Abstract
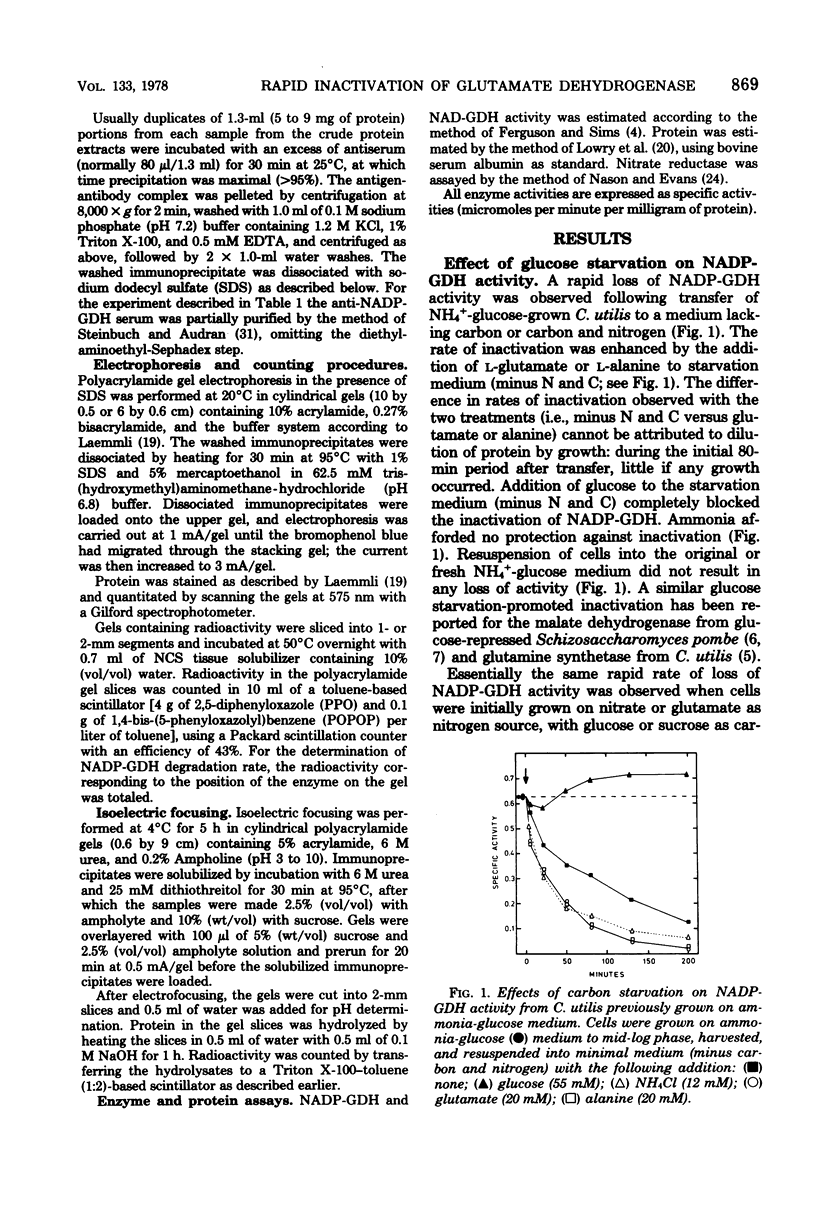
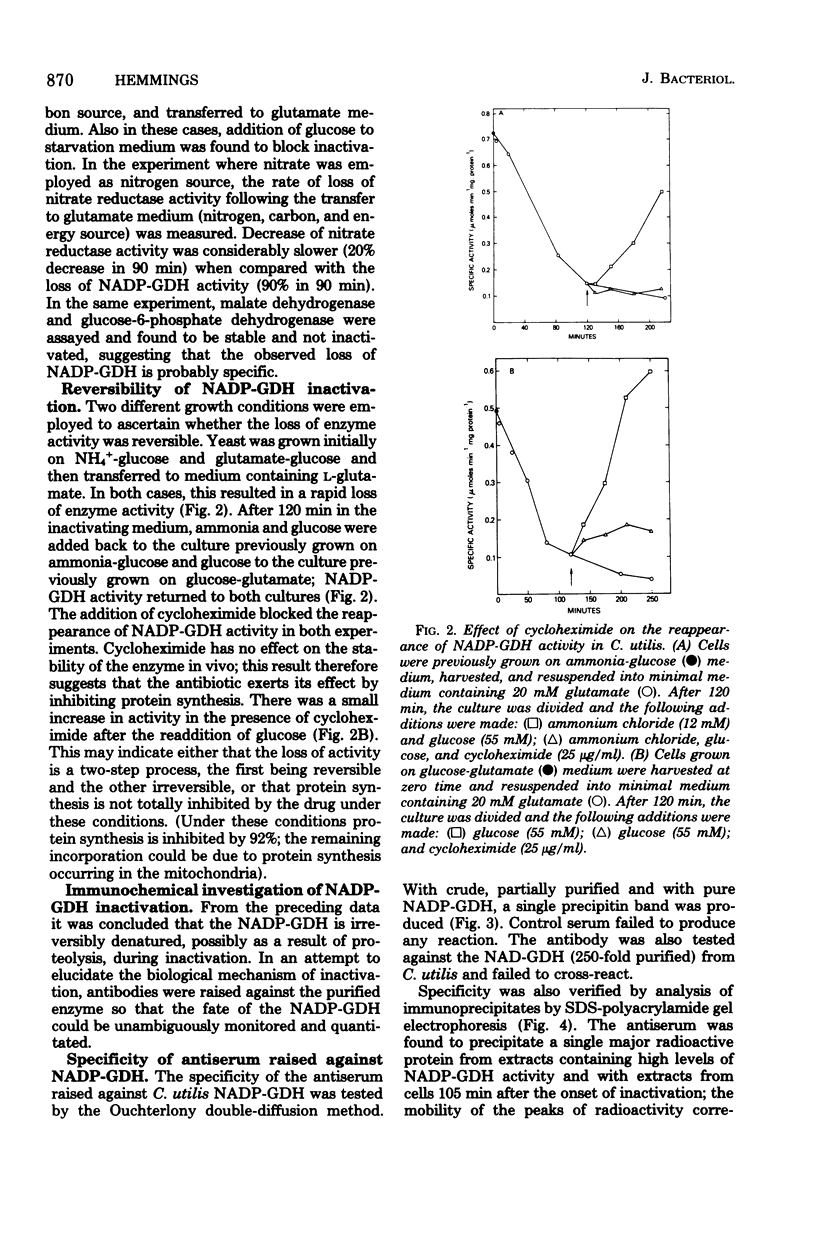
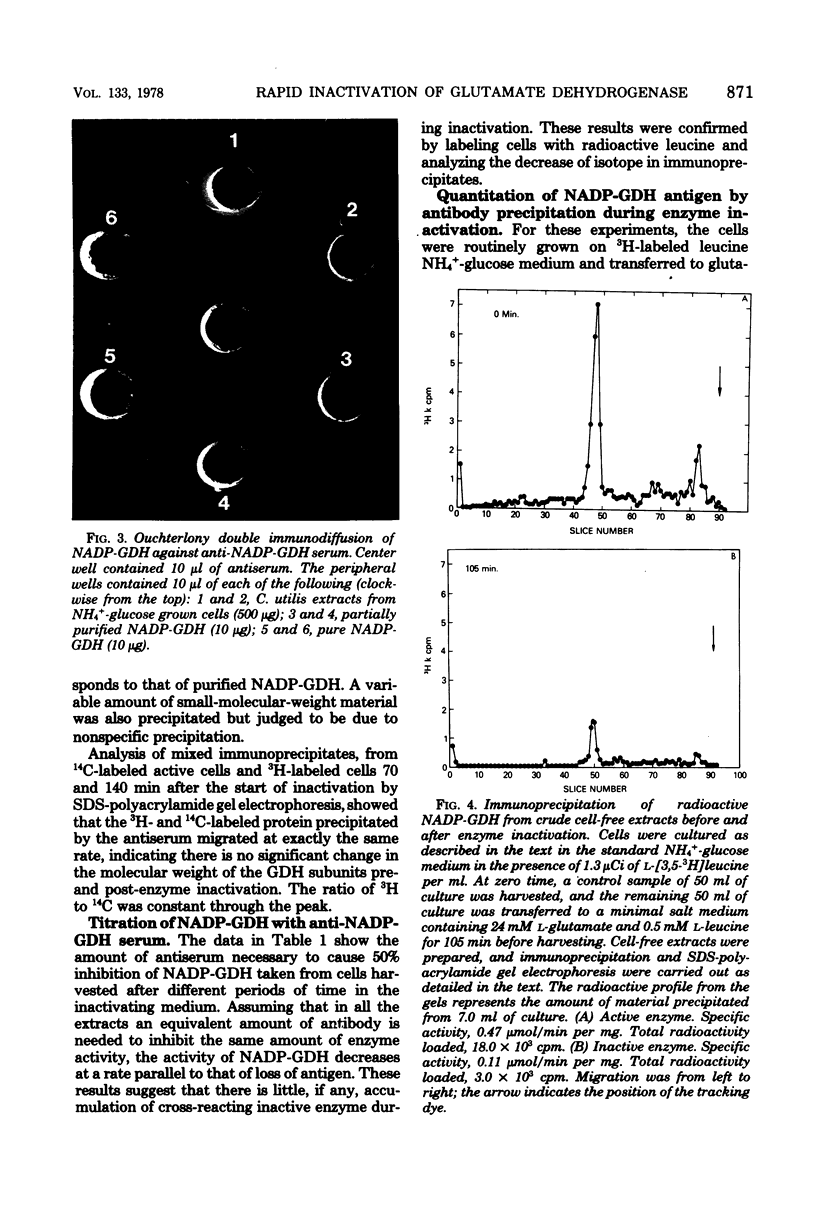
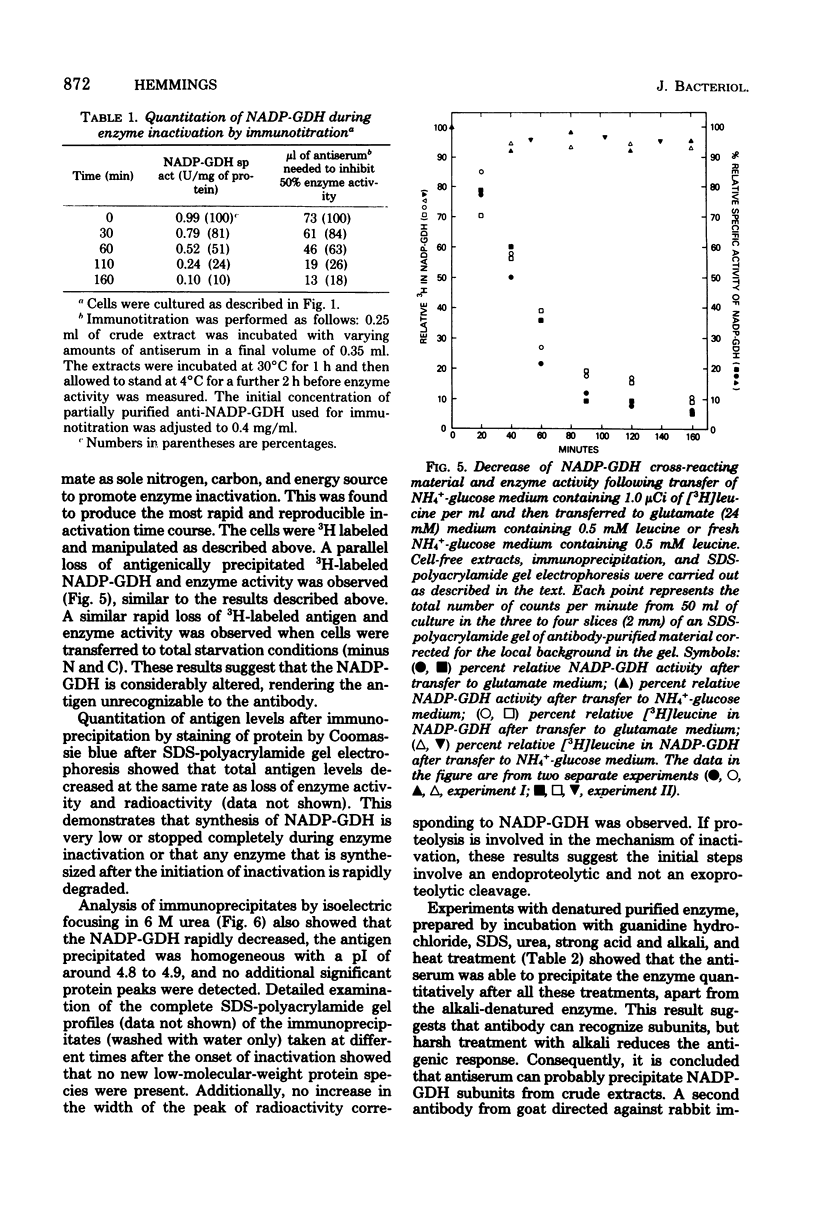
The nicotinamide adenine dinucleotide phosphate-dependent glutamate dehydrogenase (NADP-GDH) from the food yeast Candida utilis was found to be rapidly inactivated when cultures were starved of a carbon source. The addition of glutamate or alanine to the starvation medium stimulated the rate of inactivation. Loss of enzyme activity was irreversible since the reappearance of enzyme activity, following the addition of glucose to carbon-starved cultures, was blocked by cycloheximide. A specific rabbit antibody was prepared against the NADP-GDH from C. utilis and used to quantitate the enzyme during inactivation promoted by carbon starvation. The amount of precipitable antigenic material paralleled the rapid decrease of enzyme activity observed after transition of cells from NH4+-glucose to glutamate medium. No additional small-molecular-weight protein was precipitated by the antibody as a result of the inactivation, suggesting that the enzyme is considerably altered during the primary steps of the inactivation process. Analysis by immunoprecipitation of the reappearance of enzyme activity after enzyme inactivation showed that increase of NADP-GDH activity was almost totally due to de novo synthesis, ruling out the possibility that enzyme activity modulation is achieved by reversible covalent modification. Enzyme degradation was also measured during steady-state growth and other changes in nitrogen and carbon status of the culture media. In all instances so far estimated, the enzyme was found to be very stable and not normally subject to high rates of degradation. Therefore, the possibility that inactivation was caused by a change in the ratio of synthesis to degradation can be excluded.
Full text
PDF

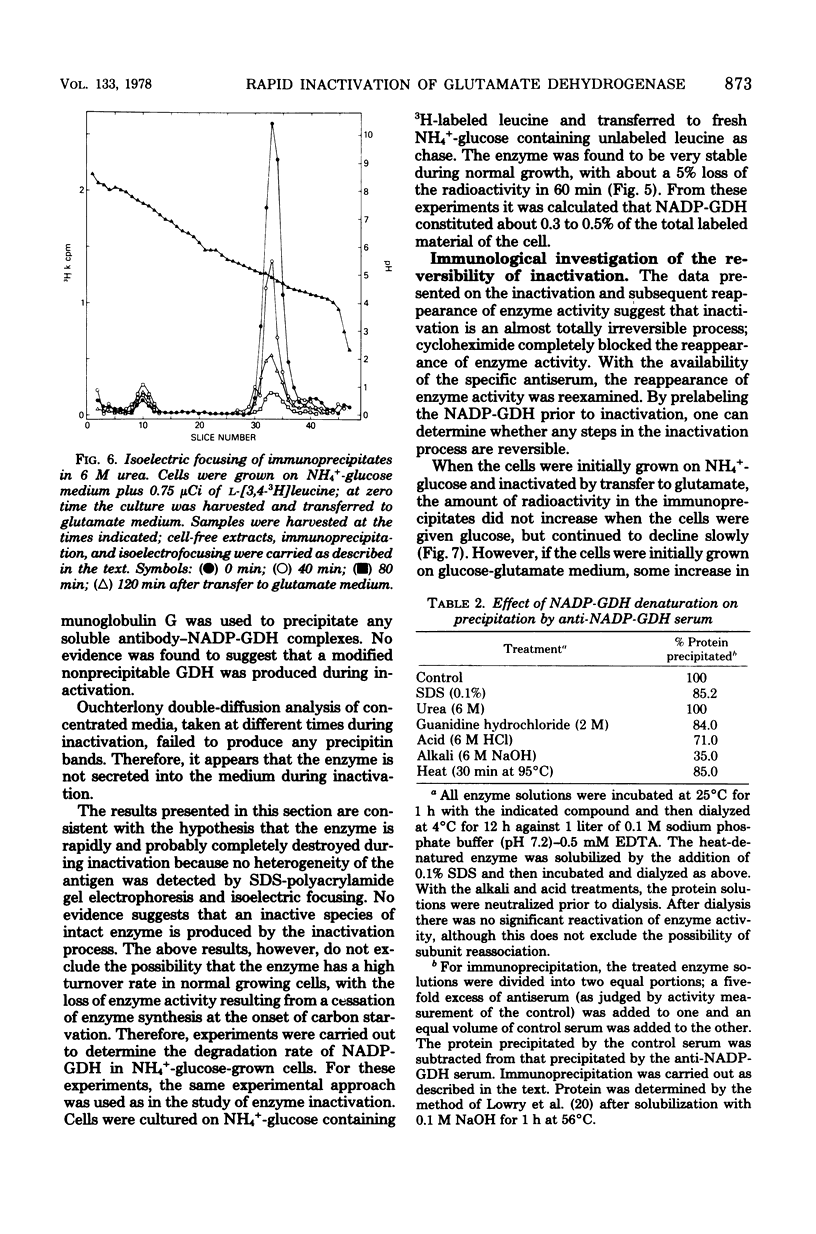
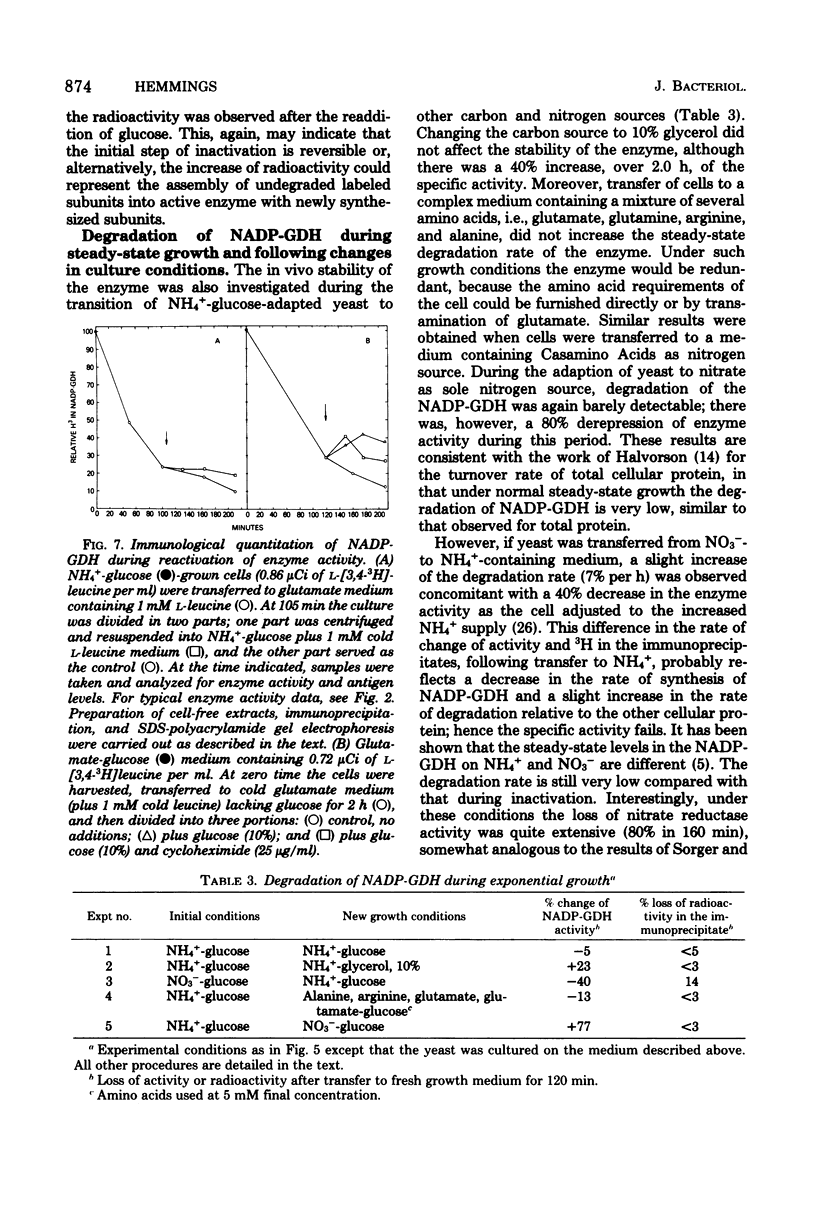



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Betz H., Hinze H., Holzer H. Isolation and properties of two inhibitors of proteinase B from yeast. J Biol Chem. 1974 Jul 25;249(14):4515–4521. [PubMed] [Google Scholar]
- Ferguson A. R., Sims A. P. Inactivation in vivo of glutamine synthetase and NAD-specific glutamate dehydrogenase: its role in the regulation of glutamine synthesis in yeasts. J Gen Microbiol. 1971 Dec;69(3):423–427. doi: 10.1099/00221287-69-3-423. [DOI] [PubMed] [Google Scholar]
- Ferguson A. R., Sims A. P. The regulation of glutamine metabolism in Candida utilis: the role of glutamine in the control of glutamine synthetase. J Gen Microbiol. 1974 Jan;80(1):159–171. doi: 10.1099/00221287-80-1-159. [DOI] [PubMed] [Google Scholar]
- Flury U., Heer B., Fiechter A. Isoenzyme pattern of malate dehydrogenase during respiratory derepression in Schizosaccharomyces pombe. Arch Microbiol. 1974 Apr 19;97(2):141–148. doi: 10.1007/BF00403053. [DOI] [PubMed] [Google Scholar]
- Flury U., Heer B., Fiechter A. Regulatory and physicochemical properties of two isoenzymes of malate dehydrogenase from Schizosaccharo-myces pombe. Biochim Biophys Acta. 1974 Apr 25;341(2):465–483. doi: 10.1016/0005-2744(74)90239-3. [DOI] [PubMed] [Google Scholar]
- Folkes B. F., Sims A. P. The significance of amino acid inhibition of NADP-linked glutamate dehydrogenase in the physiological control of glutamate synthesis in Candida utilis. J Gen Microbiol. 1974 May;82(1):77–95. doi: 10.1099/00221287-82-1-77. [DOI] [PubMed] [Google Scholar]
- Giffhorn F., Gottschalk G. Effect of growth conditions on the activation and inactivation of citrate lyase of Rhodopseudomonas gelatinosa. J Bacteriol. 1975 Dec;124(3):1046–1051. doi: 10.1128/jb.124.3.1046-1051.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giffhorn F., Gottschalk G. Inactivation of citrate lyase from Rhodopseudomonas gelatinosa by a specific deacetylase and inhibition of this inactivation by L-(+1-glutamate. J Bacteriol. 1975 Dec;124(3):1052–1061. doi: 10.1128/jb.124.3.1052-1061.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A. L., St John A. C. Intracellular protein degradation in mammalian and bacterial cells: Part 2. Annu Rev Biochem. 1976;45:747–803. doi: 10.1146/annurev.bi.45.070176.003531. [DOI] [PubMed] [Google Scholar]
- Goldschmidt R. In vivo degradation of nonsense fragments in E. coli. Nature. 1970 Dec 19;228(5277):1151–1154. doi: 10.1038/2281151a0. [DOI] [PubMed] [Google Scholar]
- HALVORSON H. Studies on protein and nucleic acid turnover in growing cultures of yeast. Biochim Biophys Acta. 1958 Feb;27(2):267–276. doi: 10.1016/0006-3002(58)90333-0. [DOI] [PubMed] [Google Scholar]
- Hasilik A., Müller H., Holzer H. Compartmentation of the tryptophan-synthase-proteolyzing system in Saccharomyces cerevisiae. Eur J Biochem. 1974 Oct 1;48(1):117–117. doi: 10.1111/j.1432-1033.1974.tb03748.x. [DOI] [PubMed] [Google Scholar]
- Hemmings B. A., Sims A. P. The regulation of glutamate metabolism in Candida utilis. Evidence for two interconvertible forms of NAD-dependent glutamate dehydrogenase. Eur J Biochem. 1977 Oct 17;80(1):143–151. doi: 10.1111/j.1432-1033.1977.tb11866.x. [DOI] [PubMed] [Google Scholar]
- Hynes M. J. The effects of carbon source on glutamate dehydrogenase activities in Aspergillus nidulans. J Gen Microbiol. 1974 Mar;81(1):165–170. doi: 10.1099/00221287-81-1-165. [DOI] [PubMed] [Google Scholar]
- Kinghorn J. R., Pateman J. A. The effect of the carbon source on ammonium regulation in Aspergillus nidulans. Mol Gen Genet. 1974;128(1):95–98. doi: 10.1007/BF00267298. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mason T. L., Poyton R. O., Wharton D. C., Schatz G. Cytochrome c oxidase from bakers' yeast. I. Isolation and properties. J Biol Chem. 1973 Feb 25;248(4):1346–1354. [PubMed] [Google Scholar]
- Milman G., Portnoff L. S., Tiemeier D. C. Immunochemical evidence for glutamine-mediated degradation of glutamine synthetase in cultured Chinese hamster cells. J Biol Chem. 1975 Feb 25;250(4):1393–1399. [PubMed] [Google Scholar]
- Morrison S. L., Zipser D. Polypeptide products of nonsense mutations. I. Termination fragments from nonsense mutations in the Z gene of the lac operon of Escherichia coli. J Mol Biol. 1970 Jun 14;50(2):359–371. doi: 10.1016/0022-2836(70)90198-1. [DOI] [PubMed] [Google Scholar]
- NASON A., EVANS H. J. Triphosphopyridine nucleotide-nitrate reductase in Neurospora. J Biol Chem. 1953 Jun;202(2):655–673. [PubMed] [Google Scholar]
- Neumann P., Markau K., Sund H. Studies of glutamate dehydrogenase. Regulation of glutamate dehydrogenase from Candida utilis by a pH and temperature-dependent conformational transition. Eur J Biochem. 1976 Jun 1;65(2):465–472. doi: 10.1111/j.1432-1033.1976.tb10362.x. [DOI] [PubMed] [Google Scholar]
- Platt T., Miller J. H., Weber K. In vivo degradation of mutant lac repressor. Nature. 1970 Dec 19;228(5277):1154–1156. doi: 10.1038/2281154a0. [DOI] [PubMed] [Google Scholar]
- Roberts J. W., Roberts C. W. Proteolytic cleavage of bacteriophage lambda repressor in induction. Proc Natl Acad Sci U S A. 1975 Jan;72(1):147–151. doi: 10.1073/pnas.72.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger G. J., Debanne M. T., Davies J. Effect of nitrate on the synthesis and decay of nitrate reductase of Neurospora. Biochem J. 1974 Jun;140(3):395–403. doi: 10.1042/bj1400395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbuch M., Audran R. The isolation of IgG from mammalian sera with the aid of caprylic acid. Arch Biochem Biophys. 1969 Nov;134(2):279–284. doi: 10.1016/0003-9861(69)90285-9. [DOI] [PubMed] [Google Scholar]
- Switzer R. L. The inactivation of microbial enzymes in vivo. Annu Rev Microbiol. 1977;31:135–157. doi: 10.1146/annurev.mi.31.100177.001031. [DOI] [PubMed] [Google Scholar]
- Thomas D. A., Wright B. E. Glycogen phosphorylase in Dictyostelium discoideum. II. Synthesis and degradation during differentiation. J Biol Chem. 1976 Mar 10;251(5):1258–1263. [PubMed] [Google Scholar]
- Waindle L. M., Switzer R. L. Inactivation of aspartic transcarbamylase in sporulating Bacillus subtilis: demonstration of a requirement for metabolic energy. J Bacteriol. 1973 May;114(2):517–527. doi: 10.1128/jb.114.2.517-527.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]




