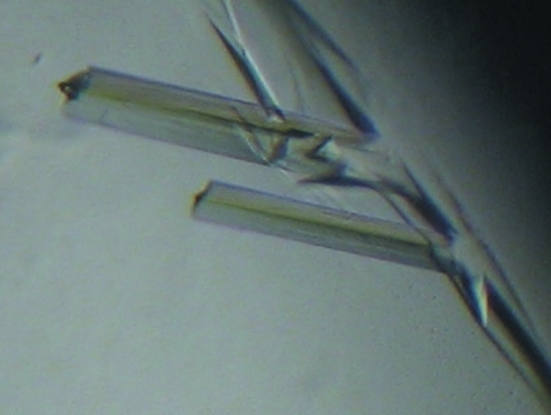
Crystals of human PRPP synthetase 1 (PRS1) in complex with Mg2+, Pi and ATP were obtained and diffraction data were collected to 2.6 Å resolution.
Keywords: human phosphoribosyl pyrophosphate synthetase 1 (PRS1), PRPP, kinetic mechanisms
Abstract
Phosphoribosyl pyrophosphate synthetase (PRS; EC 2.7.6.1) catalyzes the reaction of ribose-5-phosphate (R5P) with ATP to yield AMP and PRPP (5-phosphoribosyl-1-pyrophosphate), which is necessary for the de novo and salvage pathways of purine-, pyrimidine- and pyridine-nucleotide biosynthesis. PRPP is a metabolite that is required at all times in the cell and is thus central to life. In this study, human PRS1 was produced in Escherichia coli in soluble form and purified to homogeneity. Crystals in complex with Mg2+, inorganic phosphate (Pi) and ATP were obtained by the hanging-drop vapour-diffusion method. Diffraction data were collected to 2.6 Å resolution. The crystal belongs to space group R3, with unit-cell parameters a = b = 168.846, c = 61.857 Å, assuming two molecules in the asymmetric unit and a volume-to-weight ratio of 2.4 Å3 Da−1, which was consistent with the result calculated from the self-rotation function.
1. Introduction
5-Phosphoribosyl-1-pyrophosphate (PRPP) is an important precursor and an allosteric regulator in the synthesis of purine and pyrimidine nucleotides. The formation of PRPP is catalyzed by phosphoribosyl pyrophosphate synthetase (PRS; EC 2.7.6.1). The enzyme requires Mg2+ and Pi for its stability and activity and is subject to purine-nucleotide (ADP and GDP) feedback inhibition (Zoref et al., 1975 ▶). The family of PRPP synthetases includes representatives from a wide range of organisms from prokaryotes to eukaryotes and their high sequence similarity has been revealed by sequence alignments, indicating the conservation of PRPP synthetases (Switzer, 1969 ▶; Becker et al., 1980 ▶). Human PRPS genes encode three highly homologous PRS isoforms (PRS1, PRS2 and PRS3). PRPS1 and PRPS2 are expressed in all tissues (Taira et al., 1989 ▶) and map to the long and short arms of the X chromosome, respectively (Becker et al., 1990 ▶). PRPS1 and PRPS2 cDNAs show 79.9% nucleotide sequence identity throughout their 954 bp translated regions and 95.3% amino-acid homology of their protein products (Becker et al., 1990 ▶). Expression of the autosomal PRPS3 gene is restricted to the testes (Taira et al., 1989 ▶).
Previous studies have focused mostly on the PRPS1 gene and the structure–function relationship of PRS1. The open reading frame of human PRPS1 consists of 957 bp coding for 318 amino-acid residues. The isoelectric value was calculated to be 6.51. The human PRPS1 gene has been cloned into an expression vector and encodes a functional PRS1 as assessed by PRS1 assay (Becker et al., 1995 ▶). The functional form of the PRPP synthetase from Bacillus subtilis is a hexamer (Eriksen et al., 2000 ▶). The functional form of human PRS1 is still unclear. It is interesting that native PRPP synthetases from many sources have a tendency to exist in multiple states of aggregation (Schubert et al., 1975 ▶).
Superactivity of human PRS1 is an X-linked disorder of purine metabolism characterized by gout with uric acid overproduction (Sperling et al., 1972 ▶; Zoref et al., 1975 ▶) and neurodevelopmental impairment in some affected families (Simmonds et al., 1982 ▶; Becker et al., 1995 ▶). In 30 affected kindreds with PRS1 superactivity identified to date (Becker, 2005 ▶), the kinetic mechanisms leading to excessive enzyme activity include both regulatory defects and catalytic superactivity. PRS1 catalytic superactivity is the more common case, which is characterized by increased expression of the normal PRS1 isoform resulting from accelerated PRPS1 transcription (Becker et al., 1996 ▶), and the mechanisms remain unclear. Patients with regulatory defects of PRS1 activity are the severe clinical phenotype, with the biochemical properties of abnormal PRS1 from these patients including reduced sensitivity to the nucleotide inhibitors ADP or GDP, increased sensitivity to Pi activation and increased affinity of R5P (Becker et al., 1995 ▶). Point mutations in the translated region of the PRPS1 gene of affected individuals provide the genetic basis for regulatory defects, indicating the functional significance of these amino-acid residues in PRS1. Residues that are probably important in the catalytic site have been identified (Hove-Jensen et al., 1986 ▶; Bower et al., 1989 ▶; Harlow & Switzer, 1990 ▶; Becker et al., 1995 ▶). Eriksen et al. (2000 ▶) reported the structure of B. subtilis phosphoribosyl pyrophosphate synthetase and analyzed the structural basis of the catalytic mechanism. The studies showed that the side chains involved in binding ADP were highly conserved, revealed several important regions and identified residues important for binding substrates (ATP, R5P) and effectors (ADP, Pi, Mg2+). These studies shed light on the further study of the structure of human PRS1.
2. Materials and methods
2.1. Protein expression and purification
The complete cDNA encoding PRS1 (accession No. NM_002764) was cloned from a human brain cDNA library (BD Biosciences) into pET-22b expression vector (Novagen) as a fusion protein with a His tag at the C-terminus. The entire PRPS1 coding region was sequenced (Sangon) and the result revealed a G→C mutant at nucleotide 51 resulting in a Gln17→His amino-acid substitution in the PRS1 protein. The recombinant plasmid was transformed into Escherichia coli strain BL21 (DE3) cells and transformed cells were selected at 310 K in LB medium containing 100 µg ml−1 ampicillin overnight. Expression of the recombinant protein was induced by 0.1 mM IPTG (isopropyl β-d-thiogalactopyranoside) when the culture reached an OD600 of 0.6. Cultivation was continued at 289 K for 24 h before the cells were harvested by centrifugation at 8000g at 277 K for 8 min. The pellet was suspended in lysis buffer (20 mM Tris–HCl pH 8.5, 0.1 mM phenylmethanesulfonyl fluoride) and then lysed by sonication on ice. The lysate was centrifuged at 22 600g at 277 K for 30 min. The supernatant solution was applied onto a 5 ml Ni–NTA column (Qiagen) equilibrated with binding buffer (20 mM Tris–HCl pH 8.5). The column was first washed with ten column volumes of binding buffer and the column was then eluted with a gradient of imidazole washing buffer (20 mM Tris–HCl pH 8.5, 20, 50 and 70 mM imidazole) in ten column volumes to remove impurities from the column. Finally, PRS1 was eluted with elution buffer (20 mM Tris–HCl pH 8.5, 300 mM imidazole) in five column volumes. The fraction containing PRS1 was pooled, concentrated and desalted by ultrafiltration (Millipore, 10 kDa cutoff). The protein concentration was estimated by Bradford assay (BioRad) prior to crystallization trials and the His tag at the C-terminus of PRS1 was not removed.
2.2. Crystallization
Crystallization experiments were performed at constant temperature (295 K) by the hanging-drop vapour-diffusion method using tissue-culture plates and siliconized glass cover slips. Crystal Screens I and II (Hampton Research) were utilized to screen the initial crystallization conditions. Small crystals could be seen after 24 h in drops made by mixing 1 µl protein solution (0.5 mg ml−1 in 20 mM Tris–HCl pH 8.5, 0.1 mM magnesium acetate) with 1 µl of reservoir solution (2.0 M ammonium sulfate, 0.1 M sodium acetate pH 4.6) and the droplet was equilibrated against 200 ml reservoir solution. Further optimizations were performed and diffraction-quality crystals of PRS1 were obtained after 6 d by mixing 2 µl protein solution (0.5 mg ml−1 in 20 mM Tris–HCl pH 8.5, 0.1 mM magnesium acetate, 0.1 mM monoammonium dihydrogen phosphate and 0.1 mM ATP) with 1.2 µl reservoir solution [2.1 M ammonium sulfate, 0.1 M sodium acetate pH 4.3, 10%(v/v) glycerol] and equilibrating against 350 ml reservoir solution at a constant temperature of 295 K.
2.3. X-ray diffraction data collection and preliminary analysis
Data collection was performed under cryogenic conditions (T = 100 K) in our laboratory using an imaging plate (MAR Research dtb345) and a rotating-anode X-ray source (Cu Kα; Rigaku Micro007). Crystals were soaked in a cryoprotectant solution consisting of reservoir solution (2.1 M ammonium sulfate, 0.1 M sodium acetate pH 4.3) containing 20% glycerol prior to data collection. A total of 48 diffraction images were recorded at a crystal-to-detector distance of 220 mm. The oscillation angle and exposure time for each imaging frame were set to 1° and 20 min, respectively. The diffraction data were processed using AUTOMAR (MAR Research, v.1.4).
3. Results and discussion
PRS1 was highly expressed as a soluble protein in E. coli strain BL21 (DE3) cells and the majority of the target protein PRS1 appeared in the fraction eluted using elution buffer (20 mM Tris–HCl pH 8.5, 300 mM imidazole). The purified PRS1 showed a major band of 35 kDa on SDS–PAGE, which is in good agreement with the calculated value of 34.8 kDa (Fig. 1 ▶). However, considerable precipitation appeared during the process of desalting and condensation; we tried changing to a different buffer to improve the solubility of PRS1, but the effect was limited. We could not carry out gel-filtration experiments to determine the oligomeric state of human PRS1 because of precipitation. The final concentration of PRS1 was only 1 mg ml−1, while the concentration necessary for crystallization was only 0.5 mg ml−1. We obtained small crystals through an initial screen and after further optimization high-quality crystals (Fig. 2 ▶) suitable for X-ray diffraction were obtained using the conditions described above. The final statistics of data collection and reduction are summarized in Table 1 ▶.
Figure 1.
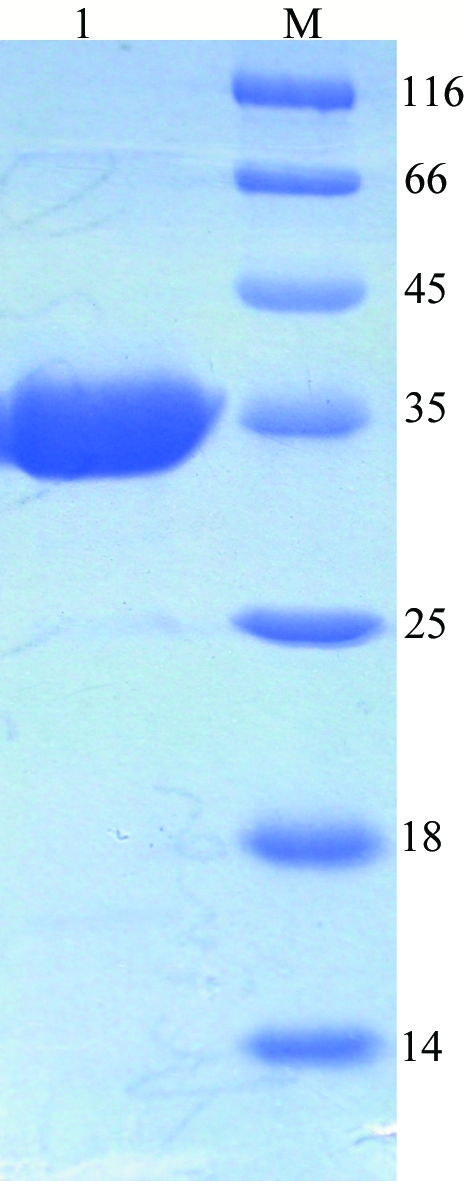
SDS–PAGE of PRS1 under non-reducing condition. Lane 1, purified fraction from Ni–NTA column (more than 95% purity); lane M, molecular-weight markers (kDa).
Figure 2.
Crystals of PRS1 in complex with ATP, Mg2+ and Pi grown by the hanging-drop method.
Table 1. Experimental conditions and data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.5418 |
| Temperature (K) | 100 |
| Space group | R3 |
| Unit-cell parameters (Å) | a = b = 168.846, c = 61.857 |
| Resolution limits (Å) | 30–2.6 |
| Total reflections | 30710 |
| Unique reflections | 19120 |
| Completeness (%) | 94.7 |
| Rmerge† (%) | 6.56 (40.63) |
| Mean I/σ(I) | 6.6 (1.3) |
| Redundancy | 1.59 |
| VM (Å3 Da−1) | 2.4 |
| Z | 2 |
| Solvent content (%) | 49.2 |
R
merge = 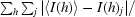
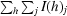 , where I(h)j is the observed reflection intensity and 〈I(h)〉 is the mean intensity of the reflection.
, where I(h)j is the observed reflection intensity and 〈I(h)〉 is the mean intensity of the reflection.
Determination of the structure of human PRS1 in complex with Mg2+, Pi and ATP will be resolved by the molecular-replacement method using phosphoribosyl pyrophosphate synthetase from B. subtilis as a search model (47% amino-acid sequence identity). Efforts to obtain crystals of PRS1 in complex with various combinations of different effectors (ADP, GDP, Pi, Mg2+ and GMP) and substrates (ATP, R5P) are under way. Once detailed structural information on these complexes has been obtained, it will lead to structure-based elucidation of the allosteric regulation of PRS1 activity and its substrate-binding specificity.
Acknowledgments
Financial support for this project to LN and MT was provided by research grants from the Chinese National Natural Science Foundation (grant Nos. 30121001, 30025012, 30130080 and 30571066) and the ‘973’ and ‘863’ Plans of the Chinese Ministry of Science and Technology (grant Nos. 2004CB520801 and 2002BA711A13).
References
- Becker, M. A. (2005). Phosphoribosyl Pyrophosphate Synthetase Superactivity. http://www.orpha.net/data/patho/GB/uk-prpp.pdf.
- Becker, M. A., Heidler, S. A., Bell, G. I., Seino, S., LeBeau, M. M., Westbrook, C. A., Neuman, W., Shapiro, L. J., Mohandas, T. K. & Roessler, B. J. (1990). Genomics, 8, 550–561. [DOI] [PubMed]
- Becker, M. A., Raivio, K. O., Bakay, B., Adams, W. B. & Nyhan, W. L. (1980). J. Clin. Invest.65, 109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, M. A., Smith, P. R., Taylor, W., Mustafi, R. & Switzer, R. L. (1995). J. Clin. Invest.96, 2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, M. A., Taylor, W., Smith, P. R. & Ahmed, M. (1996). J. Biol. Chem.271, 19894–19899. [DOI] [PubMed] [Google Scholar]
- Bower, S. G., Harlow, K. W., Switzer, R. L. & Hove-Jensen, B. (1989). J. Biol. Chem.264, 10287–10291. [PubMed] [Google Scholar]
- Eriksen, T. A., Kadziola, A., Bentsen, A.-K., Harlow, K. W. & Larsen, S. (2000). Nature Struct. Biol.7, 7303–7308. [DOI] [PubMed]
- Harlow, K. W. & Switzer, R. L. (1990). J. Biol. Chem. 265, 5487–5493. [PubMed] [Google Scholar]
- Hove-Jensen, B., Harlow, K. W., King, C. J. & Switzer, R. L. (1986). J. Biol. Chem.261, 6765–6771. [PubMed] [Google Scholar]
- Schubert, K. R., Switzer, R. L. & Shelton, E. (1975). J. Biol. Chem.250, 7492–7500. [PubMed] [Google Scholar]
- Simmonds, H. A., Webster, D. R., Wilson, J. & Lingham, S. (1982). Lancet, 2, 68–70. [DOI] [PubMed] [Google Scholar]
- Sperling, O., Boer, P., Persky-Brosh, S., Kanarek, E. & DeVries, A. (1972). Rev. Eur. Etud. Clin. Biol.17, 703–706. [PubMed] [Google Scholar]
- Switzer, R. L. (1969). J. Biol. Chem.244, 2854–2863. [PubMed] [Google Scholar]
- Taira, M., Iizasa, T., Yamada, K., Shimada, H. & Tatibana, M. (1989). Biochim. Biophys. Acta, 1007, 203–208. [DOI] [PubMed] [Google Scholar]
- Zoref, E., DeVries, A. & Sperling, O. (1975). J. Clin. Invest.56, 1093–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]