Abstract
The infected cell protein no. 0 (ICP0) of herpes simplex virus 1 is a promiscuous transactivator shown to enhance the expression of genes introduced into cells by infection or transfection. The protein interacts with several viral and cellular proteins. Earlier studies have shown that ICP0 binds and stabilizes cyclin D3 but does interfere with the phosphorylation of retinoblastoma protein, its major function. Cyclin D3 plays a key role in the transition from G1 to S phase. To define the role of cyclin D3 in productive infection, the ICP0 binding site for cyclin D3 was mapped and mutagenized by substitution of aspartic acid codon 199 with the alanine codon. We report that the substitution precluded the interaction of this protein with cyclin D3 in the yeast two-hybrid system and the stabilization of cyclin D3 in infected cells. A recombinant virus carrying this mutation could not be differentiated from wild-type parent with respect to replication in dividing cells but yielded 10-fold less progeny from infected resting cells and serum-deprived or contact-inhibited human fibroblasts. In mice, the mutant was only slightly less pathogenic than the wild-type parent by intracerebral route but was significantly less neuroinvasive after peripheral inoculation. Replacement of the mutated amino acid with aspartic acid restored wild-type phenotype. Stabilization of cyclin D3 therefore is linked to higher virus yields in nondividing cells and potentially higher virulence in experimental and natural hosts. One function of ICP0 is to scavenge the cell for proteins that could bolster viral replication.
The studies described in this report stemmed from analyses of the function of a herpes simplex virus 1 (HSV-1) protein known as the infected cell protein no. 0 (ICP0). This protein, the product of the α0 gene, is among a small number of α proteins expressed immediately after infection in the absence of de novo viral protein synthesis. The 775-aa protein is encoded by three exons (Fig. 1A, line 2) mapping in the inverted repeats flanking the unique long sequence, and therefore the gene is present in two copies per genome (Fig. 1). ICP0 plays an important role in viral pathogenesis and reactivation from latent state (reviewed in ref. 1). Genetically engineered viruses lacking ICPO (ICP0−) are avirulent in experimental animal systems. In cell culture, they multiply sluggishly and yield titers at least 100-fold lower than wild-type parent virus (2, 3). Although ICP0− mutants can be detected in sensory ganglia, the relative amounts of latent virus are diminished, and their ability to reactivate from latency is grossly reduced (4). The majority of ICP0 literature centers on the observation that it acts as a promiscuous transactivator: it enhances the expression of genes introduced into cells by transfection or infection (reviewed in ref. 5). Early studies have shown that it interacts functionally and physically with the major regulator protein ICP4 and that it localizes in infected cell nuclei in small dense bodies known as ND10 and causes their disappearance (6, 7). More recent studies have focused on the interaction of ICP0 with cellular proteins. Thus Everett and associates (8, 9) discovered that ICP0 interacts with a ubiquitin-specific protease. With the aid of a two-hybrid screen, we found that ICP0 reacted with several proteins, including the protein elongation factor EF-1δ (10), cyclin D3 (3), and a novel cellular protein designated p60 (11). The significance of the interaction of ICP0 with p60 is not yet known although p60 binds both ICP0 and ICP22, another viral regulatory protein. The interaction of ICP0 with EF-1δ garnered evidence of significance on the basis of two observations. First, several hours after the onset of infection, ICP0 translocates into the cytoplasm. Second, EF-1δ is hyperphosphorylated by the viral protein kinase encoded by the UL13 ORF (12). The significance of this interaction is underscored by the observation that EF-1δ is hyperphosphorylated in cells infected with representative members of all three subfamilies of herpesviruses (13).
Figure 1.
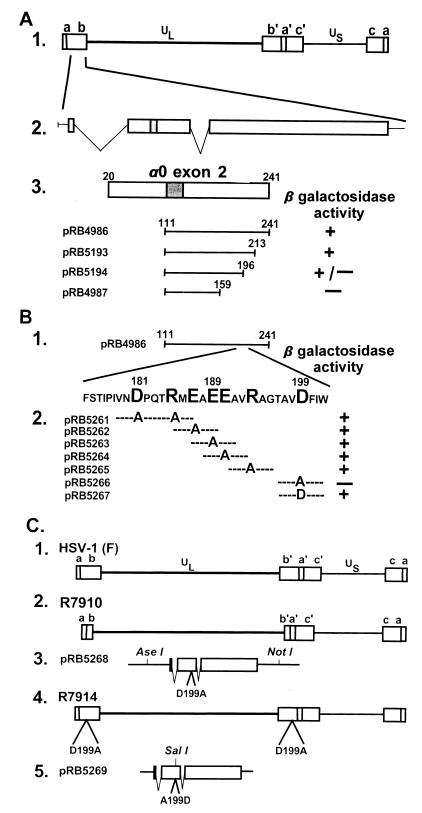
(A) Schematic diagram of the sequence arrangements of the HSV-1 genome and the location of the α0 gene. Line 1, a linear representation of the HSV-1 genome showing the unique long (UL) and short (US) sequences. The terminal repeats flanking UL and US, ab and ca, respectively, are represented as rectangles. Line 2, expansion of a portion of the repeat sequence ab showing the location of one of the copies of the α0 gene. Line 3, an expanded section of exon II (codons 20–241). The shaded area represents the ICP0 zinc ring finger. Plamsid pRB4986 (3) carries the HSV-1 strain F [HSV-1(F)] α0 coding sequence that specifically interacts with cyclin D3. Plasmids pRB5193, pRB5194, and pRB4987 carrying truncated portions of α0 binding domain from pRB4986 were analyzed in the yeast two-hybrid system as described for the ability of the expressed peptides to bind cyclin D3 (3). (B) Line 1, a representation of exon II domain cloned in pRB4986. Line 2, the amino acid sequence of the minimal binding domain mapped by truncation of the pRB4986 sequence in A. The large letters represent codons substituted with alanine codon. Also shown are the results of the yeast assays to determine whether the mutagenized fragments yields a product that interacts with cyclin D3. (C) Lines 1 and 2, schematic representation of the wild-type HSV-1(F) and R7910 (ICP0−) genomes, respectively. In R7910, both copies of the α0 gene had been deleted. The rescue of R7910 virus was effected by transfection of rabbit skin cells with the DNA cloned in pRB5268 (line 3), followed by superinfection with R7910 (line 2), which yielded the virus R7914 carrying the D199A substitution within the ICP0 reading frame. Rescue of the R7914 by cotransfection of its DNA with that of pRB5269 yielded the recombinant R7915 (line 5) in which the A199D substitution rendered the α0 gene wild type.
The interaction of ICP0 with cyclin D3 is of particular interest for several reasons. Cyclin D3 regulates the transition from G1 into S phase (reviewed in ref. 14). Although wild-type HSV-1 grows well in both resting and dividing cells, several lines of evidence suggest that the virus favors the transition of the infected cell into S phase, but not the G2/M transition (15). In reconstruction experiments in vitro, ICP0 bound cyclin D3 but did not interfere with the phosphorylation of the retinoblastonic protein (pRb) by the complex of cyclin D3 and cdk4. In the infected cells, ICP0 translocates cyclin D3 to small dense nuclear structures (ND10) and stabilizes the protein for at least 8 hr (3).
To test the significance of the interaction of cyclin D3, we mapped its binding site in ICP0 and mutagenized it. We report that the replacement of a single amino acid in the binding domain abolished the stabilization of cyclin D3, abrogated the capacity of the virus to invade the central nervous system (CNS) from a peripheral site, and reduced slightly its capacity to replicate in resting cells in vitro or cause lethal disease on inoculation directly into the CNS, but did not affect its replication in dividing cells.
MATERIALS AND METHODS
Cells and Viruses.
HEp-2, HeLa, and Vero cell lines were obtained from the American Type Culture Collection. Rabbit Skin cells were a gift of J. McClaren (University of New Mexico). Human lung fibroblasts were obtained from Aviron (Mountain View, CA). All cell lines were grown in DMEM supplemented with 5% newborn calf serum, unless otherwise stated. HSV-1 strain F [HSV-1(F)], a limited-passage isolate, is the prototype strain used in this laboratory (16). The construction and phenotypic properties of the recombinant virus R7910 lacking both copies of the α0 gene have been described (3).
Plasmids.
pRB4986 (3) carries a HSV-1(F) DNA fragment encoding α0 codons 111–241 cloned in the yeast two-hybrid vector pGBT9. To construct pRB5193, pRB4986 was digested with KpnI and PstI, treated with T4 DNA polymerase, and religated. To construct pRB5194, pRB4986 was digested with EagI and PstI, treated with T4 DNA polymerase, and religated. The sequences encoding α0 codons 111–159 represented within plasmid pRB4987 were amplified by PCR and cloned into the EcoRI and SalI sites of pGBT9. The codons of charged amino acids mapped by the deletion analysis of sequences cloned in pRB4896 were replaced with the alanine codon by site-directed mutagenesis of the α0 gene sequences in plasmids pRB5261 through pRB5267. Specifically, complementary oligonucleotides containing the specific nucleotide substitution were annealed to pRB4896 DNA according to the manufacturer’s instructions (Stratagene Quik Change, #200518). The substitutions were D181A/R185A in pRB5261, E187A in pRB5262, E189A in pRB5263, E190A in pRB5264, R193A in pRB5265, and D199A in pRB5266. The original sequence encoding D199 codon was restored in pRB5267 by site-directed mutagenesis; this plasmid was used to restore the original sequence in the D199A mutant virus. In addition to A199D substitution, a silent mutation in the wobble base of V198 codon of pRB5267 created a SalI restriction useful for differentiation of wild-type and repaired viruses. All mutations generated by site-directed mutagenesis were verified by sequencing.
The plasmid pRB5270 designed to restore ICP0 coding sequences within the ICP0 null recombinant virus R7910 was constructed by cloning the HSV-1(F) BsrGI fragment, flanking the ICP0 coding sequences, into the Acc65I site of pUC19.
pRB5268 was constructed in several steps to insert the D199A mutation into the HSV-1 genome. The MfeI–DraIII fragment containing the sequence carrying the mutation was purified and ligated to a MfeI–DraIII-digested pRB3710 plasmid (17) containing the SacI–PstI fragment that contains the ICP0 coding sequence. The resulting plasmid was digested with MfeI–Bsu36I, and the corresponding fragment was ligated into a MfeI–Bsu36I-digested pRB5270 to yield pRB5268.
Plasmid pRB5269 was constructed to restore wild-type ICP0 coding sequences to recombinant virus R7914 and thus create the R7915 repair virus. The MfeI–DraIII fragment from plasmid pRB5267 described above was cloned into the MfeI–DraIII sites of pRB3710.
Yeast Two-Hybrid Screen.
The yeast two-hybrid screen described earlier (3) was used to map the domain of exon II of the α gene capable of interacting with cyclin D3. Deletion mutants as well as alanine substitution mutants of pRB4896 were cotransformed into yeast strain Y190 with pBH1004, a previously described plasmid (3) containing the cyclin D3 cDNA fused to the GAL4 transcriptional activation domain in pACT. Cotransformants were isolated from colonies that grew on SD medium lacking tryptophan and leucine. Cotransformants then were assayed for β-galactosidase activity as reported (10).
Immunoblotting.
The antibodies used in these assays were a mAb to ICP0 (H1083, 1:500 dilution, Goodwin Institute, Plantation, FL) (18) and a mAb to cyclin D3 (1:1,000 dilution, PharMingen G107–565). The electrophoretically separated proteins transferred to nitrocellulose were reacted with antibodies as described (19) and quantified with the aid of ECL chemiluminescent reagent obtained from Amersham Pharmacia.
Determination of Viral Sequence.
To determine whether recombinant viruses R7914 and R7915 possessed the correct mutation within the ICP0 coding domains, a PCR fragment was generated from viral DNA harvested from the cytoplasm of infected cells and then subjected to sequence analysis. Primer A (5′-CACCAGCTTGGCGTTGCA-3′) and primer B (5′-CTGTTGGTGGTGGTGTTG-3′) were combined with 100 ng of purified viral DNA in the presence of 10% DMSO and subjected to standard PCR. The resulting 561-nt fragment then was purified (Wizard PCR prep DNA purification, Promega) and sequenced (20).
Construction of Recombinant Viruses.
Recombinant virus R7914 was constructed by transfecting rabbit skin cells with pRB5268 followed by superinfection of the cells with the Δα0 mutant R7910 at 0.05 plaque-forming unit (PFU)/cell. Large plaque were isolated, grown on Vero cells, and further plaque-purified and tested by immuoblotting electrophoretically separated denatured proteins for the production of ICP0. The presence of the desired amino acid substitutions was verified by sequencing of the DNA fragment amplified by PCR from viral DNA extracted from mature cytoplasmic virions.
Recombinant virus R7915 was generated by cotransfection of rabbit skin cells with plasmid pRB5269 and intact R7914 viral DNA. As noted above, plasmid pRB5269 restored amino acid D199 and in addition carried a silent mutation in V198 that created a SalI site. The progeny of the transfection of rabbit skin cells were plaque-purified and tested for the presence of the V198 silent mutation. Specifically, the DNA of virus progeny extracted after plaque purification were individually extracted, digested with SalI, electrophoretically separated on an agarose gel, transferred to a nitrocellulose sheet, and hybridized with a 32P-labeled DNA fragment contained in pRB3710 and carrying the coding sequence of ICP0. The recombinant virus of interest, R7915, exhibiting the SalI restriction endonuclease site, was verified further by sequencing a PCR-amplified fragment by using the viral DNA extracted from cytoplasmic virions as template.
RESULTS
Mapping of the Domain of the ICP0 that Binds Cyclin D3.
The interaction of ICP0 and cyclin D3 initially was discovered in a yeast two-hybrid screen and mapped between codons 111 and 241 of the second exon (Fig. 1A, line 3) (3). Fine mapping of the interactive site was done in two steps. In the first, we constructed a series of truncated portions of the 3′ half of the exon II of the α0 gene as shown in Fig. 1. These clones were tested in the yeast two-hybrid system. As indicated in Fig. 1A, line 3, the site in ICP0 that interacted with cyclin D3 mapped between codons 159 and 213. On examining amino acids 159–213, we noted a concentration of charged amino acids between codons 181 and 199 (Fig. 1B, line 1). In the next series of experiments we mutagenized the DNA fragment encoding codons 111–241 by replacing with alanine the codons 181 and 185, or single codons 187, 189, 190, 193, or 199, as described in Materials and Methods. In this experiment the product of the plasmid pRB5266 carrying the substitution D199A failed to interact with cyclin D3 in the two-hybrid system. Reconstruction of the original sequence in pRB5267 restored the interaction with cyclin D3.
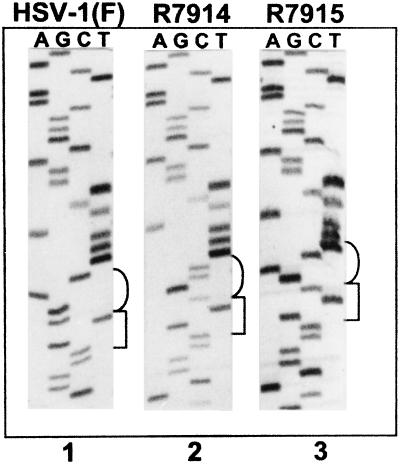
To verify the results of the yeast two-hybrid assays and to determine the phenotype of the single amino acid substitution in ICP0, we constructed two recombinant viruses. Recombinant virus R7914 was isolated from the progeny of transfection of rabbit skin cells with plasmid pRB5268 carrying the D199A substitution followed by exposure of the cells to the recombinant virus R7910 (0.05 PFU/cell) from which both copies of the α0 gene had been deleted (3). In recombinant virus R7915, the α0 gene carrying the substituted amino acid was repaired to restore the wild-type ICP0 sequence by cotransfection of rabbit skin cells with intact R7914 DNA and plasmid pRB5269 carrying an A199D substitution and a silent mutation in V198. As described in Materials and Methods, the latter mutation created a SalI restriction site designed to facilitate the screening for the repaired virus and to differentiate wild-type and repaired virus. The presence of desired mutations in the viral DNA was verified by sequencing of exon II. As shown in Fig. 2, recombinant R7914 has the desired substitution D199A whereas recombinant R7915 has the desired restoration of the original amino acid sequence and the silent mutation in the V198 codon.
Figure 2.
Autoradiographic image of ICP0 exon II sequence from wild-type HSV-1(F) and recombinant viruses R7914 and R7915. Cytoplasmic viral DNA was obtained from infected Vero cells, and a 561-bp fragment was PCR-amplified and then sequenced. The sequences of HSV-1(F) (lane 1) and recombinant viruses R7914 and R7915 (lanes 2 and 3, respectively) were identical with the exception of the desired mutations shown in brackets. The square bracket to the right of the images represents V198 (codon usage GTN), and the round brackets represent codon 199 (codon usage for aspartic acid is GAC or GAT, and codon usage for alanine is GCN).
The Recombinant Virus Carrying the D199A Substitution in ICP0 Does Not Stabilize Cyclin D3.
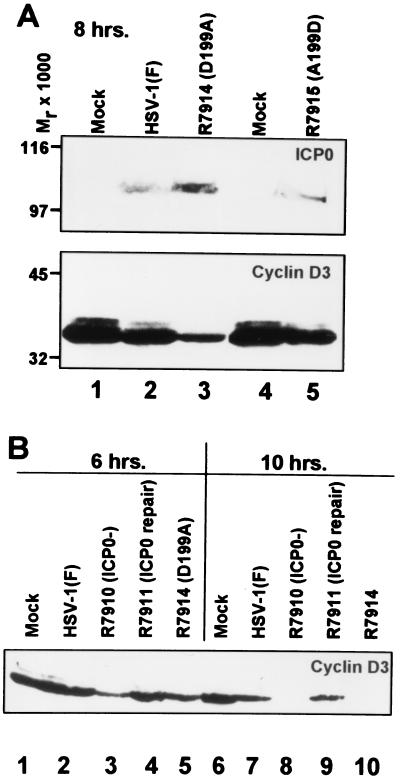
This series of studies was designed to determine whether ICP0 carrying the D199A mutation stabilized cyclin D3. In these experiments the cells were harvested at 6, 8, or 10 hr after infection, solubilized in buffer containing SDS, and subjected to electrophoresis in denaturing polyacrylamide gels. The electrophoretically separated proteins were transferred to a nitrocellulose sheet and reacted with antibody to cyclin D3. The 8-hr lysate also was reacted with antibody to ICP0. The results indicated the following: (i) Cyclin D3 was stabilized for at least 8 hr in cells infected with the wild-type virus HSV-1(F) parent or with the recombinant virus R7915 in which the D199A mutation was repaired (Fig. 3A, lanes 2 and 5). (ii) The levels of cyclin D3 were reduced at 6 and 8 hr and were not detectable at 10 hr after infection with the recombinant viruses R7910 (ICP0−, Fig. 3B, lanes 3 and 5) or R7914 (D199A, Fig. 3B, lanes 8 and 10). The mutant carrying the D199A mutation could not be differentiated from the mutant lacking ICP0. Restoration of deleted ICP0 (R7911) led to stabilization of cyclin D3 at least until 10 hr after infection (Fig. 3B, compare lanes 7 and 9).
Figure 3.
(A) Immunoblots of lysates of cells mock-infected (lanes 1 and 4) or exposed to 0.5 PFU of HSV-1(F) (lane 2), R7914 (lane 3), or R7915 (lane 5) per cell. The cells were harvested after infection, solubilized in buffer containing SDS, electrophoretically separated in a denaturing polyacrylamide gel, and probed with mAb to ICP0 (H1083, ref. 18) or the mAb to cyclin D3 (Pharmingen, #G107–565). Molecular weights are shown on the left. (B) The cells were mock-infected (lanes 1 and 6) or infected with HSV-1(F) (lanes 2 and 7), the ICP0 null recombinant R7910 (lanes 3 and 8), recombinant R7911 (repair of R7910) (lanes 4 and 9), or recombinant R7914 (lanes 5 and 10) and harvested 6 hr after infection (lanes 1–5) or 10 hr. after infection (lanes 6–10), and processed as above. The electrophoretically separated proteins were reacted with the mAb to cyclin D3.
The Replication of the Recombinant Virus Carrying the D199A Substitution in ICP0 in Resting and Dividing Cells.
These experiments were designed to characterize the replicative of properties of R7914 recombinant virus. The first series took into account the observation that the replication of ICP0− viruses was multiplicity dependent, that is, the higher ratio of virus particle per cell overcame the defect caused by the absence of the α0 gene (2). As a general principle, the replicative capacity of HSV is to some extent at least dependent on the cell line in which it is grown (21, 22). As illustrated in Table 1, there was no significant difference in the yield of wild-type parent and D199A mutant R7914 in several cell lines infected with 0.5 or 5 PFU/cell and harvested at 20 hr after infection. The second series of experiments took into account the observation that some mutants replicate better in dividing than in growth-arrested, quiescent, primary human fibroblasts. In this series of experiments human lung fibroblasts were maintained in the same medium until the cultures became totally confluent and contact-inhibited or serum-starved, maintained only in medium containing 0.25% serum. As shown in Table 2, R7914 yielded 10-fold less virus in contact inhibited or in serum-deprived human lung fibroblasts than either the wild-type parent HSV-1(F) or the recombinant R7915 in which the aspartic acid codon 199 was restored. In essence, R7914 was not impaired in dividing cells but yielded 10-fold less virus in nondividing, resting cells.
Table 1.
The yields of parent and D199A mutant virus at 20 hr after infection of various cell lines at multiplicities of infection shown
| Cell line | PFU/cell | Yield (×105)*
|
|
|---|---|---|---|
| HSV-1(F) | R7914 | ||
| Vero | 5.0 | 300 | 310 |
| 0.05 | 680 | 730 | |
| HEp-2 | 5.0 | 100 | 190 |
| 0.05 | 69 | 800 | |
| RSC | 5.0 | 3,400 | 560 |
| 0.05 | 1,600 | 420 | |
| HeLa | 5.0 | 1,800 | 6,500 |
| 0.05 | 7.7 | 9.8 | |
Virus yields were determined by standard plaque assays in Vero cells.
Table 2.
The yields of parent, D199A mutant, and rescued viruses from quiescent and serum-starved human lung fibroblasts harvested at 5 or 20 hr after infection
| HLF cells | Hr after infection | Yield*
|
||
|---|---|---|---|---|
| HSV-1(F) | R7914 | R7915 | ||
| Serum starved | 5 | 1.2 × 104 | 2.4 × 104 | 1.3 × 104 |
| 20 | 6.4 × 107 | 4.0 × 106 | 5.6 × 107 | |
| Quiescent | 5 | 6.1 × 104 | 5.5 × 104 | 4.1 × 104 |
| 20 | 1.8 × 108 | 1.1 × 107 | 1.7 × 108 | |
Diploid human lung fibroblasts (HLF) grown in 25-cm2 flasks were either serum-starved in medium containing 0.25% FCS for 5 days or allowed to become fully contact inhibited (quiescent) and maintained in same medium for 7 days. The cells were infected (0.05 PFU/cell) in the same medium in which they were maintained before infection.
The Pathogenicity of a D199A Mutant Virus.
The last series of studies reported here were based on the well-established fact that on entry into the body HSV-1 infects nerve endings and is transported by retrograde flow to the neuronal nuclei of dorsal root ganglia where HSV may multiply or establish latency. In some instances, the virus is transported to the CNS where it can cause encephalitis and death. In an experimental animal system, the capacity to invade CNS (neuroinvasiveness) and the destruction of the CNS caused by viral replication (neurotoxicity) are important and semi-independent indicators of viral virulence. Thus viruses that multiply efficiently and destroy CNS (neurotoxicity) may not be able to invade the brain from peripheral sites (neuroinvasiveness). In the first series of experiments we assayed viral neurotoxicity by intracerebral inoculation of 5-week-old mice with log dilutions of parent, mutant, and rescued viruses. As shown in Table 3, we observed a 10-fold decrease in neurotoxicity of D199A mutant as compared with wild-type viruses. In the second series of experiments, we assayed neuroinvasiveness from a peripheral site by inoculating log dilutions of each of the three viruses into 5-week-old mice. The results (Table 3) indicate a significant, 100-fold decrease in the neuroinvasiveness of the D199A mutant compared with those of the wild-type parent or rescued viruses.
Table 3.
Neurotoxicity and neuroinvasiveness of wild-type parent D199A mutant and repaired virus
| Virus | PFU ± SE*/LD50
|
|
|---|---|---|
| Intracerebral† | Intraperitoneal‡ | |
| HSV-1(F) | <100 | 135 ± 66 |
| R7914 (D199A) | 265 ± 32 | >10,000 |
| R7915 (A199D) | ND | 135 ± 71 |
LD50 ± SE was calculated by the Spearman–Karber method.
5-week-old mice in groups of six per dilution were monitored for 21 days.
5-week-old mice in groups of eight per dilution were monitored for 30 days.
DISCUSSION
Herpesviruses appear to be highly concerned with D-type cyclins. Thus, both herpes saimiri virus and its distant cousin, human herpesvirus no. 8, encode functional homologues of D-type cyclins (23–25, 28). Epstein–Barr virus encodes proteins that induce expression of a D-type cyclin (26). As shown in this and earlier reports, ICP0 encoded by HSV-1 binds and stabilizes cyclin D3 (3). The need for sustaining the activity of cyclin D3 seems a curious property of a virus that replicates in both dividing and resting cells in vitro. To define the role of cyclin D3, we mapped the binding site in ICP0 and mutagenized it. We conclude from the studies described above that the mutation D199A in ICP0 (i) leads to the more rapid disappearance of cyclin D3 in infected cells, (ii) has no effect on viral replication in dividing cells, (iii) exhibits a slight decrease in viral replication in resting cells, (iv) shows a relatively small decrease in viral neurotoxicity, and (v) has a significant effect on viral neuroinvasiveness.
One hypothesis to account for our results is that ICP0 acts as a scavenger to bind, stabilize, and activate cyclin D3 even if it is present in minuscule amounts, as may be the case in resting cells (27). That the object of ICP0-encoded function is to stabilize and activate rather than render cyclin D3 inactive stems from two considerations. First, the phosphorylation of pRb in vitro by cyclin D3 and cdk4 is not affected by large excess of the ICP0 cyclin D3 binding site (3). Second, as noted above, herpes saimiri virus and human herpesvirus no. 8 encode cyclin D homologs that bind the appropriate kinase and phosphorylate the pRb (27). These data suggest the virus benefits from the recruitment and stabilization of cyclin D3 to induce it to phosphorylate the pRb and thereby activate the E2F transcriptional factors. These, in turn, enhance the metabolic activities of the infected cell associated with the entry of the cell into S phase. The benefits are readily apparent from the observation that in the mouse, the virus capable of binding and stabilizing cyclin D3 has significant advantages over the virus that lacks this capability.
One hypothesis that explains our findings is that in an organism such as a mouse, the virus targets the central nervous system but is in direct competition with the development of an immune response. A 10-fold increase in viral yield in resting cells, as seen in this study, could readily translate into a more rapid neuroinvasiveness with the intended consequence of the destruction of CNS before the immune system can block viral spread. That this mechanism may be important even though the incremental effects of stabilization of cyclin D3 are small is apparent from the observations that some herpesviruses encode a homolog of cyclin D. Presumably in cells infected with these viruses there is no D-type cyclin to scavenge. In primary human infection, higher yields translate into more rapid spread, more extensive population of sensory neurons with latent virus, and higher frequencies of reactivation of latent virus that becomes available for transmission by direct contact to nonimmune hosts. The strategy of viral survival includes scavenging the cell for useful proteins, such as cyclin D3, and making them work for enhanced viral growth.
Recent analyses of ICP0 suggest that it consists of blocks of amino acids whose primary function appears to be interaction with cellular or viral proteins. These blocks appear to be relatively small because single amino acid substitutions effectively blocked the interaction with cyclin D3, as reported in this study, or with the ubiquitin-specific protease (9). In the few instances in which the interaction has been investigated in detail, as for example in the case of cyclin D3 and the ubiquitin-specific protease, the purpose of the interaction has been to translocate the proteins to a new location, e.g., ND10, or to stabilize cellular proteins with a relatively short half-life. The interaction of ICP0 with cyclin D3 illustrates a scavenger function of ICP0.
Acknowledgments
These studies were aided by grants from the National Cancer Institute (CA47451 and CA71933) and the U.S. Public Health Service (CA78766). C.V.S. is a predoctoral trainee aided by a grant from the National Institute of General Medical Sciences (GM07183–22).
ABBREVIATIONS
- HSV-1
herpes simplex virus 1
- HSV-1(F)
HSV-1 strain F
- ICP0
infected cell protein no. 0
- CNS
central nervous system
- PFU
plaque-forming unit
- pRb
retinoblastonic protein
References
- 1.Roizman B, Sears A E. In: Fields’ Virology. 3rd Ed. Fields B N, Knipe D M, Howley P, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. New York: Raven; 1996. pp. 2231–2295. [Google Scholar]
- 2.Sacks W, Schaffer P A. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawaguchi Y, Van Sant C, Roizman B. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai W, Schaffer P A. J Virol. 1993;67:7501–7512. doi: 10.1128/jvi.67.12.7501-7512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Everett R, Preston C, Stow N. In: Herpes Virus Transcription: Functional and Genetic Analysis of the Role of Vmw110 in Herpes Simplex Virus Replication. Wagner E K, editor. Boca Raton FL: CRC; 1991. pp. 49–76. [Google Scholar]
- 6.Gelman I, Silverstein S. J Mol Biol. 1986;191:395–409. doi: 10.1016/0022-2836(86)90135-x. [DOI] [PubMed] [Google Scholar]
- 7.Maul G, Guldner H, Spivack J G. J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 8.Everett R, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. EMBO J. 1997;16:566–577. doi: 10.1093/emboj/16.3.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everett R, Meredith M, Orr A. J Virol. 1999;73:417–426. doi: 10.1128/jvi.73.1.417-426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawaguchi Y, Bruni R, Roizman B. J Virol. 1997;71:1019–1024. doi: 10.1128/jvi.71.2.1019-1024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruni R, Fineschi B, Ogle W, Roizman B. J Virol. 1999;73:3810–3817. doi: 10.1128/jvi.73.5.3810-3817.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawaguchi Y, Van Sant C, Roizman B. J Virol. 1998;72:1731–1736. doi: 10.1128/jvi.72.3.1731-1736.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawaguchi Y, Matsumura T, Roizman B, Hirai K. J Virol. 1999;73:4456–4460. doi: 10.1128/jvi.73.5.4456-4460.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein G, Dulic V. BioEssays. 1995;17:537–543. doi: 10.1002/bies.950170610. [DOI] [PubMed] [Google Scholar]
- 15.Grand R. Virology. 1998;244:330–342. doi: 10.1006/viro.1998.9102. [DOI] [PubMed] [Google Scholar]
- 16.Ejercito P M, Keiff E D, Roizman B. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 17.McKnight J L C, Kristie T M, Roizman B. Proc Natl Acad Sci USA. 1987;84:7061–7065. doi: 10.1073/pnas.84.20.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ackermann M, Roizman B. J Virol. 1986;58:843–850. doi: 10.1128/jvi.58.3.843-850.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heine J W, Honess R W, Cassai E, Roizman B. J Virol. 1974;14:640–651. doi: 10.1128/jvi.14.3.640-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werness B A, Levine A J, Howley P. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 21.Stow E, Stow N. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 22.Cai W, Schaffer P A. J Virol. 1991;65:4058–4090. doi: 10.1128/jvi.65.8.4078-4090.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholas J, Cameron K R, Honess R W. Nature (London) 1992;355:362–365. doi: 10.1038/355362a0. [DOI] [PubMed] [Google Scholar]
- 24.Chang Y, Moore P S, Talbot I, Bosshoff C H, Zarlowska T, Godden-Kent D, Paterson H, Weiss R A, Mittnacht S. Nature (London) 1996;382:410. doi: 10.1038/382410a0. [DOI] [PubMed] [Google Scholar]
- 25.Jung J, Stoger M, Desrosiers R C. Mol Cell Biol. 1994;14:7235–7244. doi: 10.1128/mcb.14.11.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinclair A J, Palmero I, Peters G, Farrell P J. EMBO J. 1994;13:3321–3328. doi: 10.1002/j.1460-2075.1994.tb06634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherr C J. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 28.Godden-Kent D, Talbot S, Boshoff C, Chang Y, Moore P, Weiss R, Mitttnacht S. J Virol. 1997;71:4193–4198. doi: 10.1128/jvi.71.6.4193-4198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]