The crystal structure of ribose 5-phosphate isomerase from P. falciparum has been determined at 2.09 Å resolution. The structure is very similar to those of RpiA from other species, but is in some ways more similar to bacterial homologs than to those from higher eukaryotes.
Keywords: ribose 5-phosphate isomerase
Abstract
The structure of ribose 5-phosphate isomerase from Plasmodium falciparum, PFE0730c, has been determined by molecular replacement at 2.09 Å resolution. The enzyme, which catalyzes the isomerization reaction that interconverts ribose 5-phosphate and ribulose 5-phosphate, is a member of the pentose phosphate pathway. The P. falciparum enzyme belongs to the ribose 5-phosphate isomerase A family, Pfam family PF06562 (DUF1124), and is structurally similar to other members of the family.
1. Introduction
Ribose 5-phosphate isomerase (EC 5.3.1.6; Rpi) catalyzes the interconversion of the ketose ribose 5-phosphate and its isomer, the aldose ribulose 5-phosphate, in photosynthetic carbon fixation and in the pentose phosphate pathway, where it is used for purine- and pyrimidine-nucleoside synthesis and for histidine synthesis. The enzyme exists as two distinct proteins, RpiA and RpiB. Although RpiA and RpiB catalyze the same reaction, they show no sequence or overall structural homology. A number of RpiA and RpiB structures have been determined with and without ligands. RpiA is found in all three kingdoms of life and structures are available from Escherichia coli, Haemophilus influenzae, Pyrococcus horikoshii, Saccharomyces cerevisiae and Thermus thermophilus. RpiB is present in bacteria and also in the trypanosomatids (Leishmania spp., Trypanosoma) and related pathogenic eukaryotes (Entamoeba histolytica, Giardia lamblia). Structures of RpiB are only known from bacteria.
Searches against all Plasmodium spp. genomic sequences in the PlasmoDB sequence database (Bahl et al., 2002 ▶) using the TBLASTN 2.1.2 protocol and the E. coli, L. major and E. histolytica RpiB gene sequences as probes found no plausible instances of a plasmodial RpiB gene. The only hit of marginal significance (E = 0.08) was against a P. falciparum chr 10 sequence annotated as ‘rifin surface antigenic variant’. We therefore conclude that RpiA is the sole form of this enzyme present in plasmodia. The present structure determination of an RpiA from P. falciparum was undertaken as part of the SGPP (Structural Genomics of Pathogenic Protozoa Consortium; http://www.sgpp.org) effort targeting proteins from eukaryotic tropical pathogens.
2. Materials and methods
Ligase-independent cloning (Aslanidis & de Jong, 1990 ▶) was used to append a His tag to the N-terminus and a TAA stop codon to the C-terminus of the PFE0730c gene of P. falciparum, giving MAHHHHHH-orf-TAA (SGPP target identifier Pfal008434AAA). The vector has a T7 promoter for growth in E. coli with auto-induction media (without the use of IPTG). Selenomethionine protein was produced in E. coli strain BL21 DE3 Star according to the protocols of Studier (2005 ▶) and SGPP (Mehlin et al., 2006 ▶).
Pfal008434AAA (8 mg ml−1) was screened for crystallization conditions at the Hauptman–Woodward Institute high-throughput screening facility (Luft et al., 2003 ▶). The protein was combined with 1536 different crystallization cocktail solutions in a single plate under mineral oil to prevent dehydration. Experiments were set up using standard commercially available liquid-handling systems. Plates were imaged over a four-week time course. Images were reviewed and crystallization conditions were forwarded to the crystal-growth laboratory in Seattle for optimization. There, crystallization conditions from the initial screen were optimized for pH, major precipitant and additive concentrations using a vapor-diffusion sitting-drop method. Crystallization conditions for the first crystal form were 100 mM NH4SCN, 15% PEG 1000, 50 mM HEPES pH 8.2. The crystallization conditions for the crystal used for structure solution were 200 mM NaCl, 0.6 mM KAu(CN)2, 20% PEG 3000, 100 mM HEPES pH 7.5 at 277 K. Both types of crystals were flash-frozen directly in liquid nitrogen prior to shipping for data collection.
The first crystal form grown of Pfal008434AAA was in space group P1, with 8–10 monomers predicted in the asymmetric unit based on V M calculations (Matthews, 1968 ▶; later determined by searching with the refined structure to be eight monomers, giving a V M of 2.8 Å3 Da−1). Molecular replacement using the program EPMR (Kissinger et al., 1999 ▶) and monomer search models from PDB entries 1uj5 (T. thermophilus) and 1lk7 (P. horikoshii) did not succeed. Attempts to phase using SeMet MAD data from the same crystal form were also unsuccessful. In an effort to find a heavy-atom derivative, the protein was cocrystallized with KAu(CN)2 and diffraction data were collected at the Au peak wavelength at the Advanced Light Source beamline 8.2.1. The data were integrated and scaled using HKL2000 (Otwinowski & Minor, 1997 ▶). There was no evident anomalous scattering signal that would indicate incorporation of Au, but the protein had crystallized in a different space group, P21212, with two monomers in the asymmetric unit, giving a V M of 2.6 Å3 Da−1. The structure was solved by molecular replacement, using the program EPMR and a monomer from 1uj5, T. thermophilus RpiA (Hamada et al., 2003 ▶), as the search model. The asymmetric unit contains one dimer of RpiA. Alternating rounds of modelling using the program XFIT (McRee, 1999 ▶) and refinement using the program REFMAC5 (Murshudov et al., 1997 ▶) via the CCP4i interface (Potterton et al., 2004 ▶) were carried out. Non-crystallographic symmetry restraints were not used. Refinement was monitored using 5% of the data reserved for R free. Final structure validation was performed with PROCHECK (Laskowski et al., 1993 ▶), MOLPROBITY (Lovell et al., 2003 ▶) and COOT (Emsley & Cowtan, 2004 ▶). The model of the A chain consists of the last four residues of the His tag and residues 1–233 of the 236-residue open reading frame. The model of the B chain consists of residues 1–235. No density corresponding to Au atoms was seen, but a phosphate ion was clearly visible in each of the two active sites.
3. Results and discussion
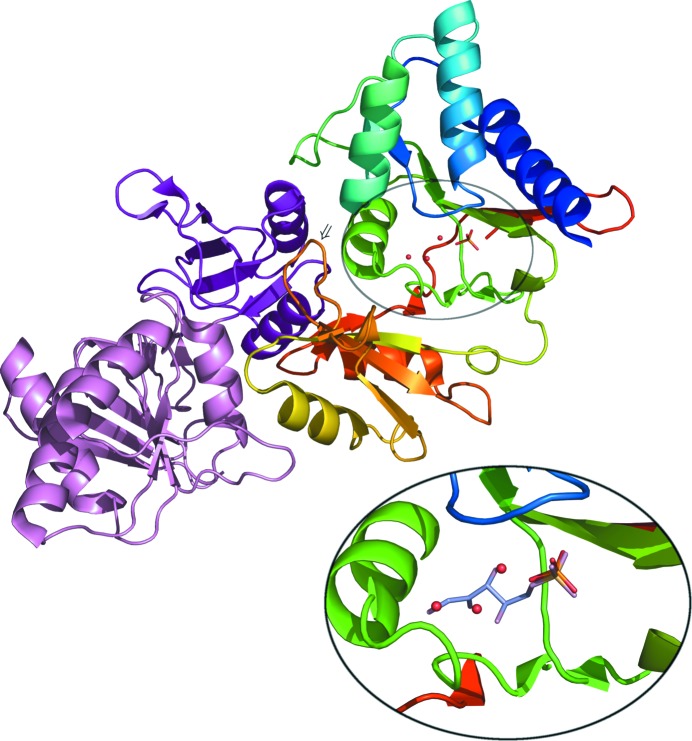
The structure of Pfal008434AAA has been solved to 2.09 Å. Data-collection statistics are shown in Table 1 ▶ and refinement statistics are shown in Table 2 ▶. The enzyme crystallizes with a dimer in the asymmetric unit; a ribbon drawing of the dimer is shown in Fig. 1 ▶. P. falciparum RpiA has 40% sequence identity over a 222-residue span with the search model from T. thermophilus; sequence identity with the other previously determined RpiA structures ranges from 31 to 36%. The human RpiA enzyme, the structure of which has not been determined, also falls in this range, with a sequence identity of 35%. As expected from the level of sequence identity with the other structures, Pfal008434AAA adopts the canonical RpiA fold. The RpiA monomer is a mixed αβ protein which consists of two domains of unequal size. The larger domain is comprised of two stretches of chain, residues 1–129 and 210–236, and the smaller domain is comprised of the intervening stretch, residues 130–209. The active site is located in the larger domain and the ‘tetramerization loop’ (see below) is part of the smaller domain. The two monomers superimpose with an r.m.s. difference of 0.41 Å for 233 Cα atoms. The largest differences between the two chains, up to 2.8 Å, are in a loop in the large domain and are seen for residues 75–78, which are involved in crystal-packing interactions.
Table 1. Data-collection statistics.
| Space group | P21212 |
| Unit-cell parameters () | a = 93.7, b = 136.2, c = 45.0 |
| Wavelength () | 1.03492 |
| Resolution range () | 502.08 (2.142.08) |
| No. unique reflections | 31605 |
| Redundancy | 5.9 (4.5) |
| Completeness (%) | 92.7 (96.6) |
| R merge | 0.073 (0.368) |
| Mean I/(I) | 26.5 (4.0) |
Table 2. Refinement and model statistics.
Values in parentheses are for the highest resolution shell.
| Resolution range () | 502.09 (2.142.09) |
| R work (18076 reflections) | 0.209 (0.280) |
| R free (926 reflections) | 0.272 (0.321) |
| R.m.s.d. bonds () (REFMAC v.5.2.005) | 0.017 |
| R.m.s.d. angles () | 1.625 |
| Residues in most favored region of / (PROCHECK) | 400 [94.8%] |
| Residues in additional allowed region of / | 22 [5.2%] |
| Residues in generously allowed region of / | 0 |
| Residues in disallowed region of / | 0 |
| Mean B factor (2), protein atoms (3555) | 28.7 |
| Mean B factor (2), water molecules (226) | 29.1 |
| Mean B factor (2), phosphate atoms (10) | 41.6 |
Figure 1.
Main figure: ribbon representation of the Pfal008434AAA dimer. The A chain is colored smoothly from blue at the N-terminus to red at the C-terminus. The B chain is colored by domain, with the larger domain in pink and the smaller domain in purple. The tetramerization loop of chain A is indicated by an arrow. Inset: enlarged view of the active site of the Pfal008434AAA A chain. The bound phosphate ion is shown in stick form and the three water molecules mentioned in the text are indicated as spheres. The substrate ribose 5-phosphate (light blue) from the T. thermophilus structure 1uj5 is shown superposed onto the Pfal008434AAA active site based on superposition of 27 Cα atoms in two stretches of chain that contain ten of the 11 residues directly in contact with the ribose 5-phosphate. The figure was generated using PyMOL (DeLano, 2002 ▶).
In the present structure, there is a phosphate ion bound in the active site of each monomer (Fig. 1 ▶, inset). Phosphate ion is a weak inhibitor (K i = 7.9 mM) of spinach chloroplast RpiA (Jung et al., 2000 ▶). Phosphate was not present in the protein buffer or crystallization solutions; presumably, it was acquired by the enzyme during protein expression. A phosphate ion was previously seen bound to the active site of RpiB from Mycobacterium tuberculosis, for which it is also a weak inhibitor (K i = 130 mM; Roos et al., 2004 ▶), but that protein was crystallized from sodium/potassium phosphate. Phosphate ions have not been seen in the active sites of previously determined RpiA structures. In our model, the phosphate ion is in the same location as the phosphate of ribose 5-phosphate in the T. thermophilus structure 1uj5 and participates in the same five direct phosphate–protein hydrogen bonds. In the 1uj5 structure, the phosphate O atoms also form hydrogen bonds to six water molecules; three of those water molecules are modelled in our structure and they interact similarly with the phosphate ion. The Pfal008434AAA active site also contains three additional water molecules. These are located at roughly the positions of the ribose hydroxyl O atoms O1, O2 and O3 and form the same hydrogen bonds to the protein as the ribose O atoms, although with slightly different geometry.
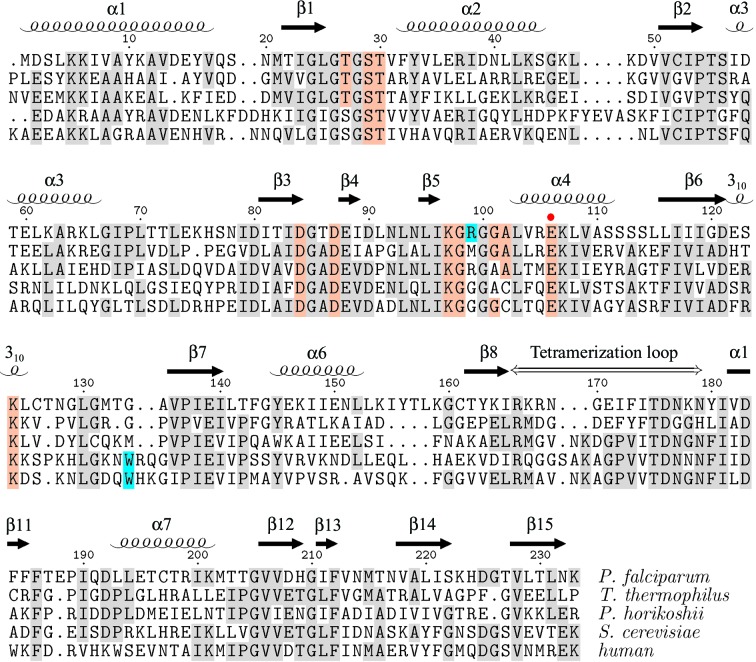
Members of the RpiA family exist in two oligomerization states, as observed in the structures determined thus far. The bacterial RpiAs (E. coli, H. influenzae, T. thermophilus) form dimers (Rangarajan et al., 2002 ▶; Zhang et al., 2003 ▶; Hamada et al., 2003 ▶; K. Das, R. Xioa, T. Acton, G. Montelione & E. Arnold, PDB code 1m0s), while the archaeal (Py. horikoshii) and eukaryotic (S. cerevisiae) enzymes form tetramers (Ishikawa et al., 2002 ▶; Graille et al., 2005 ▶). However, the P. falciparum structure presented here is observed in the crystal as a dimer. Furthermore, in the size-exclusion chromatography step of protein purification, the protein ran as a dimer (98%) with a minor 74 kDa peak (1%) that may correspond to a tetramer. Only the major peak fraction was taken into crystallization trials. The RpiA oligomerization state has been postulated (Ishikawa et al., 2002 ▶; Graille et al., 2005 ▶) to be determined by the length of what could be called the ‘tetramerization loop’. Yeast and other eukaryotes typically have 16–17 residues in this loop, as do the archaea. In previously observed structures containing this longer loop, it has a conformation which allows interaction with the same loop of a second dimer to form a tetramer. The equivalent loop is shorter in the dimeric bacterial structures. The P. falciparum loop is 14 residues long. Structure-based sequence alignment of the P. falciparum sequence with bacterial, archaeal and eukaryotic sequences shows that Pfal008434AAA has the shorter loop seen in bacterial forms of the enzyme, consistent with the dimer being the predominant form in solution (Fig. 2 ▶).
Figure 2.
Structure-based sequence alignment of ribose 5-phosphate isomerase RpiA from P. falciparum (2f8m), T. thermophilus (1uj5), Py. horikoshii (1lk5) and S. cerevisiae (1xtz). Residue numbering and secondary-structure elements are those of the present structure. The alignment was produced by the CEMC server (Guda et al., 2001 ▶, 2004 ▶). The human RpiA sequence, for which no structure has yet been reported, was added separately. Conserved residues that directly contact the ribose 5-phosphate in the 1uj5 structure are indicated by colored shading and the proposed general base/acid (Glu106 in P. falciparum) is indicated by a red dot (Ishikawa et al., 2002 ▶; Zhang et al., 2003 ▶; Hamada et al., 2003 ▶). Residues contributing to a significantly different binding site surface in the P. falciparum and human enzymes are highlighted in cyan. The figure was generated using TEXshade (Beitz, 2000 ▶).
The extent and nature of the RpiA active site are known from previous paired apo and substrate-bound structures from T. thermophilus, Py. horikoshii and E. coli. It is noteworthy that there are no significant conformational changes in active-site residue conformations between the apo (PDB code 1uj4) and substrate-bound (PDB code 1uj5) T. thermophilus structures (Hamada et al., 2003 ▶). Conformational changes at the active site upon d-4-phosphoerithronic acid binding to the Py. horikoshii enzyme (Ishikawa et al., 2002 ▶) and upon arabinose-5-phosphate binding to the E. coli enzyme are also observed to be minimal (Rangarajan et al., 2002 ▶; Zhang et al., 2003 ▶). The location of the phosphate moiety is consistent in all of these complexes and in the current structure.
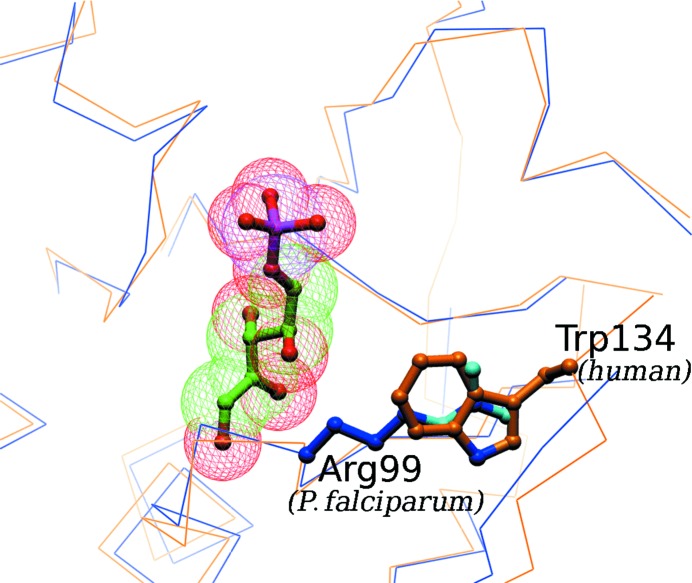
A detailed comparison of the corresponding active-site residues in the P. falciparum and human RpiA protein sequences revealed an unexpected difference. Near the 4-hydroxyl of ribose-5-phosphate, the position of which is inferred from the structure of the T. thermophilus complex (PDB code 1uj5), the active site is lined by a loop between β5 and α4. This loop has sequence KGRGGA in the Plasmodium enzyme, while the sequence is KGGGGC in the human enzyme (Fig. 2 ▶). There is no crystal structure of the human enzyme, but the S. cerevisiae enzyme structure (PDB code 1xtz) is 45% identical in sequence and provides the basis for a homology model illuminating a potentially exploitable difference in binding-site features at this point. The human and yeast enzymes contain a short insertion relative to the P. falciparum sequence near residue 135 (Fig. 2 ▶). Surprisingly, the space occupied by Arg99 in the Plasmodium enzyme structure is instead occupied by a tryptophan side chain in the yeast structure, originating from a loop between β6 and β7 (Fig. 3 ▶). Although the homologous Arg residue in P. horikoshii is not required for enzyme activity (Ishikawa et al., 2002 ▶), this Arg versus Trp difference may provide a basis for the design of selective inhibitors of the parasite enzyme. Chemical substituents that favor a polar interaction or hydrogen bond with the Arg99 side chain of the binding site of the P. falciparum enzyme will not be so favored by the purely hydrophobic face of Trp134 in the human enzyme’s binding site.
Figure 3.
Potentially exploitable difference in the ribose-5-phosphate-binding site in the P. falciparum and human RpiA proteins. The blue backbone trace in the figure is that of the P. falciparum structure presented here; the gold trace is that of a homology model for the human RpiA based on the crystal structure (1xtz) of the yeast homolog. The ribose-5-phosphate substrate is positioned as seen in the T. thermophilus enzyme–substrate complex (1uj5). The very different binding-site environment to the right of the substrate, Arg99 in P. falciparum and Trp134 in humans (Trp157 in yeast), suggests that it would be possible to design inhibitors with very different affinities for the two homologous enzymes.
The structure presented here is the second structure determination of a eukaryotic ribose 5-phosphate isomerase. This enzyme is essential in the model eukaryote S. cerevisiae (Giaever, 2002 ▶). Its role in Plasmodium biology has not been evaluated using knockout technology, but owing to its central role in the pentose phosphate pathway, RpiA is likely to be essential in P. falciparum. The pentose phosphate pathway serves two critical cellular functions: it generates NADPH for reducing power and it produces ribose-5-phosphate, the sugar component of nucleic acids. Plasmodium cells have a critical need for an abundant supply of reducing power in order to sustain their rapid growth and to detoxify heme, the product of hemoglobin digestion (Becker et al., 2003 ▶). Plasmodium also has an intense requirement for nucleic acid production to support its rapid proliferation. In fact, the ribose product of the pentose phosphate shunt (5-phospho-d-ribose 1-pyrophosphoric acid) that goes into nucleic acids is increased 56-fold in concentration in infected erythrocytes compared with uninfected erythrocytes (Roth et al., 1986 ▶). It follows that ribose 5-phosphate isomerase is likely to be a good chemotherapeutic target for Plasmodium.
Supplementary Material
PDB reference: ribose 5-phosphate isomerase, 2f8m, r2f8msf
Acknowledgments
We are grateful for the contributions of other SGPP consortium members, including Peter Myler, Elizabeth Worthey, Helen Neeley, Tracy Arakaki, Jürgen Bosch, Jonathan Caruthers, Mark A. Robien, Larry de Soto and Martin Criminale. We also thank Michael H. Gelb. Portions of this work were carried out at the Advanced Light Source, which is supported by the Director, Office of Science, Office of Basic Energy Sciences of the US Department of Energy under Contract No. DE-AC02-05CH11231. This work was supported by NIH awards GM64655 and GM62617.
References
- Aslanidis, C. & de Jong, P. J. (1990). Nucleic Acids Res. 18, 6069–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl, A., Brunk, B., Coppel, R. L., Crabtree, J., Diskin, S. J., Fraunholz, M. J., Grant, G. R., Gupta, D., Huestis, R. L., Kissinger, J. C., Labo, P., Li, L., McWeeney, S. K., Milgram, A. J., Roos, D. S., Schug, J. & Stoeckert, C. J. Jr (2002). Nucleic Acids Res. 30, 87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, K., Rahlfs, S., Nickel, C. & Schirmer, R. H. (2003). Biol. Chem. 384, 551–566. [DOI] [PubMed] [Google Scholar]
- Beitz, E. (2000). Bioinformatics, 16, 135–139. [DOI] [PubMed] [Google Scholar]
- DeLano, W. (2002). The PyMOL Molecular Graphics System. http://www.pymol.org.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Giaever, G. et al. (2002). Nature (London), 418, 387–391. [Google Scholar]
- Graille, M., Meyer, P., Leulliot, N., Sorel, I., Janin, J., Tilbeurgh, H. V. & Quevillon-Cheruel, S. (2005). Biochimie, 87, 763–769. [DOI] [PubMed] [Google Scholar]
- Guda, C., Lu, S., Scheeff, E. D., Bourne, P. E. & Shindyalov, I. N. (2004). Nucleic Acids Res. 32, W100–W103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guda, C., Scheeff, E. D., Bourne, P. E. & Shindyalov, I. N. (2001). Pac. Symp. Biocomput. 6, 275–286. [DOI] [PubMed]
- Hamada, K., Ago, H., Sugahara, M., Nodake, Y., Kuramitsu, S. & Miyano, M. (2003). J. Biol. Chem. 278, 49183–49190. [DOI] [PubMed] [Google Scholar]
- Ishikawa, K., Matsui, I., Payan, F., Cambillau, C., Ishida, H., Kawarabayasi, Y., Kikuchi, H. & Roussel, A. (2002). Structure, 10, 877–886. [DOI] [PubMed] [Google Scholar]
- Jung, C. H., Hartman, F. C., Lu, T. Y. & Larimer, F. W. (2000). Arch. Biochem. Biophys. 373, 409–417. [DOI] [PubMed] [Google Scholar]
- Kissinger, C. R., Gehlhaar, D. K. & Fogel, D. B. (1999). Acta Cryst. D55, 484–491. [DOI] [PubMed] [Google Scholar]
- Laskowski, R., MacArthur, M., Moss, D. & Thornton, J. (1993). J. Appl. Cryst. 26, 283–291. [Google Scholar]
- Lovell, S., Davis, I., Arendall, W. B. III, de Bakker, P., Word, J., Prisant, M., Richardson, J. & Richardson, D. (2003). Proteins, 50, 437–450. [DOI] [PubMed] [Google Scholar]
- Luft, J. R., Collins, R. J., Fehrman, N. A., Lauricella, A. M., Veatch, C. K. & DeTitta, G. T. (2003). J. Struct. Biol. 142, 170–179. [DOI] [PubMed] [Google Scholar]
- McRee, D. E. (1999). J. Struct. Biol. 125, 156–165. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed] [Google Scholar]
- Mehlin, C., Boni, E., Buckner, F. S., Engel, L., Feist, T., Gelb, M., Haji, L., Kim, D., Liu, C., Mueller, N., Myler, P. J., Reddy, J. T., Sampson, J. N., Subramanian, E., Van Voorhis, W. C., Worthey, E., Zucker, F. & Hol, W. G. J. (2006). In the press.
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Potterton, L., McNicholas, S., Krissinel, E., Gruber, J., Cowtan, K., Emsley, P., Murshudov, G. N., Cohen, S., Perrakis, A. & Noble, M. (2004). Acta Cryst. D60, 2288–2294. [DOI] [PubMed] [Google Scholar]
- Rangarajan, E. S., Sivaraman, J., Matte, A. & Cygler, M. (2002). Proteins, 48, 737–740. [DOI] [PubMed] [Google Scholar]
- Roos, A. K., Andersson, C. E., Bergfors, T., Jacobsson, M., Karlen, A., Unge, T., Jones, T. A. & Mowbray, S. L. (2004). J. Mol. Biol. 335, 799–809. [DOI] [PubMed] [Google Scholar]
- Roth, E. F. Jr, Ruprecht, R. M., Schulman, S., Vanderberg, J. & Olson, J. A. (1986). J. Clin. Invest. 77, 1129–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier, F. W. (2005). Protein Expr. Purif. 41, 207–234. [DOI] [PubMed] [Google Scholar]
- Zhang, R., Andersson, C. E., Savchenko, A., Skarina, T., Evdokimova, E., Beasley, S., Arrowsmith, C. H., Edwards, A. M., Joachimiak, A. & Mowbray, S. L. (2003). Structure, 11, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: ribose 5-phosphate isomerase, 2f8m, r2f8msf