Abstract
Legionella pneumophila is the cause of Legionnaires’ disease, which is a form of potentially fatal pneumonia. To identify genes required for virulence of the bacterium, a library of 1,386 L. pneumophila signature tagged transposon mutants was studied for guinea pig virulence. The mutants were screened in pools of 96 each in a guinea pig model of L. pneumophila pneumonia. Sixteen unique mutant clones were determined to have attenuated virulence after being screened twice in the animal model. All 16 mutants failed to multiply in both lungs and spleens. Four of the sixteen had no apparent defect for intracellular multiplication in macrophages. Partial DNA sequences of the interrupted genes adjacent to the transposon insertions showed that six of them had mutations in five known L. pneumophila virulence genes: dotB, dotF/icmG, dotO/icmB, icmX, and proA. Three of the sequenced clones contained mutations in genes without known homology to other published bacterial genes, and seven clones appeared to be homologous to five different known bacterial genes but are still being characterized. With this methodology, we demonstrate the existence of L. pneumophila genes responsible for non-macrophage-related virulence. The discovery of L. pneumophila virulence genes indicates the utility of the signature tagged mutagenesis technique for pulmonary pathogens.
Legionella pneumophila is the most common etiologic agent of Legionnaires’ disease, a type of pneumonia affecting immunocompromised and immunocompetent humans (1). This Gram-negative bacterium is a facultative intracellular parasite of mononuclear cells in vivo and in vitro and resides within nonfused and nonacidified phagosomes within macrophages (Mφ) (2, 3). Several L. pneumophila virulence factors facilitating intracellular infection or growth have been identified in screens using Mφ or Mφ-like cell lines but not animals. Thus, prior L. pneumophila virulence factors have been selected with a bias for deficient growth within Mφ (4–9). An excellent guinea pig pneumonia model of Legionnaires’ disease exists that closely mimics the human disease in immunocompromised hosts (10, 11). In this animal model, bacteria are introduced into the trachea, causing pneumonia and later septicemia. However, the difficulty and expense of the animal model makes screening of hundreds of bacterial mutants prohibitive.
Recently, Holden and others have described the use of signature tagged mutagenesis (STM) to find new bacterial virulence genes (12–16). Here, bacteria are mutagenized by transposon mutagenesis by using a pool of individually unique transposons that each contain an oligonucleotide with common outer arms and a unique random central sequence. Each mutant has a unique molecular signature, which can be specifically detected by using PCR with primers specific to the outer common arms. Animals are infected with pools of mutant clones, and the clones present in the infecting inoculum, but absent in the organ or organs of interest, are identified by a colony blot technique. This negative selection technique provides the ability to screen large numbers of mutants at one time in one animal. Use of this technique also affords the ability to determine the targeting and growth of specific clones (or their absence) within different animal organs. It reduces the potential selection bias inherent in screening mutants in just one type of in vitro test of virulence. Use of the STM technique in Salmonella typhimurium, Vibrio cholerae, Staphylococcus aureus, and Streptococcus pneumoniae in various animal models has resulted in the discovery of novel virulence genes for each bacterium (12–16).
The STM method has not been previously used to study the pathogenesis of an intracellular pulmonary pathogen. We demonstrate here that this method can be used successfully to study L. pneumophila guinea pig pneumonia pathogenesis, and we also report the identification of several L. pneumophila virulence genes.
METHODS
Bacterial Strains, Plasmids, and Growth Conditions.
L. pneumophila serogroup 1 strain AA100jm is a spontaneous streptomycin resistant mutant of strain 130b, which is virulent in guinea pigs, Mφ, and amoebae (11, 17, 18). L. pneumophila strain AP2.B5 is a spontaneous, salt-resistant, avirulent mutant of strain AA100jm that also contains an oligonucleotide signature tag (ST) inserted by transposon mutagenesis as described below. L. pneumophila strain AP3 is a mip mutant of strain AA100jm and contains a ST. AP3 and AP2.B5 were gifts from Andrea Polesky (Stanford Univ., Stanford, CA). The growth of strain AP3 in amoebae is reduced ≈10-fold compared with AA100jm. L. pneumophila was grown at 35°C in a humidified incubator, unless stated, on either Mops-buffered charcoal yeast extract agar medium supplemented with α-ketoglutarate (BCYEα) or in N-[2-acetamido]-2-aminoethanesulfonic acid-buffered yeast extract broth supplemented with α-ketoglutarate (BYEα) (19). Escherichia coli strains DH5αλpir, SM10λpir, and XL-1 blue (Stratagene) were grown at 37°C in air on either Luria–Bertani (LB) agar or broth. E. coli was grown in 2 × YT broth after electroporation. Selective antimicrobial agents were added to the growth media when appropriate and included kanamycin (Km), 30 μg/ml; ampicillin (Ap), 50 μg/ml; and streptomycin, 200 μg/ml .
Plasmids pCVD442 (20) and pKD368 (21) were gifts from Michael Donnenberg (Univ. of Maryland, Baltimore) and Keith Derbyshire (David Axelrod Institute for Public Health, Albany, NY), respectively. pCVD442 is composed of the mobRP4, oriR6K, and bla regions from pGP704, a multiple cloning site, and the sacB gene. pKD368 is derived from pBR322 and contains a defective IS903, the kanamycin resistance (KmR) gene from IS903, oriM13, and lacZ; IS903 transposes efficiently, singly, and randomly into L. pneumophila (22).
Primers.
PCR primers P3 and P5 were used to amplify genomic DNA containing the STs as described (16). Primers P2 and P4 were used to make 32P-labeled DNA for probing colony blots of input and output pools, as described; 27 PCR cycles were used (16). Sequencing primers M13 forward and M13 reverse were used for sequencing DNA inserted into the multiple cloning site of pUC18 (Life Technologies, Gaithersburg, MD). Primer PE101 (5′ ttttgccgctatttctctgttc 3′) was used to sequence DNA upstream of nucleotide 101 of pKD368.
Nucleic Acid Manipulations.
All nucleic acid manipulations were accomplished according to standard molecular biology techniques (23).
Transposon Shuttle Vector.
A transposon shuttle vector was constructed from pKD368. The 4.5-kilobase EcoRV-digested and dephosphorylated fragment of pCVD442 containing ori6K and mobRP4 was gel purified. An 8-kilobase EagI/XmnI-digested fragment of pKD368 was gel purified and was made blunt by using the Klenow reagent; this fragment contains IS903. The two fragments were ligated and transformed by electroporation into E. coli DH5αλpir, and KmR transformants were selected, resulting in pPE5. A KpnI linker was ligated to the EcoRV site of pPE5, and the resulting plasmid was KpnI digested, gel purified, and recircularized by using T4 DNA ligase. This then was transformed into E. coli DH5αλpir by electroporation and KmR ApR transformants selected, creating pC6.
Library Construction.
A library of random STs contained in pUTmini-Tn5Km2 was a gift from D. W. Holden (Royal Postgraduate Medical School, London). These STs have common outer arms and a random inner 40-bp sequence (NK)20, with a diversity of 106. The STs were amplified by using PCR with primers P3 and P5 (16). The resulting ≈90 bp product was gel-purified, was digested with KpnI, and was ligated into KpnI digested, dephosphorylated, and gel-purified pC6. The ligation product was transformed in DH5αλpir by electroporation, and ≈2 × 105 KmR ApR transformants were pooled. About 104 of these cells were grown in LB broth (1 liter), from which plasmid DNA was purified by a CsCl gradient method; this plasmid DNA was designated pC6HT.
The STs were placed into L. pneumophila AA100jm by plate conjugation mating. E. coli SM10λpir was transformed with pC6HT by electroporation and was grown in 2 × YT broth (1 ml) for 1 h at 37°C; this (0.4 ml) then was used to inoculate LB broth (250 ml) containing Km and Ap, which was incubated overnight on a 30°C shaker. The culture then was incubated at 21°C for 0.5 h before being washed three times with PBS (100 ml, 4°C). Finally, the transformants were resuspended in sterile distilled water to a final concentration of 2 × 106 colony-forming units (cfu)/ml. L. pneumophila AA100jm was grown overnight in BYEα broth in a 37°C shaker, was washed once with sterile distilled water, was adjusted to a final concentration of 2 × 108 cfu/ml in the water, and then was heated to 42°C for 10 min. The E. coli transformant suspension (0.5 ml), which had been processed in parallel, was mixed with the L. pneumophila suspension (1 ml), and the mixture was plated on a single large (150 mm) BCYEα plate for 6 h at 37°C. The bacteria were washed from the plate with sterile distilled water (2 ml) and then were plated on large BCYEα plates containing Km and streptomycin. The plates were incubated for 4 days, and the transconjugants were screened for l-cysteine growth dependence and susceptibility to Ap. The transposition frequency was ≈10−5. About 2,300 ApS KmR streptomycinR L. pneumophila transconjugants were obtained. Poorly growing bacteria were deleted from the library, and the 2,200 individual clones were assembled into 24 different 96-well microtiter trays containing BYEα broth supplemented with Km.
Characterization and Assembly of Pools.
To identify clones containing STs that hybridized well enough to use in a screening assay, colony blots were performed (16). Charged nylon membranes (Hybond N+, Amersham Pharmacia) were replica-stamped from the microtiter wells of each bacterial pool and were placed on BCYEα with Km, and the bacteria were grown on the membranes for 2–3 days. The DNAs from the bacterial colonies on the membranes then were denatured by alkaline lysis and were neutralized, and the membranes were washed extensively in 2 × standard saline citrate (SSC). The dried membranes were incubated with 32P-labeled probes made from homologous ST DNAs by using standard methods. About half of the clones did not hybridize well enough with their homologous probes to be visualized unambiguously on autoradiographs of the hybridized membranes, leaving 1,386 clones in 14 newly assembled microtiter trays.
The frequency and relative location of transposon insertion into L. pneumophila AA100jm was determined for 11 randomly selected clones. Genomic DNA from the clones was digested with HindIII and was used in a Southern blot probed with 32P randomly labeled pC6HT DNA (Rediprime DNA labeling system, Amersham Pharmacia). The Southern blot revealed that all 11 clones had a single transposon insertion, each one at a different location.
Clones from five different tray pools were probed with a probe made only to the ST DNAs of the first column of clones from each respective tray, using the colony blot method described above. There was no heterologous hybridization detected within each tray, and strong hybridization to the first column of each tray by its homologous DNAs was detected.
Animal Model.
The guinea pig model of L. pneumophila pneumonia was used as described (24). L. pneumophila was grown in BYEα broth under the appropriate selective conditions and was diluted in sterile water at a concentration of 1 × 107 cfu/ml. This was injected into the surgically exposed tracheas of Hartley strain male guinea pigs weighing ≈250 g. The animals were killed 2 days later. The right lower lung lobe and spleen were removed aseptically, were weighed and ground in Mueller-Hinton broth, and were diluted in the same broth type in decimal dilutions. The tissue homogenates were plated onto multiple large BCYEα + Km plates to obtain individual colonies used to extract genomic DNA. Dilution plating was performed on the same medium to determine organ bacterial concentrations. In some experiments, organ homogenates were plated onto both BCYEα and BCYEα + Km to differentiate between KmS parent (AA100jm) and KmR mutant (AP2.B5 or AP3) strains. The quantity of parent strain was determined by the difference in bacterial concentrations of those bacteria growing on BCYEα and BCYEα + Km. To detect revertants, plating of tissue homogenates from one animal from each experiment was performed on BCYEα plates made with and without Km.
Sequencing Methods.
Genomic DNA from clones of interest was digested with restriction enzymes known not to cut the transposon insert upstream of the Km cassette. Southern blots of the electrophoresed, digested DNAs were probed with a digoxigenin-labeled probe to the Km cassette. This probe was made from the 1.7-kilobase fragment of a BamHI digest of pUTmini-Tn5Km2 and was labeled with digoxigenin by using the DIG DNA labeling kit (Boehringer Mannheim); an alkaline-phosphatase antibody to digoxigenin and chromogenic substrates were used to detect DNA hybridizing with the probe (Boehringer Mannheim). Km cassette-containing fragments that were from 3.8 to 7 kilobases in size were gel-purified and ligated into appropriately digested pUC18. E. coli strain XL-1 blue was transformed with the ligation product introduced by electroporation. Plasmid DNA of KmR transformants was restriction digest-mapped to confirm proper insertion of the desired DNA fragment into the plasmid. Plasmid DNA was purified by using a Qiagen spin filter and was sequenced with M13 reverse, M13 forward, or PE101 sequencing primers, depending on the orientation and size of the L. pneumophila DNA in the new plasmid. The ABI Big Dye Taq FS terminator sequencing kit (Applied Biosystems), was used to synthesize the dye-terminated DNA, which then was sequenced by using an ABI 377 automated sequencer (University of Pennsylvania Sequencing Facility). GenBank sequence database searching was performed with the blastx and blastn search algorithms (http://www.ncbi.nlm.gov/BLAST).
Determination of Auxotrophy.
A modification of the CAA medium described by Mintz et al. (25) and colleagues was used to determine the ability of the mutant clones to multiply on a minimal medium. Starch was replaced by activated charcoal; full growth of the bacteria on this medium required 10–14 days of incubation. Decimal dilutions of the bacterial suspensions were plated onto both the modified CAA and BCYEα media.
Mφ Virulence Assay.
L. pneumophila clones of interest were tested for their ability to grow in freshly explanted guinea pig alveolar Mφ as described (26). L. pneumophila bacteria were grown on BCYEα agar for 2 days and then were harvested in water. The inoculum density was adjusted to a concentration of 108 cfu/ml and then was further diluted in tissue culture medium to achieve a final cell density of 105 cfu/ml. This was added to tissue culture wells containing 105 adherent Mφ per well, so that the final bacterial concentration was 104 cfu per well. Tissue culture supernatants were cultured quantitatively for L. pneumophila 1 h, 2 days, and 3 days after inoculation. L. pneumophila does not grow in the tissue culture medium alone, so increases in the bacterial concentrations in the tissue culture supernatant are caused solely by intracellular growth of the bacterium. A major defect in Mφ multiplication was classified by one that resulted in at least a 5-fold lower bacterial concentration than that observed for the parent strain.
RESULTS
Characterization of the Performance of the Guinea Pig Model.
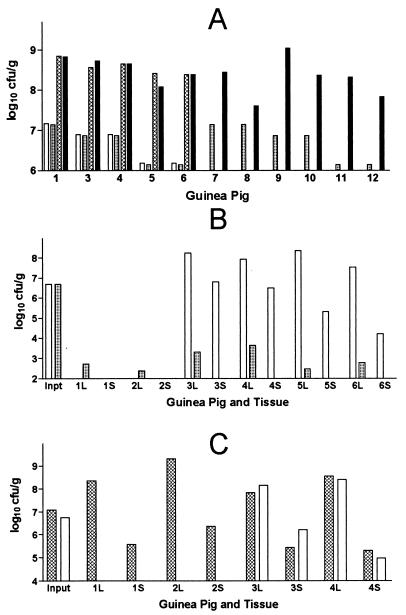
Three different experiments were conducted to determine whether the animal model of L. pneumophila pneumonia detected bacterial mutants that do not multiply in the animal. First, one randomly selected pool of 96 transposon-mutated clones (pool 36) was used to infect guinea pigs by itself and, when mixed with the parent strain, AA100jm. This experiment was designed to determine the optimal bacterial inoculum and duration of infection. The experiment also was used to determine whether the population of mutant clones that multiplied in the lung was representative of the input population or was confined to just a few. Three different bacterial inocula were used: 1 × 107, 5 × 106, or 1 × 106 cfu/animal; this made the delivered inoculum ≈1 × 105, 5 × 104, or 1 × 104 cfu of each mutant/animal. Six animals received approximately equal amounts of both pool 36 and AA100jm, and six animals received the same amount of pool 36 alone as was given in the mixed infection studies. One animal died immediately postoperatively. Nine of the animals were killed 45 h postinfection whereas two were killed 70 h postinfection. Differential lung concentrations of the bacteria were determined by use of KmR of the pool 36 clones and KmS of AA100jm. No significant differences in animal weights or temperatures were detected for the pool 36 alone and pool 36 mixed with AA100jm animal groups, regardless of infecting inoculum or observation period. The parental strain did not outcompete the mutant clone pool, as approximately equal concentrations of parent and mutant pool bacteria were recovered from the animal lungs for those animals that received a mixed inoculum (Fig. 1). The lung bacterial concentrations of the mutant pool bacteria was approximately the same for those animals receiving the mutant pool alone and those receiving a mixture of parent and mutant pool. Ninety of the ninety-six clones in the pool 36 input pool could be detected in the output pool of bacteria obtained from the lungs of the six animals receiving pool 36 alone, demonstrating that the mutant clone population in the infected lungs was not confined to a minority of introduced clones. Finally, the experiment demonstrated that the transposon insertion was not an inherent cause of avirulence.
Figure 1.
Recovery of L. pneumophila (log10 cfu/ml) from guinea pig organs. (A) Recovery from lungs of strain AA100jm and a pool of mutant clones (pool 36) for 11 guinea pigs. Animals 1–6 received both AA100jm and pool 36, and animals 7–12 received only pool 36 bacteria. All animals except 6 and 12 were killed 2 days after infection; 6 and 12 were killed 3 days after infection. Delivered bacterial inocula are shown by the clear bar (AA100jm) and the light cross hatched bar (pool 36); the L. pneumophila lung concentrations are shown by the heavy cross hatched (AA100jm) and black bars (pool 36); e.g., for animal 1, the bars from left to right are, respectively, AA100jm input, pool 36 input, AA100 output, and pool 36 output. (B) Results for animals inoculated with the salt tolerant mutant AP2.B5 (animals 1 and 2), or both AA100jm and AP2.B5 (animals 3–6). AA100 input and output is designated by the clear bars, and AP2.B5 is designated by the cross hatched bars. L, lung; S, spleen. Animals 5 and 6 were killed 3 days after infection, and animals 1–4 were killed killed 2 days after infection. (C) Results for animals inoculated with the mip− mutant AP3 (animals 1 and 2) or with AP3 and AA100jm (animals 3 and 4). AA100 input and output is designated by the clear bars, and APB3 is designated by the cross hatched bars. All animals were killed 2 days after infection.
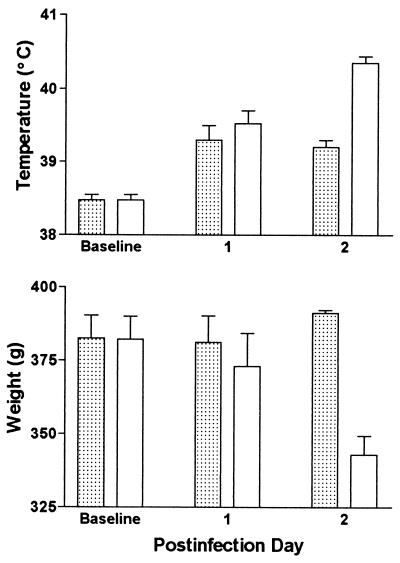
To determine the sensitivity and specificity of the animal model for the detection of L. pneumophila mutants with attenuated virulence, a L. pneumophila salt avirulent mutant containing a signature tag (AP2.B5) was tested in the animal model. Two guinea pigs were infected with AP2.B5 alone, and four animals received a combination of AP2.B5 and AA100jm; the delivered inocula were 5.7 × 106 and 5.3 × 106 cfu/animal for AP2.B5 and AA100jm, respectively. The animals were killed two or three days postinoculation, and the lungs and spleens were cultured quantitatively for KmS (AA100jm) and KmR (AP2.B5) L. pneumophila bacteria. In contrast to dramatic weight loss and high fever in the animals inoculated with both AP2.B5 and AA100jm, AP2.B5 given by itself did not cause significant disease in the animal, aside from the slight fever normally seen with inoculation with dead bacteria (Fig. 2). Rapid lung and spleen clearance of AP2.B5 was detected, with at least a 4 log10 decrease in bacterial lung concentrations over 2–3 days (Fig. 1). Over the same time periods, there was ≈2 log10 multiplication of AA100jm in the lungs. Spleen concentrations of AA100jm were lower in the two animals killed 3 days postinfection, as opposed to those killed after 2 days, so it was elected to standardize the postinfection period to 50 h.
Figure 2.
Guinea pig temperatures and body weights are shown for one experiment. Guinea pigs were inoculated with L. pneumophila strains AA100jm and AP2.B5. Stippled bars, only mutant strain inoculated (two animals); clear bars, both mutant and parent strains inoculated (four animals). Error bars represent standard error of the mean.
Genomic bacterial DNA was harvested from 5,000–16,000 colonies growing on nonselective BCYEα plates plated with lung suspensions of all four animals inoculated with both L. pneumophila strains. DNA probes could not be made from these genomic DNA preparations because no PCR products were obtained by using primers P2 and P4 as described. This result was confirmed in three additional experiments while at the same time demonstrating the expected PCR product for a positive control. Absent target DNA was the reason for failure to detect the PCR product because the ratio of the signature tagged AP2.B5 to the non-signature tagged AA100jm bacterial colonies in the lungs was ≈1:105 to 1:106. Thus, no AP2.B5 colonies would be expected in the 5,000–16,000 bacterial colonies harvested from the animal lungs. This experiment confirmed the avirulence of AP2.B5 and demonstrated that we could easily detect the disappearance of an avirulent bacterium in our screening method.
Lastly, the sensitivity and specificity of the animal model for the detection of a L. pneumophila mutant with subtle virulence defects was determined by using AP3, which is mip− and contains a ST. AP3 was fully virulent in the animal model (Fig. 1). Two guinea pigs were inoculated with 1 × 107 cfu of AP3 alone, and two animals received the same dose of AP3 and 6 × 106 cfu of AA100jm. No significant differences were detected in animals weights, temperatures, or L. pneumophila bacterial concentrations in spleens or lungs. No attempt was made to use the ST method to detect AP3 on blots because of the very high concentrations of this bacterium in lung and spleens. These results showed that the animal model was insensitive to subtle virulence differences and that mip− mutants would not be detected in our screen.
Screening of L. pneumophila Transposon Mutant Pools in the Guinea Pig Model.
L. pneumophila mutants (1,386 mutants) were screened in guinea pigs in pools of ≈96 clones each. Each pool was used to inoculate two animals each; the average inoculum size was 4.9 × 106 cfu/animal, with a 95% confidence interval of 4.2 × 106 to 5.6 × 106 cfu/animal. A clone was scored as absent in the output pool when three of the four animal organs examined (two spleens and two lungs each) did not contain the clone by colony blots. A total of 35 clones, or 2.5% of the clones tested, were not represented in the output pool. These 35 clones were reassembled into a new pool and were tested again in six guinea pigs. A clone was considered to be attenuated in virulence when it was not found in at least three of the six organs (three lungs and spleens each) tested. A total of 16 clones appeared to be attenuated in virulence. Fifteen clones were not present in all six organs examined, and one was absent in four of six organs tested. These 16 clones represented the attenuated virulence clones that were further characterized. No revertants were detected in any animal study.
Partial Sequencing of Interrupted Genes.
Six of the 16 attenuated virulence mutant clones had transposon insertions interrupting known L. pneumophila virulence genes, five of which were in the dot-icm locus and one of which was in the proA gene (Table 1). An additional mutant clone had a transposon insertion 608 bp upstream of the start of the icmF gene. Three mutations (including the one adjacent to icmF) had no known DNA sequence homologies in the GenBank database, and seven had mutations in known genes of other bacteria. Mutations in three different genes were found in six of the clones. Two of these six clones had mutations in the same region, the pts homolog. The transposon insertion sites were 434 bp apart and in opposite directions, which means that these are two independent clones. Two other mutants had transposon insertions 106 bp apart and in opposite directions within the aroB homolog and therefore were also independent mutants. Another two mutants both had transposon insertions in the icmX gene, 615 bp apart, and were independent mutants.
Table 1.
Characterization of clones
| Clone | Transposon insertion site | Mφ growth*
|
Animal model† | GenBank no. | Identity/Similarity, e value‡ | |
|---|---|---|---|---|---|---|
| 2 days | 3 days | |||||
| 6b | L. pneumophila icmX | −3.7 | −4.9 | LS | 3602929 | 86/97, e−59 |
| 3c | L. pneumophila dotB | −3.6 | −5.5 | LS | AF026533 | 100/100, e−10 |
| 2f | L. pneumophila dotO/icmB | −3.1 | −4.5 | LS | AF026534 | 100/100, e−75 |
| 6c§ | L. pneumophila icmX | −4.0 | −5.2 | LS | 3602929 | 91/94, e−117 |
| 8a | L. pneumophila dotF/icmG | −1.1 | 0.2 | LS | AF026534 | 97/97, e−14 |
| 8g | NH¶ (608 bp upstream of L. pneumophila icmF) | −2.3 | −2.1 | LS | ||
| 5b | L. pneumophila proA | −1.3 | −0.2 | LS | M31884 | 100/100, e−46 |
| 4a | E. coli phosphoenolpyruvate phosphotransferase (pts) homolog | −1.8 | −0.5 | LS | 1255724 | 40/61, e−36 |
| 3h‖ | E. coli phosphoenolpyruvate phosphotransferase (pts) homolog | −1.7 | −0.5 | LS | 1255724 | 45/67, e−31 |
| 4b | Haemophilus influenzae 3-dehydroquinate synthetase (aroB) homolog | −2.9 | −2.7 | LS | 1073786 | 57/72, e−71 |
| 5f** | H. influenzae 3-dehydroquinate synthetase (aroB) homolog | −1.5 | −0.2 | LS | 1073786 | 34/51, e−10 |
| 4d | Quenine tRNA-ribosyltranferase (tgt) homolog | −0.5 | −0.1 | LS | Q55983 | 42/58, e−77 |
| 4c | Helicobacter pylori β-alanine synthetase homolog | −0.2 | 0.2 | LS | 2313883 | 46/57, e−16 |
| 8b | Hypothetical 107-kDa protein in argR-cafA region homolog | −0.3 | 0.0 | ls | 2851641 | 28/51, e−6 |
| 3b | NH | −0.8 | 0.1 | LS | ||
| 1e | NH | −0.3 | 0.1 | LS | ||
Difference (log10 cfu/ml) between concentration of clone and parent strain grown in guinea pig alveolar macrophages, after 2 and 3 days. Data represent mean of two independent experiments. The median differences and 95% Cls between the two experiments for each strain were log10 cfu/ml 0.09 (0.07–0.19), 0.26 (0.16–0.35), and 0.34 (0.22–0.46) for days 0, 2, and 3, respectively.
Organs not containing detectable clone by colony blot. LS, missing from all three lungs and spleens from three animals; ls, missing from the lungs and spleens of two of three animals.
blastx search identity and similarity percent, and e value.
The transposon insertion sites of clones 6b and 6c were 615 bp apart in the same direction.
NH, no homology found in blastx search of the GenBank database.
The transposon insertion sites of clones 4a and 3h were 434 bp apart and in opposite directions.
The transposon insertion sites of clones 4b and 5f were 106 bp apart and in opposite directions.
Determination of Auxotrophy.
Clones with mutations in homologs of aroB and prs failed to grow on the minimal medium, which represented at least a 3 log10 decrease in bacterial concentration when compared with growth on the complete (BCYEα) medium.
Intracellular Multiplication in Mφ of the 16 Unique Attenuated Virulence Clones.
To determine whether the 16 unique attenuated virulence clones were defective in macrophage multiplication, we tested each individually in the Mφ multiplication assay described above. These experiments could not determine whether a defect in intracellular multiplication was caused by poor Mφ internalization or other factors. Twelve of the sixteen unique attenuated virulence clones had major defects in Mφ multiplication that were most marked after 2 days of incubation with the Mφ (Table 1). However, after 3 days of incubation with the Mφ, 6 of these 12 clones multiplied almost as well as AA100jm. The other four clones had minor to no defects in Mφ multiplication after both 2 and 3 days of incubation with the Mφ.
DISCUSSION
We have demonstrated that use of the STM method identified a broad range of potential L. pneumophila virulence genes, far in excess of those discovered to date by using screening of transposon mutants in Mφ cell lines or targeted mutagenesis (6–8, 27–30). The key to obtaining the broad spectrum of mutants appears to be the use of an animal pneumonia model for screening the mutants, rather than Mφ assays.
Three different classes of Mφ virulence phenotypes were discovered in these experiments. One class of six mutants had a markedly reduced ability to multiply within Mφ and included mutants of the already known dot-icm complex (8, 31). Detection of the dot-icm complex mutants through use of the STM technique serves to validate our experimental approach. Another class of six mutants had an initial defect in Mφ multiplication but were able to multiply in Mφ as well as the parent strain after prolonged incubation. These mutants may have a defect in Mφ uptake, but those that enter the cell may be fully virulent. Alternatively, the bacteria may multiply more slowly within the Mφ. Such a finding has been made before for a L. pneumophila mip− mutant (32).
We identified a novel class of mutants whose phenotype is independent of Mφ virulence. In this class of four mutants, which had little if any defect in Mφ multiplication, was one of the three mutants with partial sequences not matching known genes in the GenBank database. This group of mutants may have some alternative reason for reduced virulence, such as killing or inhibition by serum, intraalveolar components, or other host cells, including alveolar epithelial cells (33–36). Some members of this mutant group may be cleared more efficiently by nonspecific local host defenses. Alternatively, only very minor defects in Mφ multiplication may provide enough host advantage to prevent L. pneumophila multiplication within the lungs. Because the only prior screening of L. pneumophila mutants has been for those bacteria unable to invade or multiply in Mφ, there have been no reports of reduced virulence mutants able to multiply within Mφ.
Our experiments confirm a previous study (11) showing that the L. pneumophila major secretory protein (proA), a zinc metallo protease, is a virulence factor of this bacterium. The role of this protease in disease pathogenesis has been controversial because other studies have not demonstrated a defect in virulence for L. pneumophila proA mutants (37–39).
Three of the sixteen (19%) of the partially sequenced L. pneumophila clones had no sequence homology with known bacterial genes. This number of unidentified loci has been a general theme in other studies using the STM method. Unidentified loci accounted for 4–26% of putative virulence genes discovered by the STM method in S. aureus (13, 14), 10% in V. cholerae (12), and 32% in S. typhimurium (16).
There are probably many virulence factors that were not detected by us because we did not screen a sufficient number of mutants to cover the genome. We did not detect all of the known dot-icm locus genes, and our methods were unable to discriminate between a mip− mutant and the parent strain in the animal model. Furthermore, we may have discounted some strains with reduced virulence by using strict criteria for interpreting the colony blots. Finally, there are L. pneumophila virulence factors that we could not have detected in an animal model. These include factors important for survival in aerosols (40), multiplication within amoebae and subsequent spread to humans (18, 41), phase variation (42), and those that may modulate a growth phase, temperature or osmotic change induced enhancement of virulence (43, 44). The less-than-absolute sensitivity of the STM method has been observed in other STM studies.
We have demonstrated that the STM method can be used to study the pathogenesis of a pulmonary pathogen in an animal model and predict that it can be used successfully to study the pathogenesis of other pulmonary pathogens. The animal model lends itself to the discovery of non-Mφ-dependent virulence factors and could be important for differentiating between pulmonary and nonpulmonary virulence mechanisms. Either intracellular or extracellular pathogens could be studied by using this method, which is of major benefit in the absence of accurate in vitro measures of the pulmonary disease caused by extracellular bacteria.
Acknowledgments
Daniel Martin formulated the construction of the shuttle vector and provided helpful suggestions for many aspects of this project. Andrea Polesky generously provided L. pneumophila strains AP2.B5 and AP3 and helpful advice. Joan Mecsas, Brendan Cormack, Lalita Ramakrishnan, Raphael Valdivia, Bärbel Raupach, Nina Salama, and Jacqueline Shea provided technical suggestions and instruction in various laboratory techniques. Lalita Ramakrishnan and Joan Mecsas also provided excellent editorial assistance in their critical readings of the manuscript. Jianjun Ren, David Moon, Thao Truong, and Takayuki Miyara provided excellent technical assistance. Part of this work was funded by National Institutes of Health Grant AI30618 to Lucy Tompkins. Futoshi Higa was supported by a study grant from the Japanese Ministry of Education, Science, and Culture.
ABBREVIATIONS
- Ap
ampicillin
- Mφ
macrophage
- ST
signature tag
- STM
signature tagged mutagenesis
- Km
kanamycin
- cfu
colony-forming unit
References
- 1.Edelstein P H, Meyer R D. In: Advances in Pathology and Laboratory Medicine. Weinstein R S, Graham A R, Anderson R E, Benson E S, Cotran R S, Jarett L, Wick M R, Zumwalt R E, editors. St. Louis: Mosby; 1995. pp. 149–167. [Google Scholar]
- 2.Horwitz M A. J Exp Med. 1983;158:2108–2126. doi: 10.1084/jem.158.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horwitz M A, Maxfield F R. J Cell Biol. 1984;99:1936–1943. doi: 10.1083/jcb.99.6.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hickey E K, Cianciotto N P. Infect Immun. 1997;65:133–143. doi: 10.1128/iai.65.1.133-143.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirby J E, Vogel J P, Andrews H L, Isberg R R. Mol Microbiol. 1998;27:323–336. doi: 10.1046/j.1365-2958.1998.00680.x. [DOI] [PubMed] [Google Scholar]
- 6.Kirby J E, Isberg R R. Trends Microbiol. 1998;6:256–258. doi: 10.1016/s0966-842x(98)01310-9. [DOI] [PubMed] [Google Scholar]
- 7.Purcell M, Shuman H A. Infect Immun. 1998;66:2245–2255. doi: 10.1128/iai.66.5.2245-2255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Segal G, Shuman H A. Trends Microbiol. 1998;6:253–255. doi: 10.1016/s0966-842x(98)01308-0. [DOI] [PubMed] [Google Scholar]
- 9.Swanson M S, Isberg R R. Infect Immun. 1996;64:2585–2594. doi: 10.1128/iai.64.7.2585-2594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winn W C, Jr, Davis G S, Gump D W, Craighead J E, Beaty H N. Lab Invest. 1982;47:568–578. [PubMed] [Google Scholar]
- 11.Moffat J F, Edelstein P H, Regula D P, Jr, Cirillo J D, Tompkins L S. Mol Microbiol. 1994;12:693–705. doi: 10.1111/j.1365-2958.1994.tb01057.x. [DOI] [PubMed] [Google Scholar]
- 12.Chiang S L, Mekalanos J J. Mol Microbiol. 1998;27:797–805. doi: 10.1046/j.1365-2958.1998.00726.x. [DOI] [PubMed] [Google Scholar]
- 13.Mei J M, Nourbakhsh F, Ford C W, Holden D W. Mol Microbiol. 1997;26:399–407. doi: 10.1046/j.1365-2958.1997.5911966.x. [DOI] [PubMed] [Google Scholar]
- 14.Coulter S N, Schwan W R, Ng E Y W, Langhorne M H, Ritchie H D, Westbrock-Wadman S, Hufnagle W O, Folger K R, Bayer A S, Stover C K. Mol Microbiol. 1998;30:393–404. doi: 10.1046/j.1365-2958.1998.01075.x. [DOI] [PubMed] [Google Scholar]
- 15.Polissi A, Pontiggia A, Feger G, Altieri M, Mottl H, Ferrari L, Simon D. Infect Immun. 1998;66:5620–5629. doi: 10.1128/iai.66.12.5620-5629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 17.Cianciotto N P, Eisenstein B I, Mody C H, Engleberg N C. J Infect Dis. 1990;162:121–126. doi: 10.1093/infdis/162.1.121. [DOI] [PubMed] [Google Scholar]
- 18.Cirillo J D, Falkow S, Tompkins L S. Infect Immun. 1994;62:3254–3261. doi: 10.1128/iai.62.8.3254-3261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edelstein P H, Edelstein M A C. J Clin Microbiol. 1993;31:3329–3330. doi: 10.1128/jcm.31.12.3329-3330.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donnenberg M S, Kaper J B. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derbyshire K M. Gene. 1995;165:143–144. doi: 10.1016/0378-1119(95)00512-5. [DOI] [PubMed] [Google Scholar]
- 22.Wiater L A, Sadosky A B, Shuman H A. Mol Microbiol. 1994;11:641–653. doi: 10.1111/j.1365-2958.1994.tb00343.x. [DOI] [PubMed] [Google Scholar]
- 23.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- 24.Edelstein P H, Calarco K, Yasui V K. Am Rev Respir Dis. 1984;130:849–856. doi: 10.1164/arrd.1984.130.5.849. [DOI] [PubMed] [Google Scholar]
- 25.Mintz C S, Chen J X, Shuman H A. Infect Immun. 1988;56:1449–1455. doi: 10.1128/iai.56.6.1449-1455.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edelstein P H, Edelstein M A C. Epidemiol Infect. 1993;111:499–502. doi: 10.1017/s095026880005723x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cianciotto N, Eisenstein B I, Engleberg N C, Shuman H. Mol Biol Med. 1989;6:409–424. [PubMed] [Google Scholar]
- 28.Hacker J, Ott M, Wintermeyer E, Ludwig B, Fischer G. Int J Med Microbiol Virol Parasitol Infect Dis. 1993;278:348–358. doi: 10.1016/s0934-8840(11)80851-0. [DOI] [PubMed] [Google Scholar]
- 29.Isberg R R, Rankin S, Roy C R, Swanson M S, Berger K H. Infect Agents Dis. 1993;2:220–223. [PubMed] [Google Scholar]
- 30.Pope C D, O’Connell W, Cianciotto N P. Infect Immun. 1996;64:629–636. doi: 10.1128/iai.64.2.629-636.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berger K H, Merriam J J, Isberg R R. Mol Microbiol. 1994;14:809–822. doi: 10.1111/j.1365-2958.1994.tb01317.x. [DOI] [PubMed] [Google Scholar]
- 32.Cianciotto N P, Eisenstein B I, Mody C H, Toews G B, Engleberg N C. Infect Immun. 1989;57:1255–1262. doi: 10.1128/iai.57.4.1255-1262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horwitz M A, Silverstein S C. J Exp Med. 1981;153:386–397. doi: 10.1084/jem.153.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinn F D, Weinberg E D. Curr Microbiol. 1988;17:111–116. [Google Scholar]
- 35.Fitzgeorge R B, Featherstone A S, Baskerville A. Br J Exp Pathol. 1988;69:105–112. [PMC free article] [PubMed] [Google Scholar]
- 36.Mody C H, Paine R, Shahrabadi M S, Simon R H, Pearlman E, Eisenstein B I, Toews G B. J Infect Dis. 1993;167:1138–1145. doi: 10.1093/infdis/167.5.1138. [DOI] [PubMed] [Google Scholar]
- 37.Blander S J, Szeto L, Shuman H A, Horwitz M A. J Clin Invest. 1990;86:817–824. doi: 10.1172/JCI114779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mintz C S, Miller R D, Gutgsell N S, Malek T. Infect Immun. 1993;61:3416–3421. doi: 10.1128/iai.61.8.3416-3421.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szeto L, Shuman H A. Infect Immun. 1990;58:2585–2592. doi: 10.1128/iai.58.8.2585-2592.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dennis P J, Lee J V. J Appl Bacteriol. 1988;65:135–141. doi: 10.1111/j.1365-2672.1988.tb01501.x. [DOI] [PubMed] [Google Scholar]
- 41.Brieland J, Mcclain M, Legendre M, Engleberg C. Infect Immun. 1997;65:4892–4896. doi: 10.1128/iai.65.11.4892-4896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lüneberg E, Zähringer U, Knirel Y A, Steinmann D, Hartmann M, Steinmetz I, Rohde M, Köhl J, Frosch M. J Exp Med. 1998;188:49–60. doi: 10.1084/jem.188.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Byrne B, Swanson M S. Infect Immun. 1998;66:3029–3034. doi: 10.1128/iai.66.7.3029-3034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liles M R, Viswanathan V K, Cianciotto N P. Infect Immun. 1998;66:1776–1782. doi: 10.1128/iai.66.4.1776-1782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]