Abstract
Mycoplasma insons sp. nov., first cultured from the choanae and tracheae of healthy green iguanas (Iguana iguana) from El Salvador, was readily distinguished from all previously described mollicutes and assigned to the Mycoplasma fastidiosum phylogenetic cluster by 16S rRNA gene sequence comparisons. Growth inhibition assays distinguished the isolates serologically from the other two members of that cluster. Many M. insons cells exhibit a remarkable twisted rod morphology despite lacking a cell wall. The organism is nonmotile, produces acid from glucose, but does not hydrolyze arginine, esculin, or urea. Mycoplasma insons 16S rRNA gene was also detected by PCR in packed blood cells from culture-negative iguanas. The type strain I17P1T has been deposited with the Mollicutes Collection at Purdue University and with the American Type Culture Collection (ATCC BAA-1435) in the USA. A limited number of cultures generated by the authors have also been deposited with the Culture Collection, University of Göteborg, in Sweden (CCUG 53461).
Keywords: novel species, mycoplasma, mollicutes, fastidiosum cluster, twisted morphology, hemotropic
Introduction
Reptiles are susceptible to infection with diverse mycoplasmas, and experience a wide range of effects of mycoplasmosis, similar to other vertebrates (Brown, 2002). A single species, Mycoplasma iguanae, is currently known to infect green iguanas, and was presumptively associated with spinal lesions (Brown et al., 2006). Subsequent infection studies failed to demonstrate pathogenic potential for M. iguanae (Brown et al., 2007). Mycoplasmal isolates recovered from healthy iguanas during infection studies were found to differ from M. iguanae and all known mollicutes in the 16S rRNA gene sequence.
This study aims to characterize five clonal isolates of this novel species. The name Mycoplasma insons, reflecting its apparent commensalism with iguanas, is proposed. Numerous genetic, biochemical, and morphological assessments have been conducted of the type strain I17P1T and four additional isolates (I17P2–I17P5).
Materials and methods
Bacterial culture conditions
Tracheal and choanal swabs from green iguanas were assessed for the presence of mycoplasmas. Primary isolation of M. insons was on American Type Culture Collection (ATCC) medium 988 (SP-4) agar, pH 7.6–7.8, containing 105 U L−1 penicillin G, 105 U L−1 polymyxin B, 65 mg L−1 cefoperazone, and 20% v/v fetal bovine serum, incubated at 30°C in 5% CO2. Five triple filter-cloned (Tully, 1983) strains (I17P1T and I17P2–I17P5) were characterized. Assessment of arginine hydrolysis was performed by growth of M. insons in SP-4 medium supplemented with 0.21% w/v L-arginine. Similarly, esculin hydrolysis was detected by culturing of M. insons on SP-4 agar supplemented with 1 mg mL−1 esculin. The urease production was assessed by growth in 10-B medium. The requirement for cholesterol was confirmed by attempted passage in SP-4 medium without fetal bovine serum. The temperature range of M. insons was determined by broth culture at 4, 25, 30, 37, and 44°C.
DNA amplification and sequencing
Genomic DNA was extracted from M. insons using Easy DNA reagents according to the manufacturer’s specifica-tions (Invitrogen, Carlsbad, CA). Mycoplasmal 16S rRNA gene was amplified using established mollicutes primers (Brown et al., 1995). Nucleotide sequencing of the 16S rRNA gene was performed at the Interdisciplinary Center for Biotechnology Research at the University of Florida according to their standard operating procedure.
Phylogenetic analysis
The 16S rRNA gene sequence of M. insons was aligned with the prokaryotic small subunit rRNA sequences in the Ribosomal Database Project release 8.1 using SEQUENCE_ MATCH version 2.7 (Cole et al., 2003). A Kimura 2-parameter distance matrix (Felsenstein, 1989) was then generated to establish the relationship of strain I17P1T with the most closely related species, Mycoplasma fastidiosum and Mycoplasma cavipharyngis.
Identification of M. insons in iguana tissues
The recovery of mycoplasmas was attempted by culture from the trachea, choanae, conjunctivae, lungs, liver, spleen, heart, brain, spinal cord, limb joint synovial fluid, and whole peripheral blood in SP-4 medium. The detection of M. insons in cultures obtained from tracheal and choanal swabs and in packed blood cells of green iguanas was performed by amplification of the 16S rRNA gene, followed by restriction fragment length polymorphism (RFLP) analysis using EcoRI to distinguish between M. insons and M. iguanae. The genetic homogeneity of M. insons clones was demonstrated by random amplification of polymorphic DNA (RAPD) fingerprinting as described previously (Geary et al., 1994; Mettifogo et al., 2006), and protein gel electrophoresis under denaturing conditions.
Growth inhibition studies
The requirement for sterols was further assessed by digitonin sensitivity assays as described previously (Tully, 1995). Sensitivity to hyperimmune antisera raised against M. insons and its two closest relatives, M. fastidiosum and M. cavipharyngis, was determined as described by Clyde (1983). Mycoplasma insons I17P1T antiserum was prepared as follows: each of two rabbits was immunized with 250 μg I17P1T whole-cell lysate mixed with complete Freund’s adjuvant on day 1 of a 98-day protocol (Lampire Biological Laboratories, Pipersville, PA). Each rabbit was subsequently immunized with 250 μg I17P1T whole-cell lysate mixed with incomplete Freund’s adjuvant on days 7, 14, 28, 56, and 84.
Microscopy
Cultures of M. insons were grown attached to coverslips at 30°C in SP-4 broth for scanning electron microscopy. Cells were fixed in 2% formaldehyde/1.5% glutaraldehyde/0.1 M Na cacodylate for 30 min, dehydrated in a series of increasing concentrations of EtOH to 100%, critical point dried, and gold coated. Transmission electron microscopy was performed at the Interdisciplinary Center for Biotechnology Research at the University of Florida according to their standard operating procedure. Microcinematography to assess gliding motility was performed as described previously (Hatchel et al., 2006).
Results
Growth characteristics of M. insons
Five isolates were shown to be genetically and phenotypically indistinguishable by RAPD and denaturing protein gel electrophoresis before characterization. Colonies of the organism had a typical umbonate morphology and were variable in size from 100 to 300 μm diameter following growth for 4–7 days. Their sharply defined margins and the absence of satellite colony formation on SP-4 agar indicated that the organisms were nonmotile (International Committee on Systematic Bacteriology Subcommittee on the Taxonomy of Mollicutes, 1995). The maximal growth rate occurred at 30°C, as indicated by acidification of SP-4 broth supplemented with glucose and incubated in ambient air without agitation. The organisms also grew in broth at 25, 30, and 37°C, but did not grow at 4 or 44 °C. All five strains produced acid from glucose (Razin & Cirillo, 1983), but did not hydrolyze arginine, esculin, or urea (Aluotto et al., 1970). All grew in SP-4 broth containing as little as 0.2% v/v fetal bovine serum, but none could be passaged in serum-free medium (Tully, 1995). For diagnostic purposes, a comparative table indicating the similarities and differences between M. iguanae and M. insons is provided (Table 1).
Table 1.
Comparison of Mycoplasma iguanae* with Mycoplasma insons
| Mycoplasma iguanae | Mycoplasma insons | |
|---|---|---|
| Habitat | Green iguana | Green iguana |
| Suspected tropisms | Central nervous system | Respiratory tract, blood cells |
| Colonial morphology | Umbonate | Umbonate |
| Cellular morphology | Pleomorphic | Pleomorphic, twisted rod |
| Respiration | Facultative anaerobe | Facultative anaerobe |
| Optimum (range) growth temperature | 37 (25–42) °C | 30 (25–37) °C |
| Motility | – | – |
| Glucose fermentation | + | + |
| Arginine hydrolysis | – | – |
| Urea hydrolysis | – | – |
| Serum requirement | + | + |
| Digitonin sensitivity | + | + |
| Esculin hydrolysis | – | – |
| 16S rRNA gene sequence | GenBank AY714305 | GenBank DQ522159 |
| rRNA gene intergenic spacer sequence | GenBank DQ840509 | GenBank DQ840508 |
Phylogenetic analysis
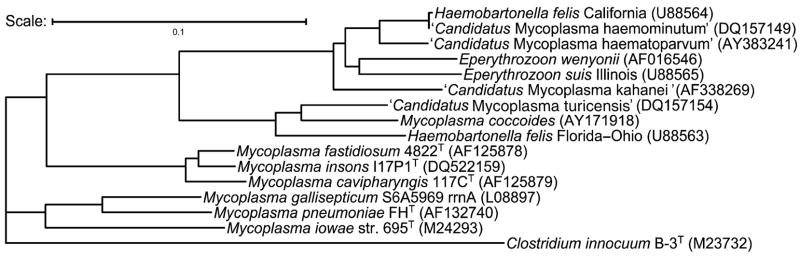
Alignment of the 16S rRNA gene sequence of M. insons (GenBank accession no. DQ522159) revealed that the best match (score = 0.814) was to M. fastidiosum strain 4822T, supporting taxonomic assignment to the M. fastidiosum phylogenetic cluster within the genus Mycoplasma (Johansson & Pettersson, 2002). The Kimura two-parameter distance matrix (Felsenstein, 1989) established the relationship of the isolates with M. fastidiosum and M. cavipharyngis, the only other species in the cluster (matrix similarity scores = 0.971 and 0.950, respectively), and closely related species (Fig. 1). The next closest scores were to Mycoplasma iowae, Mycoplasma gallisepticum, and Mycoplasma pneumoniae (similarity scores = 0.836, 0.834, and 0.828, respectively). Signature nucleotides, including T250, T296, A307, T441, A637, T886, T1287, A1317, and G1352, and the higher order structural characteristics of M. fastidiosum and M. cavipharyngis 16S rRNA gene shared with the hemotropic mollicutes (Johansson et al., 1999) were also conserved in the 16S rRNA gene sequence of strain I17P1T, although it differed from those species by the unique motif TGCA in positions 1188–1191.
Fig. 1.
Unrooted neighbor-joining phylogram of 16S rRNA gene sequences of species within the Mycoplasma fastidiosum cluster and closely related species most similar to the 16S rRNA gene sequence of Mycoplasma insons I17P1T. The phylogram was generated using the PHYLIP program neighbor with default parameters (Felsenstein, 1989). Scale bar = 0.1 substitutions site−1.
Identification of M. insons in iguana tissues
Mycoplasma insons was recovered by culture from swabs of the choanae or tracheae of seven out of 15 total clinically normal iguanas examined. Mycoplasma iguanae, the only other mycoplasma known from iguanas (Brown et al., 2006), was not recovered from any of those iguanas. No mycoplasmas were recovered by culture of conjunctivae, lungs, liver, spleen, heart, brain, spinal cord, limb joint synovial fluid, or whole peripheral blood specimens, and histologic examinations of these tissues did not reveal any lesions. However, the M. insons 16S rRNA gene was readily detected in packed blood cells from six out of the seven upper respiratory tract culture-positive and eight culture-negative iguanas by PCR, followed by RFLP analysis. No association between blood cell PCR results and mean packed cell volumes was evident.
Growth inhibition studies
All examined isolates were sensitive to digitonin (Tully, 1995) with 23–27-mm-diameter zones of inhibition. The growth of M. insons colonies was not inhibited by neat antisera to M. fastidiosum or M. cavipharyngis, but homologous antiserum yielded 3 mm zones of inhibition.
Microscopy
Scanning electron micrographs of strain I17P1T in the exponential phase of growth showed cells that were pleomorphic; however, many appeared remarkably similar to M. fastidiosum, a wall-less twisted filamentous rod measuring up to 2 μm in length (Lemcke & Poland, 1980). Parallel raised ridges following a helical path along the highly textured surface were sometimes prominent (Fig. 2). The cells were surrounded by a single-unit membrane but had no wall. Also similar to M. fastidiosum, diagonal striations across many of the cell sections, and an extracellular matrix of low electron density closely applied to the cell membrane were striking features evident in transmission electron micrographs (Fig. 3). The absence of gliding motility suggested by the lack of satellite colonies was confirmed by microcinematography.
Fig. 2.

Scanning electron micrograph of Mycoplasma insons I17P1Tcells showing a twisted rod morphology (inset), absence of the coiled symmetry of spiroplasmas, and a highly textured surface. Raised ridges following a helical path along the surface are prominent on some cells (arrows).
Fig. 3.
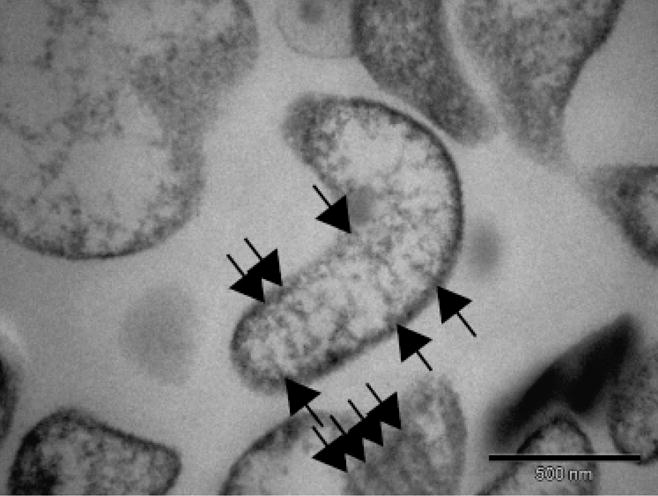
Transmission electron micrograph of a thin section of Mycoplasma insons I17P1T cells showing a single cytoplasmic membrane, absence of cell wall, and absence of a tip structure. Diagonal striations across the cut cell sections (arrows) were similar to Mycoplasma fastidiosum (Lemcke & Poland, 1980).
Discussion
The broad range of host environments capable of supporting mollicutes is quite remarkable, given the high degree of host specificity for many individual species. Here, the second mycoplasma demonstrated to live in association with green iguanas has been presented. The isolation of M. insons from the tracheae and choanae of healthy animals shows that it can exist as an innocent commensal among their upper respiratory tract flora. The detection of M. insons DNA in packed blood cells suggests that the organism may be facultatively hemotropic; however, the clinically normal packed cell volumes of infected iguanas suggest that the presence of M. insons is not a predisposing factor for infectious anemia.
The overall matrix similarity scores provide evidence that the M. insons isolates represent a distinct species (Stackebrandt & Goebel, 1994; Stackebrandt et al., 2002) that falls within the M. fastidiosum phylogenetic cluster. The 16S rRNA gene retained unique signature nucleotides and the higher order structural characteristics of M. fastidiosum and M. cavipharyngis. These qualities are also found within the hemotropic mollicutes (Johansson et al., 1999). The taxonomic association of M. insons with both the hemotropic mollicutes and M. fastidiosum is significant in that it supports two phenotypic characteristics: the detection of the organism in packed blood cells and the highly unusual morphology.
Although they did not display the coiled symmetry of spiroplasmas, M. insons cells appeared elongated and twisted, and at times raised ridges and diagonal striations (visible in cross-sections) were prominent. This morphology is highly unusual for mollicutes, which tend to be extremely pleomorphic due to the lack of a cell wall. Among mollicutes, only M. fastidiosum resembles M. insons in this regard.
The properties of M. insons strain I17P1T described herein that fulfill the criteria for assignment to the class Mollicutes (International Subcommittee on the Taxonomy of the Mollicutes, 1995) include absence of a cell wall, filterability, and penicillin resistance. Facultative anaerobic growth in artificial media and the necessity of sterols for growth as evidenced by serum requirement and digitonin sensitivity exclude assignment to the orders Anaeroplasmatales or Acholeplasmatales. Nonspiral cellular morphology, lack of motility, and regular association with a vertebrate host support exclusion from the families Spiroplasmataceae and Entomoplasmataceae, and indicate placement in the order Mycoplasmatales and family Mycoplasmataceae. The inability to hydrolyze urea, and the presence of conserved 16S rRNA gene sequences indicate that the organism belongs in the genus Mycoplasma. The 16S rRNA gene sequence, which exhibits only 97% similarity to that of any previously recognized species (Stackebrandt & Goebel, 1994), and twisted rod morphology associate the organism with the M. fastidiosum phylogenetic cluster. Growth inhibition was a discriminating serological test in support of the 16S rRNA gene analyses to demonstrate its stature as a unique species in that cluster (Roselló-Mora & Amann, 2001). In light of these observations, the designation M. insons is proposed for this organism, in consideration of its initial recovery from a healthy green iguana.
Description of M. insons species nova
Mycoplasma insons sp. nov. (in’sons) L. neut. adj. insons, guiltless, innocent. Cells are pleomorphic, but many have a highly atypical shape for a mycoplasma, often resembling a twisted rod. Cells devoid of cell wall, surrounded only by cytoplasmic membrane often with a highly textured extra-cellular matrix of low electron density closely applied. Nonmotile. Filterable through 220 nm membranes. Colonies on solid medium with 0.8% agar exhibit umbonate morphology and are variable in size. Chemoorganotroph. Acid produced from glucose. Does not hydrolyze esculin, arginine, or urea. Serum or sterol required for growth. Temperature range 25–37 °C, with the maximum growth rate at 30 °C. Unique 16S rRNA gene sequence (GenBank accession no. DQ522159) distinct from the most closely related species in the M. fastidiosum phylogenetic cluster. First isolated from the trachea and choanae of a healthy green iguana (Iguana iguana) from El Salvador. The type strain is I17P1T (= ATCC BAA-1435). Cultures prepared by the authors can also be obtained from the culture collection of the University of Göteborg, Sweden, under accession number CCUG 53461.
Acknowledgments
The authors thank Jennifer Hatchel and the Miami University Electron Microscopy Facility for help in obtaining scanning electron micrographs. Researchers interested in access to the reference collection of antisera that originated at the US National Institute of Allergy and Infectious Diseases Laboratory should contact the Mollicutes Collection at Purdue University, West Lafayette, IN. Mycoplasma insons strain I17P1T and homologous antiserum have been deposited in that collection. This work was supported by Morris Animal Foundation grant D05ZO-066 (L.D.W., D.S.R., D.R.B.), Public Health Service grant 1R01GM076584-01A1 from the National Institute of General Medical Sciences (D.R.B.), and funding from Miami University (M.F.B.).
References
- Aluotto BB, Wittler RG, Williams CO, Faber JE. Standardized bacteriologic techniques for the characterization of Mycoplasma species. Int J Syst Bacteriol. 1970;20:35–38. [Google Scholar]
- Brown DR. Mycoplasmosis and immunity of fish and reptiles. Front Biosci. 2002;7:1338–1346. doi: 10.2741/A844. [DOI] [PubMed] [Google Scholar]
- Brown DR, Crenshaw BC, McLaughlin GS, Schumacher IM, McKenna CE, Klein PA, Jacobson ER, Brown MB. Taxonomic analysis of the tortoise mycoplasmas Mycoplasma agassizii and Mycoplasma testudinis by 16S rRNA gene sequence comparison. Int J Syst Bacteriol. 1995;45:348–350. doi: 10.1099/00207713-45-2-348. [DOI] [PubMed] [Google Scholar]
- Brown DR, Demcovitz DL, Plourdé DR, Potter SM, Hunt ME, Jones RD, Rotstein DS. Mycoplasma iguanae sp. nov., from a green iguana (Iguana iguana) with vertebral disease. Int J Syst Evol Microbiol. 2006;56:761–764. doi: 10.1099/ijs.0.63852-0. [DOI] [PubMed] [Google Scholar]
- Brown DR, Wendland LD, Rotstein DS. Mycoplasmosis in green iguanas (Iguana iguana) J Zoo Wildlife Med. 2007;38:348–351. doi: 10.1638/1042-7260(2007)038[0348:MIGIII]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Clyde WA. Growth inhibition tests. In: Razin S, Tully JG, editors. Methods in Mycoplasmology. Vol. 4. Academic Press; New York: 1983. pp. 405–410. [Google Scholar]
- Cole JR, Chai B, Marsh TL, et al. The ribosomal database project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 2003;31:442–443. doi: 10.1093/nar/gkg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP – Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Geary SJ, Forsyth MH, Aboul Saoud S, Wang G, Berg DE, Berg CM. Mycoplasma gallisepticum strain differentiation by arbitrary primer PCR (RAPD) fingerprinting. Mol Cell Probes. 1994;8:311–316. doi: 10.1006/mcpr.1994.1042. [DOI] [PubMed] [Google Scholar]
- Hatchel JM, Balish RS, Duley ML, Balish MF. Ultrastructure and gliding motility of Mycoplasma amphoriforme, a possible human respiratory pathogen. Microbiology. 2006;152:2181–2189. doi: 10.1099/mic.0.28905-0. [DOI] [PubMed] [Google Scholar]
- International Committee on Systematic Bacteriology Subcommittee on the Taxonomy of Mollicutes. Revised minimum standards for the description of new species of the class Mollicutes (division Tenericutes) Int J Syst Bacteriol. 1995;45:605–612. [Google Scholar]
- Johansson K-E, Pettersson B. In: Taxonomy of Mollicutes. Molecular Biology and Pathogenicity of Mycoplasmas. Razin S, Herrmann R, editors. Kluwer Academic/Plenum; New York: 2002. pp. 1–30. [Google Scholar]
- Johansson K-E, Tully JG, Bölske G, Pettersson B. Mycoplasma cavipharyngis and Mycoplasma fastidiosum, the closest relatives to Eperythrozoon spp. and Haemobartonella spp. FEMS Microbiol Lett. 1999;174:321–326. doi: 10.1111/j.1574-6968.1999.tb13585.x. [DOI] [PubMed] [Google Scholar]
- Lemcke RM, Poland J. Mycoplasma fastidiosum: a new species from horses. Int J Syst Bact. 1980;30:151–162. [Google Scholar]
- Mettifogo E, Buzinhani M, Buim MR, Piantino Ferreira AJ, Kleven SH, Timenetsky J. Molecular characterization of MG isolates using RAPD and PFGE isolated from chickens in Brazil. J Vet Med B Infect Dis Vet Public Health. 2006;53:445–450. doi: 10.1111/j.1439-0450.2006.00978.x. [DOI] [PubMed] [Google Scholar]
- Razin S, Cirillo VP. Sugar fermentation. In: Razin S, Tully JG, editors. Methods in Mycoplasmology. Vol. 1. Academic Press; New York: 1983. pp. 337–344. [Google Scholar]
- Roselló-Mora R, Amann R. The species concept for prokaryotes. FEMS Microbiol Rev. 2001;25:39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in Bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- Stackebrandt E, Frederiksen W, Garritty GM, et al. Taxonomic note: report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol. 2002;52:1043–1047. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- Tully JG. Cloning and filtration techniques for mycoplasmas. In: Razin S, Tully JG, editors. Methods in Mycoplasmology. Vol. 1. Academic Press; New York: 1983. pp. 173–177. [Google Scholar]
- Tully JG. Determination of cholesterol and polyoxyethylene sorbitan growth requirements of mollicutes. In: Razin S, Tully JG, editors. Molecular and Diagnostic Procedures in Mycoplasmology. Vol. 1. Academic Press; San Diego, CA: 1995. pp. 381–389. [Google Scholar]