%(w/v) PEG 4000]. Upon optimization, the best crystals were grown using pool B protein (13 mg ml−1 in buffer B) and reservoir solutions consisting of 0.2 M MgCl2, 0.1 M Tris–HCl pH 8.1–8.5 and 28–36%(w/v) PEG 4000. These crystals appeared as rectangular blocks with dimensions 0.06 × 0.06 × 0.16 mm (Fig. 2 ▶).
Figure 2.
Crystals of H. influenzae rP4. The smallest division of the ruler corresponds to 0.02 mm.
2.3. Data collection and processing
The crystals were prepared for cryogenic data collection by soaking them in 0.2 M MgCl2, 0.1 M Tris–HCl pH 8.5, 40%(w/v) PEG 4000 and 15%(v/v) PEG 200. The cryoprotected crystals were picked up with Hampton mounting loops and plunged into liquid nitrogen. Diffraction to 2.6 Å resolution was observed using an in-house Cu rotating-anode system and autoindexing calculations suggested a primitive tetragonal lattice with unit-cell parameters a = 66, c = 101 Å.
We note that attempts to grow this crystal form using pool A protein resulted in clusters of fused crystals rather than single crystals and the resulting diffraction patterns could not be reliably indexed. Thus, isolation of the 28 509 Da form of rP4 was critical for the growth of high-quality single crystals.
Native X-ray diffraction data sets were collected at beamline 4.2.2 of the Advanced Light Source at Lawrence Berkeley National Laboratory using a NOIR-1 CCD detector. The best data set consisted of a 90° wedge of data collected with an oscillation angle of 1°, an exposure time of 10 s per degree of oscillation, a crystal-to-detector distance of 125 mm and a detector 2θ angle of 0°. The data were integrated with MOSFLM (Leslie, 2006 ▶) and scaled with SCALA (Evans, 2006 ▶). The data exhibited acceptable processing statistics to 1.7 Å resolution (Table 1 ▶). The space group is P42212 and the refined unit-cell parameters are a = 65.6, c = 101.4 Å. Matthews calculations suggested that this crystal form has one molecule in the asymmetric unit, 37% solvent content and a Matthews coefficient of 1.95 Å3 Da−1 (Matthews, 1968 ▶).
Table 1. Data-collection and processing statistics.
Values in parentheses are for the outer resolution shell.
| Beamline | ALS 4.2.2 |
| Wavelength (Å) | 1.0359 |
| Space group | P42212 |
| Unit-cell parameters (Å) | a = 65.6, c = 101.4 |
| Resolution limits (Å) | 42–1.70 (1.79–1.70) |
| No. of observations | 158442 |
| Unique reflections | 25076 |
| Average redundancy | 6.3 (3.6) |
| Completeness (%) | 99.8 (98.3) |
| Average I/σ(I) | 17.5 (2.6) |
| Rmerge(I) | 0.071 (0.410) |
Since the apparent Laue class is 4/mmm, the possibility of merohedral twinning was investigated. The cumulative intensity distribution for acentric reflections did not display the sigmoidal shape characteristic of twinned data (Rees, 1980 ▶). The average value of 〈I 2(h)〉/〈I(h)〉2 was 2.0 for acentric reflections, which equals the expected value for nontwinned data (Redinbo & Yeates, 1993 ▶). Note that the expected value of 〈I 2(h)〉/〈I(h)〉2 for the case of perfect hemihedral twinning is 1.5 (Redinbo & Yeates, 1993 ▶). Thus, difficulties arising from twinning are not anticipated.
Analysis of the rP4 amino-acid sequence with BLAST (Altschul et al., 1990 ▶) failed to detect a homolog in the Protein Data Bank (Berman et al., 2000 ▶) that could serve as a suitable search model for molecular-replacement phasing. Therefore, the structure of rP4 is being determined using single isomorphous replacement with anomalous scattering.
Acknowledgments
This research was supported by National Institutes of Health grant U54 AI057160 to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (MRCE, to JJT and TJR) and the University of Missouri Research Board (to JJT and TJR). The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division of the US Department of Energy under Contract No. DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory.
References
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990). J. Mol. Biol.215, 403–410. [DOI] [PubMed] [Google Scholar]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res.28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed] [Google Scholar]
- Granoff, D. M. & Munson, R. S. Jr (1986). J. Infect. Dis.153, 448–461. [DOI] [PubMed] [Google Scholar]
- Green, B. A., Baranyi, E., Reilly, T. J., Smith, A. L. & Zlotnick, G. W. (2005). Infect. Immun.73, 4454–4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, B. A., Farley, J. E., Quinn-Dey, T., Deich, R. A. & Zlotnick, G. W. (1991). Infect. Immun.59, 3191–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed] [Google Scholar]
- Loeb, M. R. & Smith, D. H. (1980). Infect. Immun.30, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Rao, V. K., Krasan, G. P., Hendrixson, D. R., Dawid, S. & St Geme, J. W. III (1999). FEMS Microbiol. Rev.23, 99–129. [DOI] [PubMed] [Google Scholar]
- Redinbo, M. R. & Yeates, T. O. (1993). Acta Cryst. D49, 375–380. [DOI] [PubMed] [Google Scholar]
- Rees, D. C. (1980). Acta Cryst. A36, 578–581. [Google Scholar]
- Reilly, T. J., Chance, D. L. & Smith, A. L. (1999). J. Bacteriol.181, 6797–6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly, T. J. & Smith, A. L. (1999). Protein Expr. Purif.17, 401–409. [DOI] [PubMed] [Google Scholar]
- Rossolini, G. M., Schippa, S., Riccio, M. L., Berlutti, F., Macaskie, L. E. & Thaller, M. C. (1998). Cell. Mol. Life Sci.54, 833–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachon, V., Lyew, D. J. & Coulton, J. W. (1985). J. Bacteriol.162, 918–924. [DOI] [PMC free article] [PubMed] [Google Scholar]