Abstract
PapD is an immunoglobulin-like chaperone that mediates the assembly of P pili in uropathogenic strains of Escherichia coli. It binds and caps interactive surfaces on pilus subunits to prevent their premature associations in the periplasm. We elucidated the structural basis of a mechanism whereby PapD also interacts with itself, capping its own subunit binding surface. Crystal structures of dimeric forms of PapD revealed that this self-capping mechanism involves a rearrangement and ordering of the C2–D2 and F1–G1 loops upon dimerization which might ensure that a stable dimer is not formed in solution in spite of a relatively large dimer interface. An analysis of site directed mutations revealed that chaperone dimerization requires the same surface that is otherwise used to bind subunits.
The assembly of P pili in uropathogenic strains of Escherichia coli requires the periplasmic chaperone PapD (1–4). Genes important in pilus biogenesis, papA–papG, are linked in the pap operon and their expression is coordinately controlled (3). P pili are composite fibers consisting of a thin open helical tip fibrillum joined to the distal end of a thicker cylindrical rod. The distal end of the tip fibrillum contains an adhesin, PapG, which mediates binding to specific receptors in the kidney. This binding event has been shown to be critical in the ability of E. coli to cause pyelonephritis (5–7). PapD is a member of a superfamily of periplasmic immunoglobulin-like chaperones that facilitates the assembly of over 26 architecturally diverse adhesive surface organelles in Gram-negative bacteria (8). The crystal structure of monomeric PapD was previously solved and refined to 2.0-Å resolution (9) (A. Holmgren and D. J. Ogg, personal communication). The structure of PapD consists of two globular domains with a deep cleft between them. Each domain has a β-barrel structure formed by two antiparallel β-pleated sheets with an overall topology similar to an immunoglobulin fold (9, 10). The cleft of the entire chaperone superfamily contains two positively charged residues, Arg-8 and Lys-112, that form a critical part of the subunit binding site (5, 11–13). Mutations in these residues abolish the ability of the chaperone to bind subunits and facilitate their assembly into pili (12, 13). The G1 β strand of PapD makes a β zippering interaction with a conserved C-terminal β strand present in all of the subunits (12, 14), which otherwise participates in head-to-tail subunit–subunit interactions in the pilus fiber (14, 15). Thus, when bound to a chaperone, the interactive C-terminal tail remains directly capped. This capping event is thought to be coupled with the folding of the subunits directly on the chaperone template (14, 15) in a process that also facilitates the release of subunits from a membrane-tethered state into the periplasm as soluble chaperone–subunit complexes (2, 16–19). The chaperone–subunit complexes are targeted to outer membrane ushers, where the chaperone is dissociated and the subunit–chaperone interactions are exchanged for subunit–subunit interactions that drive the assembly of pili (20, 21).
We discovered that when PapD is not engaged in binding to subunits, it is capable of interacting transiently with itself to form a weakly but specifically bound dimer species. In this report, the crystal structures of two dimeric forms of PapD were solved to gain insight into the molecular basis of PapD dimer formation. The structure–function analysis revealed that PapD interacts with itself by means of the same interactive surfaces that it uses to bind subunits, possibly representing a self-capping mechanism that may mimic how PapD interacts with subunits.
MATERIALS AND METHODS
Strains and Plasmids.
DH5α (22) was the E. coli strain used in all experiments. pDH1 was constructed as described (23).
Protein Purification and Crystallization.
Wild-type PapD, R8A PapD, Q108C PapD, and F168R PapD were purified as described for wild-type (WT) PapD (4). For crystallization, the Q108C PapD dimer was separated from the monomer by loading the fractions containing Q108C PapD onto a Hi-propyl (Beckman) column and eluting with a gradient of 1.0–0.0 M ammonium sulfate. All other Q108C PapD dimer species were separated from the monomer by gel filtration chromatography on a Superdex 75 (Pharmacia) column. Wild-type PapD, R8A PapD, and Q108C PapD were crystallized by using the hanging drop vapor diffusion method by mixing 2 μl of PapD (10 mg/ml) in 20 mM KMes buffer (pH 6.5) with 2 μl of well solution containing 16–22% PEG 4000, 14% isopropyl alcohol, and 0.1 M sodium acetate buffer (pH 5.2), and equilibrating at room temperature.
Structure Determination and Refinement.
R8A PapD data were collected on a MAR Research imaging plate at the Deutsches Elektronen Synchrotron (DESY)/X31 beamline (c/o European Molecular Biology Laboratory, Hamburg, Germany). Data were integrated and scaled by using denzo and scalepack (24). The R8A PapD crystals belong to space group P21212 with unit cell dimensions a = 176.7 Å, b = 55.3 Å, c = 46.6 Å, with two R8A PapD molecules in the asymmetric unit. The structure was solved by molecular replacement using as the search model the polyalanine backbone of WT PapD refined to 2.0-Å resolution (A. Holmgren and D. J. Ogg, personal communication). Molecular replacement calculations were done with AMoRe (25) and tffc (26). Refinement was carried out by using x-plor (27) and refmac (28). Bulk solvent correction was applied with x-plor. Between rounds of refinement, remodeling was done by using o (29). The current model consists of residues 1–96 and 104–214 for subunit A and residues 1–122 and 125–214 for subunit B. Wild-type PapD was crystallized under conditions identical to those used for R8A PapD, giving crystals that were isomorphous to the R8A PapD crystals. Data to 3.8-Å resolution were collected by using an Raxis I imaging plate system equipped with a rotating anode Cu Kα source. The structure was solved by molecular replacement using the same search model as that used for the R8A PapD structure. The asymmetric unit contains a dimer that is essentially the same as the R8A PapD dimer. Crystals of Q108C PapD grown under conditions similar to those used for R8A PapD belong to space group P212121 with cell dimensions a = 53.1 Å, b = 75.4 Å, c = 115.2 Å. Data were collected by using a Raxis I imaging plate system with a Cu Kα rotating anode source and processed with denzo and scalepack. The structure was solved by molecular replacement using the B subunit from the R8A PapD dimer structure, with the Gln-108 side chain omitted, as the search model. The two chaperone molecules found in the asymmetric unit form a dimer that is distinct from the R8A PapD dimer although essentially the same surfaces are involved in dimerization. Bulk solvent correction and refinement were carried out with x-plor by using standard protocols. The current model includes residues 1–95 and 106–215 in subunit A, residues 8–95 and 103–215 in subunit B, and 70 water molecules.
Mutagenesis.
Site-directed mutagenesis was performed as described (23, 30) using the following primers to introduce mutations in the papD gene: Q108C, coding strand 5′-ATAGCCTTATGCACCAAAATA-3′, noncoding strand 5′-TATTTTGGTGCATAAGGCTAT-3′; N89C, coding strand 5′-TTTTATTTTTGCCTCAGGGAA-3′, noncoding strand 5′-TTCCCTGAGGCAAAAATAAAA-3′; K110C, coding strand 5′-TTACAGACCTGCATAAAGCTT-3′, noncoding strand 5′-AAGCTTTATGCAGGTCTGTAA-3′; and F168R, coding strand 5′-GAAGGTGAGCGCGAAACCGTG-3′, noncoding strand 5′-CACGGTTTCGCGCTCACCTTC-3′. Mutations in papD were confirmed by sequencing. The resulting plasmids, pQ108C, pN89C, pK110C, and pF168R, encode PapD with point mutations changing Gln-108 to Cys, Asn-89 to Cys, Lys-110 to Cys, and Phe-168 to Arg, respectively. The mutants I105A PapD, I105E PapD, L107A PapD, and L107E PapD were constructed as described (23).
Pulse–Chase Analysis and Immunoprecipitation.
Pulse–chase experiments were done as described (21). The level of induction of the papD− pap operon, pDH1, was 0.01 mM isopropyl β-d-thiogalactoside (IPTG) and the level of induction of PapD (WT or mutant) was 0.0025% arabinose (Sigma) unless noted otherwise. PapH, PapC, PapJ, PapK, PapE, PapF, and PapG are all encoded on pDH1. Periplasms were isolated from cells at each time point as described for WT PapD (13) and radioimmunoprecipitated as described (23). The samples were analyzed either by SDS/PAGE in the presence or absence of 2-mercaptoethanol in the sample buffer or by acidic native PAGE (18).
Native PAGE.
Periplasmic extracts (13) and purified PapD (WT, Q108C, N89C, and K110C) were analyzed by acidic native PAGE (18) on Gradient 10–15 (Pharmacia) gels and silver stained on a Pharmacia Phast.
ELISA.
Maltose-binding protein (MBP) fused to the C-terminal residues 175–314 of PapG (MBP/G175–314) was purified as described (31). Serial dilutions of purified WT PapD, Q108C PapD, F168R PapD, and reduced/alkylated Q108C PapD (IAA-Q108C PapD) were incubated with immobilized MBP/G175–314 as described (31). Binding was detected with polyclonal mouse anti-PapDK antiserum (provided by Medimmune, Gaithersburg, MD) and quantitated as described (31) by reading the absorbance at 405 nm.
Alkylation of Q108C PapD.
Q108C PapD was reduced and alkylated as described for WT PapD (19).
Glutaraldehyde Crosslinking.
Glutaraldehyde crosslinking was performed by adding 0.1 vol of 0.125% glutaraldehyde (Sigma, grade I) to radiolabeled periplasm and incubated at room temperature for 30 min. The reaction was stopped by adding 4.5 vol of stop solution (20 mM sodium phosphate, pH 7.0/1.0% 2-mercaptoethanol/0.1% SDS) and heating at 100°C for 2 min. Crosslinked periplasms were pre-cleaned on 40 μl of protein A-Sepharose CL-4B (Pharmacia) beads and radioimmunoprecipitated (23) with polyclonal mouse anti-PapDK antiserum provided by Medimmune.
RESULTS
Crystal Structure of the PapD Dimer.
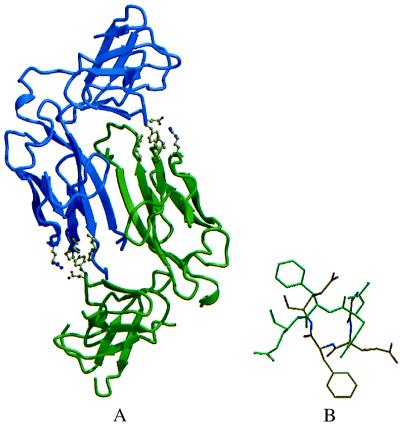
The structure of R8A PapD was solved and refined to 2.6-Å resolution (Table 1). There were no gross overall changes in the structure of the mutant as compared with the WT chaperone (9). Failure of the R8A PapD to bind subunits is thus a direct result of the missing arginine side chain, showing that the anchoring interaction between the Arg-8 side chain and the C terminus of subunits is critical for productive pilus subunit binding (12, 13). In the R8A PapD crystal, the two PapD monomers in the asymmetric unit form a dimer through interactions involving mainly residues in the G1 strand and the F1–G1 and C2–D2 loops (Fig. 1A). The G1 β strand is also required for the binding of the chaperone to a subunit (12, 14, 15, 17). The structure of an essentially identical dimer of WT PapD has also been solved to 3.8-Å resolution, showing that dimerization is not an effect of the mutated Arg-8 residue in the binding cleft (data not shown). In this report, the R8A PapD dimer model will be used to describe the structure of the dimer because it has been solved to the highest resolution. Chaperone molecules in the dimer will be referred to as chaperone subunits to distinguish them from pilus subunits.
Table 1.
Data collection and refinement statistics
| Measurement | Value
|
|
|---|---|---|
| R8A PapD | Q108C PapD | |
| Data collection | ||
| Resolution limits, Å | 15.0–2.6 | 15.0–2.4 |
| (outer shell) | (2.64–2.60) | (2.44–2.40) |
| Measured reflections | 53,166 | 107,688 |
| Unique reflections | 12,852 | 18,542 |
| Completeness (outer shell), % | 87.7 (91.1) | 99.3 (93.8) |
| Overall I/σ(I) (outer shell) | 12.6 (4.9) | 16.7 (4.1) |
| R*sym (outer shell), % | 7.2 (27.7) | 6.9 (27.0) |
| Refinement | ||
| Resolution limits, Å | 15.0–2.6 | 15.0–2.4 |
| Rfactor†/Rfree‡, % | 20.3/28.6 | 20.2/27.4 |
| rms deviation bond lengths, Å | 0.006 | 0.007 |
| rms deviation bond angles, deg | 1.328 | 1.397 |
Rsym = Σ|I − 〈I〉/Σ〈I〉, where I is the observed intensity, and 〈I〉 is the average intensity of multiple observations of symmetry-related reflections.
Rfactor = Σ|FO − FC|/ΣFO, where FO is the observed structure factor and FC is the structure factor calculated from the model.
Rfree (32) is the Rfactor for a subset of reflections that were not included in the crystallographic refinement. For the R8A PapD refinement this subset constituted 10% of the reflections; for the Q108C PapD refinement 5% of the data were used for Rfree.
Figure 1.
Structure of the R8A PapD dimer. (A) molscript (33) ribbon drawing of the R8A PapD dimer. The view is looking down the dimer twofold axis. One chaperone subunit is shown in blue and the second subunit in green. In the dimer, contacts between the two N-terminal domains are mediated mostly by the two G1 edge strands across the dimer twofold axis. The interactions between two residues in the C2–D2 loop, Glu-167 and Phe-168, of one subunit with residues Pro-30, Leu-32, Ile-93, Pro-95, and Arg-58 at the lip of the second subunit are shown as ball-and-stick models. (B) Comparison of the C2–D2 loop conformation in monomeric PapD (green) and in the R8A PapD dimer (yellow). The figure was generated after superpositioning of C-terminal domains in monomeric WT PapD and dimeric R8A PapD with an rms deviation of 0.565 Å for 88 Cα atoms.
The contact surfaces between the chaperone subunits in the dimer have the following features. The G1 strand from one chaperone subunit forms a short stretch of antiparallel β-sheet with the G1 strand from the second chaperone subunit in the dimer. The Gln-108 residue in the G1 strand of one chaperone subunit interacts with the Gln-108 residue of the opposite chaperone subunit via hydrogen bonds between the main chain O and N of each residue and extensive side-chain-to-side-chain packing. Because of the twist of the two G1 strands, only limited main-chain hydrogen bonding occurs between the two antiparallel β-strands. Additional hydrogen bonding is provided by the hydroxyl group of Thr-109 in each chaperone subunit, which forms a hydrogen bond to the main-chain carbonyl oxygen atom of Ala-106 in the opposite chaperone subunit.
The F1–G1 loop packs in the binding cleft of the neighboring chaperone subunit, where it interacts mostly with residues from the C2–D2 loop, causing considerable conformational changes in these loops compared with the monomeric PapD structure. In the monomeric structure of PapD, the F1–G1 and C2–D2 loops are flexible and disordered as indicated by high temperature factors for residues in these loops (9). In the chaperone dimer structure, as well as in chaperone–peptide complexes (12, 14), the F1–G1 loop becomes more ordered. In the PapD monomeric structure, Phe-168 from the C2–D2 loop is at the surface of the hydrophobic core of the C-terminal domain with the plane of the phenyl ring more or less parallel to the subunit surface. In the dimer structure, Phe-168 has flipped around and rotated almost 180° (Fig. 1B) so that the side chain now points away from the surface of the chaperone subunit and packs into a shallow hydrophobic groove formed by residues Pro-30, Leu-32, Ile-93, and Pro-95 in the neighboring subunit (Fig. 1A). Residue Glu-167 is also in a very different conformation in the dimer and forms a salt link to Arg-58 in the neighboring chaperone subunit. The total surface area of each chaperone subunit that is buried in the dimer interface is approximately 1100 Å2, which constitutes about 10% of the total surface area of a PapD monomer. This interface is of similar magnitude to contact areas in known dimeric proteins (34, 35) and covers most of the conserved surface in the cleft of PapD-like chaperones. In PapD, this conserved region has been shown to mediate binding of PapD to pilus subunits (5, 11, 12, 14, 17, 23).
Specificity of PapD Dimerization.
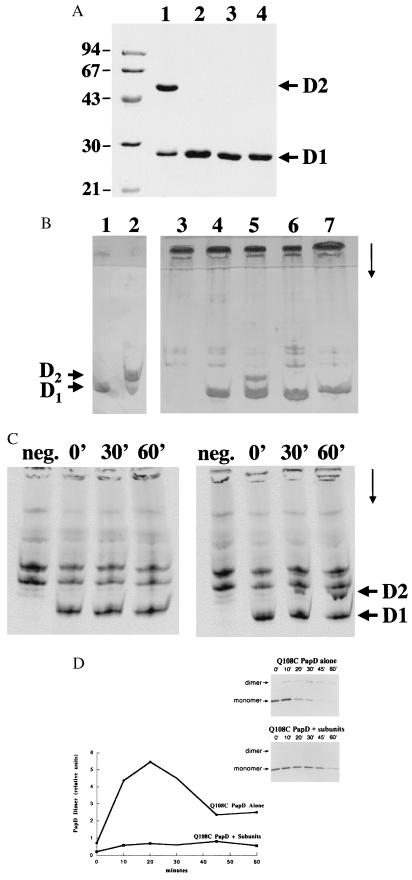
Because the Gln-108 residues from each chaperone subunit in the dimer interact with one another, this residue was mutagenized to a cysteine and the mutated Q108C PapD was tested for its ability to form a disulfide-bonded dimer. Neighboring residues, Asn-89 and Lys-110, which are not involved in any interaction in the R8A PapD dimer, were also independently changed to cysteines to investigate the specificity of disulfide-bonded dimer formation (N89C PapD and K110C PapD, respectively). On the basis of pili preparations and hemagglutination titers, WT PapD, N89C PapD, and K110C PapD all were able to complement a papD− pap operon to a fully piliated phenotype (data not shown). In contrast, Q108C PapD assembled a greatly reduced amount of pili. Q108C PapD was purified to homogeneity, and the dimer species was separated from the monomer by gel filtration chromatography. Analysis of the material by SDS/PAGE revealed that most of the material ran as a dimer under nonreducing conditions, whereas the WT PapD ran as a monomer under these conditions (Fig. 2A, lanes 1 and 3, respectively). Under reducing conditions, the Q108C PapD dimer was dissociated into PapD monomers (Fig. 2A, lane 2). Dimer formation was also analyzed on acidic native PAGE (Fig. 2B). Unlike most periplasmic proteins, PapD has an unusually high isoelectric point (9.4) which facilitates its migration into acidic native polyacrylamide gels (18) (Fig. 2B, lane 4). Purified WT PapD (monomeric PapD) migrates faster on the acidic native PAGE (Fig. 2B, lane 1, labeled D1) than the purified Q108C PapD dimer (Fig. 2B, lane 2, labeled D2). Thus, acidic native PAGE (18) is a good assay for analyzing the presence of PapD monomers and dimers in periplasmic extracts. Analysis of periplasmic extracts from cells expressing Q108C PapD revealed that more than 30% of the chaperone existed in the dimeric state (Fig. 2B, lane 5). In contrast, neither N89C PapD nor K110C PapD formed disulfide-bonded dimers as determined by acidic native PAGE (Fig. 2B, lanes 6 and 7, respectively), arguing that the chaperone–chaperone interaction in the dimer is specific.
Figure 2.
Specificity of chaperone–chaperone interaction. (A) Coomassie blue-stained SDS/12.5% PAGE showing purified Q108C PapD and WT PapD proteins run in nonreducing conditions (lanes 1 and 3, respectively) and in reducing conditions (lanes 2 and 4, respectively). (B) Silver-stained acidic native PAGE gels of purified WT PapD protein (lane 1), purified Q108C PapD protein (lane 2), and periplasmic extracts expressing the following: no chaperone (lane 3), WT PapD (lane 4), Q108C PapD (lane 5), N89C PapD (lane 6), and K110C PapD (lane 7). D1 indicates where WT PapD (monomer) migrates and D2 indicates where dimeric PapD migrates. Arrow indicates the direction of migration. (C) Autoradiogram showing radiolabeled periplasmic extracts of cells expressing WT PapD (Left) and Q108C PapD (Right) after various times of chase analyzed by acidic native PAGE (18) on Gradient 10–15 (Pharmacia) gels and silver stained on a Pharmacia Phast. The first lane of each gel (labeled neg.) is radiolabeled periplasmic extract from cells expressing no chaperone at 30 min into the chase. D1, D2, and the arrow are as in B. (D) (Inset) Autoradiogram showing radiolabeled Q108C PapD immunoprecipitated after various times of chase from periplasmic extracts of cells expressing only Q108C PapD (pQ108C) (upper gel) and in cells expressing Q108C PapD (pQ108C) in trans with a papD− pap operon, pDH1 (lower gel). The samples were analyzed on nonreducing SDS/12.5% PAGE gels. For quantification, triplicate gels of each experiment were exposed to the Bio-Rad Molecular Imaging Screen-CS of the Bio-Rad Molecular Imager System, model GS-363, and the amount of PapD dimer was quantified by using Molecular Analyst/Macintosh Software, Version 2.0. The averages are graphed. The y axis equals the density of the PapD dimer band.
In a pulse–chase analysis, over 30% of the labeled Q108C PapD monomers could be chased into disulfide-bonded dimers over time. No dimer species was seen with the labeled WT PapD (Fig. 2C). We hypothesized that chaperone–pilus subunit complex formation should compete for chaperone–chaperone dimer formation because the same G1 β strand at the exposed edge of the conserved sheet in PapD is involved in both interactions. To test this hypothesis, chaperone dimer formation was analyzed after expressing Q108C PapD alone or in trans with subunits (expressed from a papD− pap operon). The dimer species was undetectable on an acidic native polyacrylamide gel when subunits were coexpressed with Q108C PapD (data not shown). In a pulse–chase analysis, more than 30% of the labeled Q108C PapD participated in dimer formation when it was expressed alone. In contrast, coexpression of subunits titrated PapD into chaperone–subunit complexes, thus reducing the level of PapD dimer to below 3% (Fig. 2D). The amount of PapD dimer formed was inversely proportional to the level of induction of the subunits (data not shown). These results argue that chaperone–subunit complex formation is favored over chaperone–chaperone dimer formation.
Crystal Structure of Disulfide-Linked Dimer.
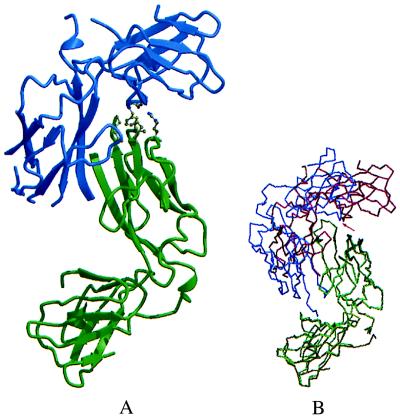
To characterize the structure of the disulfide-linked Q108C PapD dimer, the protein was purified, analyzed, and crystallized (Table 1). Subunits in the Q108C PapD dimer interacted at essentially the same surfaces as in the R8A PapD dimer. However, the packing between the two G1 strands in the Q108C PapD dimer had been shifted by two residues, presumably to meet the requirements for optimal stereochemistry of the disulfide bond (Fig. 3A). Comparison of the R8A and Q108C PapD dimers revealed that one subunit in the Q108C PapD dimer was rotated by about 30° around the lip of the N-terminal domain of the second subunit (Fig. 3B), thus breaking the twofold symmetry of the dimer. Remarkably, in spite of this large rotation, residues in the C2–D2 loop of the second chaperone subunit of the Q108C PapD dimer remained perfectly aligned with the corresponding R8A PapD residues. In both dimers, the Phe-168 side chain flipped 180° relative to its position in monomeric PapD to interact identically with the neighboring subunit.
Figure 3.
Structure of the Q108C PapD dimer and comparison of R8A PapD and Q108C PapD dimers. (A) molscript ribbon drawing of the Q108C PapD dimer with one chaperone subunit shown in blue and the second subunit in green. The orientation of the green subunit is the same as for the green R8A PapD subunit in Fig. 1A. (B) Comparison of R8A PapD (green and blue) and Q108C PapD (light green and red) PapD dimers. N-terminal domains in one subunit from each dimer were superimposed by using the lsq options in o (29). The pivotal point for the rotation is at the tip of the C2–D2 loop (indicated by a red arrow), which remains in essentially the same position in both dimers relative to the superimposed N-terminal domains.
Probing the PapD Dimer Interface.
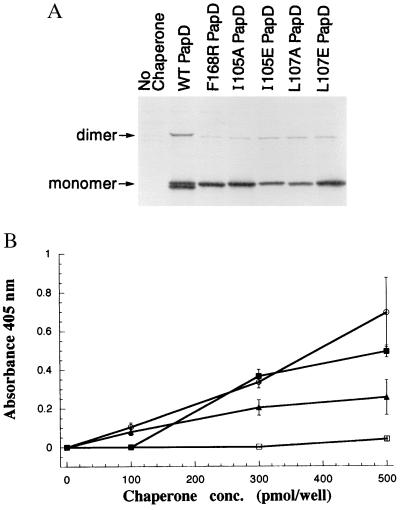
Dimer interface residues were identified in the crystal structures and mutagenized to analyze the specificity of dimer formation. The C2–D2 loop residue Phe-168 was changed to Arg (F168R) and the alternating hydrophobic residues Ile-105 and Leu-107 along the G1 β strand dimer interface were changed independently to an alanine (I105A and L107A) or glutamic acid (I105E and L107E) (23). The ability of the mutant and WT PapDs to be crosslinked into a dimer by glutaraldehyde was investigated. Twenty percent of WT PapD could be crosslinked into a dimer species. No higher oligomers were observed (Fig. 4A). The I105A, I105E, L107A, and L107E mutations in the G1 β strand of PapD all abolished the ability of PapD to be crosslinked into a dimer (Fig. 4A). These same mutations have also been shown to interfere with PapD’s ability to bind subunits in vivo and have been shown to be part of the subunit binding cleft (23). The F168R mutation in the C2–D2 loop also abolished the ability of PapD to form a dimer (Fig. 4A). These data argue that residues found along the dimer interface in the crystal structures are necessary for PapD dimer formation in solution.
Figure 4.
Effect of mutants on chaperone–subunit interactions and chaperone dimerization. (A) Effect of mutations on PapD dimerization. WT PapD, F168R PapD, and G1 β strand mutants I105A PapD, I105E PapD, L107A PapD, and L107E PapD were induced for expression 5 min prior to pulse-labeling and then chased for 20 min. Periplasm was isolated from the cells and then subjected to glutaraldehyde crosslinking, and PapD was immunoprecipitated with anti-PapDK antiserum. The immunoprecipitates were subjected to electrophoresis on reducing SDS/12.5% polyacrylamide gels. Triplicate gels of each experiment were quantified as in Fig. 2D. (B) Curve showing the binding of purified WT PapD (○), F168R PapD (▴), native (□), or alkylated Q108C PapD (IAA-Q108C) (■) to immobilized MBP/G175–314 protein quantified by ELISA.
Chaperone Dimerization Caps Subunit Binding Surface.
The structure–function analyses described above argue that chaperone dimerization may represent a self-capping event. This hypothesis was investigated by testing the ability of the Q108C PapD dimer to bind to PapG in both in vivo and in vitro chaperone binding assays. PapD has been shown previously to be able to bind to a chimeric protein containing MBP fused to the C-terminal residues 175–314 of PapG (MBP/G175–314) in an ELISA (31). Thus, we tested the ability of purified WT PapD, F168R PapD, and the disulfide-locked Q108C PapD dimer to bind to MBP/G175–314 in the ELISA (31). WT PapD bound MBP/G175–314 the best (Fig. 4B). F168R PapD also bound but not as well as WT PapD. Virtually no binding was detected for the Q108C PapD dimer. These results suggested that PapD’s subunit binding surface is capped when it is locked as a disulfide-linked dimer. To test this hypothesis, we analyzed whether dissociation of the dimer restored its ability to bind to MBP/G175–314 in the ELISA. The Q108C PapD dimer was dissociated with the reducing agent DTT, and the free sulfhydryl groups were alkylated with iodoacetic acid (19) to prevent re-dimerization (IAA-Q108C). Previous work demonstrated that WT PapD binds MBP/G175–314 equally well when alkylated (19). The IAA-Q108C PapD was able to bind MBP/G175–314 at nearly WT levels (Fig. 4B), arguing that only monomeric PapD can bind subunits. In vivo, only monomeric Q108C PapD was found complexed to tip fibrillum subunits in immunoprecipitations using anti-tip fibrillum antiserum (anti-PapGFEK), arguing that dimeric Q108C PapD was unable to participate in chaperone–subunit complex formation (data not shown). The decreased ability of the Q108C PapD dimer to bind to subunits and its propensity to become locked into a dimer in the periplasm explain why the Q108C mutation reduces the hemagglutination (HA) titer and pilus assembly. In addition, the Gln-108 residue is positioned on the G1 β strand, which is known to be critical for subunit binding (12, 14, 15, 17). The F168R PapD bound subunits efficiently (Fig. 4B and data not shown) and produced a higher HA titer than did WT PapD. The higher HA titer correlated with the presence of an increased amount of pili as determined by SDS/PAGE of purified pili (data not shown).
DISCUSSION
PapD is required in P pilus biogenesis to bind to pilus subunits in the periplasm. Specifically, PapD makes a β zippering interaction with a conserved C-terminal β strand of pilus subunits as they emerge from the cytoplasmic membrane in semi-unfolded conformations (12, 14, 15, 17). This interaction has been shown to facilitate the release of subunits from the cytoplasmic membrane into the periplasm as chaperone–subunit complexes and may provide the proper environment or template for the subunit to acquire its assembly-competent (native) fold (12, 14, 17). PapD then takes part in one-to-one interactions with the pilus subunits, protecting them from proteolytic degradation in the periplasm (1–4, 16–18). Assembly-competent pilus subunits bound to PapD are predominately β-sheet proteins, reminiscent of PapD’s structure (9), as determined by crystallization of the PapD-PapK complex (F. G. Sauer, K. Fütterer, S. Pinkner, K. Dodson, S.J.H., and G. Waksman, unpublished results). Here, we have shown that PapD can interact transiently with itself, using the same interactive surface used to bind the C terminus of subunits in what may represent a self-capping mechanism.
Formation of the PapD dimer in the periplasm was demonstrated by crosslinking. Immunoprecipitation of the crosslinked dimer revealed no higher oligomeric species, and no other proteins were crosslinked to the dimer. The molecular interaction between the two chaperones in the WT and R8A PapD dimers buries a substantial portion of the conserved subunit binding surface (8, 11). Mutagenizing the hydrophobic residues in the dimer interface along the G1 β strand and in the C2–D2 loop abolished dimer formation as detected in our crosslinking studies. One of the dimer interface residues in the G1 β strand, Gln-108, interacts with Gln-108 on the neighboring chaperone subunit. When it was mutagenized to Cys, PapD formed disulfide-bonded dimers. However, changing nearby residues that were not part of the dimer interface to cysteines did not result in disulfide-bonded dimers. These results strongly suggest that the interactions in the dimer crystal structures are specific.
The interaction between the C2–D2 loop and the neighboring subunit involves a 180° flip of the Phe-168 side chain relative to its position in monomeric PapD. In the monomer, the phenyl ring is at the edge of the hydrophobic core (9). Upon dimerization, it flips 180° to become surface exposed, where it packs into a shallow hydrophobic groove on the other chaperone subunit. In the Q108C PapD dimer, the dimeric interface along the G1 β strand was shifted, resulting in a rotation of one chaperone subunit about 30° in relation to the other. In spite of this large rotation, the placement and packing of the Phe-168 side chain was virtually unchanged. This finding suggests a very strong tendency to form these interactions, since they occur despite very different quaternary structures in the two dimers and different crystal packing in the two dimer crystal forms. Mutation of the Phe-168 residue to an arginine (F168R PapD) abolished our ability to detect PapD dimers by means of crosslinking, arguing that the flipping of this residue and the subsequent interactions in which it takes part are critical for dimer formation. The F168R PapD was stable as a monomer. Under conditions where an identical pool of pilus subunits was expressed, the F168R PapD assembled 2- to 3-fold more pili than did WT PapD. The inability of the F168R PapD to form a dimer presumably results in an increased ratio of monomer chaperones available to bind and assemble subunits, accounting for the increase in piliation.
In the structure of monomeric WT PapD (9) both the F1–G1 and the C2–D2 loops are flexible and have among the highest temperature factors in the model. The F1–G1 loop, which forms a large part of the dimer interface, is known to be able to adopt a number of different conformations as seen in the various crystal structures available (9, 12, 14). Furthermore, the C2–D2 loop, with which the F1–G1 loop interacts, undergoes a dramatic conformational change upon dimerization. The rearrangement and ordering of the C2–D2 and F1–G1 loops upon dimerization might ensure that a stable dimer is not formed in solution in spite of a relatively large dimer interface. In the case of PapD, formation of a stable dimer throughout assembly is not desirable because this would block binding of pilus subunits.
The dimeric species of PapD may be either more stable and/or less aggregative than monomer PapD, since the hydrophobic G1 β strand is buried in the PapD dimer. Thus, chaperone dimerization may represent a self-capping mechanism. The recently solved three-dimensional structures of the PapD–PapK (F. G. Sauer, K. Fütterer, S. Pinkner, K. Dodson, S.J.H., and G. Waksman, unpublished results) and the FimC–FimH complexes (D. Choudhury, A. Thompson, V. Stojanoff, S. Langermann, J. Pinkner, S.J.H., and S.D.K., unpublished results) have revealed that pilin subunits have the same immunoglobulin-like topology as the subunit-binding N-terminal domain of the chaperone. Interestingly, the structural basis of chaperone–chaperone, chaperone–subunit, and subunit–subunit interactions all involve the same highly conserved immunoglobulin-like surfaces. Thus, molecular mimicry of a subunit’s structure may give the chaperone a subunit-like template that facilitates chaperone–subunit interactions in a mechanism that may mimic subunit–subunit interactions, thereby ensuring that the interactive assembly surfaces of the subunits are directly capped throughout pilus biogenesis.
Acknowledgments
We thank Dr. Matt Mulvey and Dr. David Thanassi for their work on Q108C PapD. This work was supported by National Institutes of Health Training Grant AI07172-16 (D.L.H.), a grant from the Swedish Research Council (S.D.K.), and National Institutes of Health Grants RO1AI29549 and ROIDK51406 (S.J.H.).
ABBREVIATIONS
- WT
wild-type
- MBP
maltose-binding protein
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1QPP for R8A PapD and 1QPX for Q108C PapD).
References
- 1.Normark S, Baga M, Goransson M, Lindberg F P, Lund B, Norgren M, Uhlin B E. In: Microbial Lectins and Agglutinins: Properties and Biological Activity. Mirelman D, editor. New York: Wiley Interscience; 1986. pp. 113–143. [Google Scholar]
- 2.Hultgren S J, Lindberg F, Magnusson G, Kihlberg J, Tennent J M, Normark S. Proc Natl Acad Sci USA. 1989;86:4357–4361. doi: 10.1073/pnas.86.12.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hultgren S J, Abraham S N, Caparon M G, Falk P, St. Geme J W, III, Normark S. Cell. 1993;73:887–901. doi: 10.1016/0092-8674(93)90269-v. [DOI] [PubMed] [Google Scholar]
- 4.Lindberg F, Tennent J M, Hultgren S J, Lund B, Normark S. J Bacteriol. 1989;171:6052–6058. doi: 10.1128/jb.171.11.6052-6058.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuehn M J, Heuser J, Normark S, Hultgren S J. Nature (London) 1992;356:252–255. doi: 10.1038/356252a0. [DOI] [PubMed] [Google Scholar]
- 6.Lindberg F, Lund B, Johansson L, Normark S. Nature (London) 1987;328:84–87. doi: 10.1038/328084a0. [DOI] [PubMed] [Google Scholar]
- 7.Roberts J A, Marklund B-I, Ilver D, Haslam D, Kaack M B, Baskin G, Louis M, Mollby R, Winberg J, Normark S. Proc Natl Acad Sci USA. 1994;91:11889–11893. doi: 10.1073/pnas.91.25.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung D L, Knight S D, Woods R M, Pinkner J S, Hultgren S J. EMBO J. 1996;15:3792–3805. [PMC free article] [PubMed] [Google Scholar]
- 9.Holmgren A, Brändén C-I. Nature (London) 1989;342:248–251. doi: 10.1038/342248a0. [DOI] [PubMed] [Google Scholar]
- 10.Williams A F, Barclay A N. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 11.Holmgren A, Kuehn M J, Brändén C-I, Hultgren S J. EMBO J. 1992;11:1617–1622. doi: 10.1002/j.1460-2075.1992.tb05207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuehn M J, Ogg D J, Kihlberg J, Slonim L N, Flemmer K, Bergfors T, Hultgren S J. Science. 1993;262:1234–1241. doi: 10.1126/science.7901913. [DOI] [PubMed] [Google Scholar]
- 13.Slonim L N, Pinkner J S, Brändén C-I, Hultgren S J. EMBO J. 1992;11:4747–4756. doi: 10.1002/j.1460-2075.1992.tb05580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soto G E, Dodson K W, Ogg D, Liu C, Heuser J, Knight S, Kihlberg J, Jones C H, Hultgren S J. EMBO J. 1998;17:6155–6167. doi: 10.1093/emboj/17.21.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones C H, Bullitt E, Striker R, Soto G, Jacob-Dubuisson F, Pinkner J, Wick M J, Makowski L, Hultgren S J. Proc Natl Acad Sci USA. 1996;93:12890–12895. doi: 10.1073/pnas.93.23.12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones C H, Danese P N, Pinkner J S, Silhavy T J, Hultgren S J. EMBO J. 1997;16:6394–6395. doi: 10.1093/emboj/16.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuehn M J, Normark S, Hultgren S J. Proc Natl Acad Sci USA. 1991;88:10586–10590. doi: 10.1073/pnas.88.23.10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Striker R, Jacob-Dubuisson F, Frieden C, Hultgren S J. J Biol Chem. 1994;269:12233–12239. [PubMed] [Google Scholar]
- 19.Jacob-Dubuisson F, Pinkner J, Xu Z, Striker R, Padmanhaban A, Hultgren S J. Proc Natl Acad Sci USA. 1994;91:11552–11556. doi: 10.1073/pnas.91.24.11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dodson K W, Jacob-Dubuisson F, Striker R T, Hultgren S J. Proc Natl Acad Sci USA. 1993;90:3670–3674. doi: 10.1073/pnas.90.8.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacob-Dubuisson F, Striker R, Hultgren S J. J Biol Chem. 1994;269:12447–12455. [PubMed] [Google Scholar]
- 22.Hanahan D. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 23.Hung D L, Knight S D, Hultgren S J. Mol Microbiol. 1999;31:773–783. doi: 10.1046/j.1365-2958.1999.01216.x. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z. In: Proceedings of the CCP4 Study Weekend: Data Collection and Processing. Sawyer L, Isaacs N, Bailey S, editors. Daresbury, U.K.: SERC Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- 25.Navazza J G. Acta Crystallogr A. 1994;50:157–169. [Google Scholar]
- 26.Collaborative Computational Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 27.Brünger A T. x-plor v3.1, A System for X-Ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 28.Murshudov G, Vagin A, Dodson E. Proceedings of the CCP4 Study Weekend: Refinement of Protein Structures. Daresbury, U.K.: SERC Daresbury Laboratory; 1996. [Google Scholar]
- 29.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 30.Morrison H G, Desrosiers R C. BioTechniques. 1993;14:454–457. [PubMed] [Google Scholar]
- 31.Xu Z, Jones C J, Haslam D, Pinkner J S, Dodson K, Kihlberg J, Hultgren S J. Mol Microbiol. 1995;16:1011–1020. doi: 10.1111/j.1365-2958.1995.tb02326.x. [DOI] [PubMed] [Google Scholar]
- 32.Brünger A T. Nature (London) 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 33.Kraulis P. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 34.Janin J, Miller S, Chothia C. J Mol Biol. 1988;204:155–164. doi: 10.1016/0022-2836(88)90606-7. [DOI] [PubMed] [Google Scholar]
- 35.Miller S, Lesk A M, Janin J, Chothia C. Nature (London) 1987;328:834–836. doi: 10.1038/328834a0. [DOI] [PubMed] [Google Scholar]