Abstract
Comparative genomic analysis at the genetic-map level has shown extensive conservation of the gene order between the different grass genomes in many chromosomal regions. However, little is known about the gene organization in grass genomes at the microlevel. Comparison of gene-coding regions between maize, rice, and sorghum showed that the distance between the genes is correlated with the genome size. We have investigated the microcolinearity at Lrk gene loci in the genomes of four grass species: wheat, barley, maize, and rice. The Lrk genes, which encode receptor-like kinases, were found to be consistently associated with another type of receptor-like kinase (Tak) on chromosome groups 1 and 3 in Triticeae and on chromosomes homoeologous to Triticeae group 3 in the other grass genomes. On Triticeae chromosome group 1, Tak and Lrk together with genes putatively encoding NBS/LRR proteins form a cluster of genes possibly involved in signal transduction. Comparison of the gene composition at orthologous Lrk loci in wheat, barley, and rice revealed a maximal gene density of one gene per 4–5 kb, very similar to the gene density in Arabidopsis thaliana. We conclude that small and large grass genomes contain regions that are highly enriched in genes with very little or no repetitive DNA. The comparison of the gene organization suggested various genome rearrangements during the evolution of the different grass species.
The extensive contiguous genomic sequencing of the dicot model species Arabidopsis thaliana (1) has shown that the gene density is of about one gene every 4–5 kb (2). However, A. thaliana has a small genome and low amount of repetitive DNA compared with other plant species with more complex genomes. Grass genomes are highly variable in size, ranging from diploid species with 415 Mb in rice to 16,000 Mb in the hexaploid wheat. The accumulation of repetitive sequences, which can represent up to 80%, such as in wheat (3), raises the question of the gene distribution in these complex genomes. Comparative mapping studies have established syntenic relationships between the different chromosome groups in the grass genomes (4). Most of these studies have been performed at the genetic map level, and little is known about the consequences of the evolutionary divergence on the gene composition and organization at the molecular level. Recent analysis of large, orthologous genomic fragments from maize, rice, and sorghum genomes demonstrated that the gene density varies in correlation with the genome size. Indeed, at the Sh2/A1 region, the intergenic regions were about 7 times longer in maize compared with the orthologous regions in rice and sorghum (5). At the Adh1/u22 locus, the genes were separated by more than 120 kb in maize, whereas the orthologous genes in sorghum were only 50 kb apart (6). Molecular analysis of 280 kb at the maize Adh1 locus demonstrated the presence of 10 retroelement families representing 60% of the DNA (7). These studies suggested that the discrepancy in the size of the intergenic regions in different grasses correlated with the accumulation of repeated elements between the genes in large genome species. Very little data are available on the gene distribution in large and complex genomes of barley and wheat. In barley, a gene density of one gene every 20 kb was found in the region surrounding the mlo gene (8). In wheat, Gill et al. (9) demonstrated by deletion mapping that the genes are distributed nonrandomly in gene-rich regions at the distal end of chromosome group 1. Recently, the analysis of a genomic fragment at a starch-branching enzyme I locus on chromosome 7D in Triticum tauschii (10) showed the presence of three tandemly repeated genes within 16 kb.
Here, we isolated and compared genomic fragments up to 23 kb in size surrounding the receptor-like kinase gene Lrk10 (11) on Triticeae homoeologous chromosome groups 1 and 3 in wheat, barley, maize, and rice. Comparison of the gene composition and organization at both chromosomal locations revealed at the microlevel the presence of gene-rich regions in the different grass genomes and indicated the emergence of different receptor-like kinase gene families during evolution. The finding of high-gene-density regions independent of the genome size is of importance for map-based cloning in large genomes.
MATERIALS AND METHODS
λ and Yeast Artificial Chromosome (YAC) Library Screening.
Genomic libraries from wheat (Triticum aestivum, var. ThatcherLr10) and barley (Hordeum vulgare, var. Igri) (Stratagene) were screened with the 32P-labeled pLrk10-A wheat probe (12) under high (65°C) stringency. The barley genomic library was also screened under high stringency with a 32P-labeled 610-bp HindIII/EcoRV fragment of the wheat clone 3ASLrk. Genomic libraries from rice (Oryza sativa, var. IR36) and maize (Zea mays, var. B73) (CLONTECH) were screened with the 32P-labeled pLrk10-A probe under low (50°C) stringency. A barley (H. vulgare, var. Franka) YAC library (MALTAGEN) was screened by PCR by using primers (5′-GGAAGTTAGCATGCTCGG-3′ and 5′-CATACCGCCAGGCATACAG-3′) designed in conserved sequences of the extracellular domain of the Hv1Lrk1 and Hv1Lrk2 genes. The YAC sublibrary was constructed in λZAPII (Stratagene) according to Whittaker and Rakesh (13). The library was screened with the 32P-labeled probe pLrk10-A as well as probes corresponding to the nucleotide-binding site (NBS) regions of HV1LRR1 and HV1LRR2. The NBS region of HV1LRR1 was obtained by PCR amplification on Hv1Lrr1 (AF108008) with the primers 5′-CACTCTTGTTGATCATGTGT-3′ and 5′-AAGCTGACCTTTAGGATC-3′. The NBS region of HV1LRR2 consisted of a EcoRI/NotI fragment of 1.4 kb isolated from Hv1Lrr2 (AF108010).
Sequence Analysis.
Sequencing reactions were performed with the Thermo Sequenase cycle sequencing kit (Amersham) according to the instructions of the manufacturer and subsequently were run on an automatic DNA sequencer. The sequences were analyzed with GCG software (Wisconsin Package), bcm genefinder software, and the blast program of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/).
Genetic Mapping.
Linkage analysis in wheat and barley was performed as described previously (14). Mapping in maize was performed on 56 F2 plants from a cross between the inbred lines Tx303 and CO159 at the University of Missouri–Columbia.
RESULTS
A Cluster of Receptor-Like Kinase Genes Is Located in a Gene-Rich Region on Wheat and Barley Chromosome Group 1.
To investigate the gene density and gene organization at the Lrk10 locus (11) in wheat, a genomic fragment of 13,887 bp (λW1AS) containing the Lrk10 gene was isolated and characterized. Two additional genes were found upstream from Lrk10 (Fig. 1a). At a 620-bp distance from the Lrk10 translation start site, a second gene encoding another receptor-like kinase (Tak10, U78762) starts in reverse orientation. The TAK10 protein showed high homology (68%) to LRK10 in the Ser/Thr kinase domain. In addition, the Tak10 gene contains two introns at positions similar to those of Lrk10, i.e., the first intron at the end of the extracellular domain and the second one within the transmembrane domain. No extensive homology was detected at the nucleotide and amino acid sequence levels between the extracellular domains, the transmembrane, and the charged domains of LRK10 and TAK10. However, a number of amino acids were conserved at similar positions between the two proteins. The best conservation was found in a stretch of 10 residues (GXYXVS/TAIXY) between positions 87 and 96 of the LRK10 sequence and positions 83 and 92 in TAK10. This suggests that Lrk10 and Tak10 have evolved from a common ancestor gene.
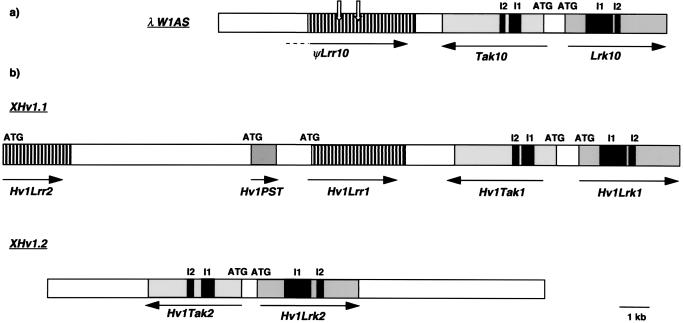
Figure 1.
Physical maps of the wheat and barley loci orthologous to Lrk10. (a) Wheat genomic fragment (λW1AS) containing the three genes, Lrk10, Tak10, and ψLrr10. (b) Two barley loci, XHv1.1 and XHv1.2, containing the Hv1Lrk1, Hv1Lrk2, Hv1Tak1, Hv1Tak2, Hv1Lrr1, and Hv1Lrr2 genes homologous to the wheat Lrk10, Tak10, and ψLrr10, respectively. The Hv1PST gene corresponds to an ORF of 226 aa detected by bcm genefinder. The solid arrows start at the putative translation start sites (ATG) and indicate the direction of transcription. The black boxes represent the introns (I1 and I2) found in the receptor-like kinase genes whereas the open arrows indicate interruptions in ORFs.
At 618 bp upstream from Tak10, we found a gene of 2,630 bp (ψLrr10) (Fig. 1a), which showed 50% homology at the nucleotide sequence level with RPM1, a resistance gene from A. thaliana against a bacterial disease (15). RPM1 belongs to the resistance gene class encoding intracellular NBS/LRR types of proteins. However, ψLrr10 (U76215) is probably a pseudogene because the coding sequence for the first 70 aa at the N-terminal sequence is lacking compared with RPM1. In addition, there is a frameshift mutation at position 943 and an in-frame stop codon at position 1327. Thus, within 14 kb, at the locus encoding the leaf rust-resistance gene Lr10 (11) on chromosome 1AS, there are two genes encoding receptor-like kinases and one pseudogene homologous to resistance genes.
To compare the conservation of gene organization between the Triticeae genomes, we isolated the corresponding region from barley. Under high stringency, the extracellular domain of Lrk10 (pLrk10-A) hybridized with two unique XbaI fragments that cosegregated on barley chromosome 1HS (14). We screened a barley genomic library and isolated fragments corresponding to the two loci (XHv1.1 and XHv1.2) detected by Southern hybridization (Fig. 1b). DNA stretches of 23 and 5 kb were sequenced at the XHv1.1 and XHv1.2 loci, respectively. At the XHv1.1 locus, we detected two genes, Hv1Lrk1 and Hv1Tak1, homologous to Lrk10 and Tak10, respectively. Similarly, two genes, Hv1Lrk2 and Hv1Tak2, were found at the XHv1.2 locus (Fig. 1b). The relative orientation of the Hv1Lrk and Hv1Tak genes was identical to the gene organization found on wheat chromosome 1AS (Fig. 1a), i.e., Hv1Lrk and Hv1Tak genes were found in opposite orientation (Fig. 1b). Comparison of both barley Hv1LRK1/Hv1LRK2 and Hv1TAK1/Hv1TAK2 proteins with the wheat LRK10 and TAK10, respectively, showed up to 79% homology at the amino acid level.
Three additional genes (Hv1Lrr1, Hv1Lrr2, and Hv1PST) were found at the XHv1.1 locus, upstream from Hv1Tak1 (Fig. 1b). Hv1Lrr1 and Hv1Lrr2 encode proteins with sequences characteristic for NBS/LRR proteins. Hv1Lrr1 had an ORF of 865 aa and showed 85% homology with ψLRR10 at the amino acid level. Hv1Lrr2, which had an ORF of 591 aa, was only 45% homologous to HV1LRR1 and showed the best homology with R6, a resistance gene analogue isolated by PCR from rice by Leister et al. (16). Because of the presence of two successive stop codons at position 1832, Hv1LRR2 had a shorter LRR domain compared with Hv1LRR1 and RPM1. Alternatively, Hv1Lrr2 could be a pseudogene because the 890 bp after the stop codons showed homology at the amino acid level to the LRR domains of Hv1LRR1 and RPM1. A genefinder analysis with the nucleotide sequence between Hv1Lrr1 and Hv1Lrr2 detected an ORF of 226 aa (Hv1PST), showing no homology with known proteins in the databases. A DNA stretch of 1.5 kb between Hv1PST and Hv1Lrr2 showed 50% homology with a putative reverse transcriptase of A. thaliana. These data demonstrated a very good conservation of the gene organization between wheat and barley in a region with a high gene density (five genes in 23 kb) in the distal part of the short arm of chromosome group 1.
The gene density was studied in an extended region around the XHv1 loci. A barley YAC library was screened by PCR by using primers corresponding to conserved sequences of the extracellular domain of Hv1Lrk1 and Hv1Lrk2. A YAC clone (Y102) of about 160 kb was isolated and analyzed. Restriction digests and subsequent hybridization with Lrk10 and Tak10 as probes demonstrated that the Hv1Lrk1, Hv1Tak1, Hv1Lrk2, and Hv1Tak2 genes were all present on this YAC. Moreover, PCR amplification products were obtained from Y102 DNA by using primers that were specific for either Hv1Lrk1 or Hv1Lrk2. To confirm these data at the sequence level, a λ library was constructed from Y102 DNA and screened with the pLrk10-A probe. Sequencing of both λ clones and specific PCR products confirmed that the Hv1Lrk1/Tak1 and Hv1Lrk2/Tak2 gene clusters were present on this fragment. Thus, the XHv1.1 and XHv1.2 loci are not separated from each other by more than 150 kb.
Southern hybridization of restricted Y102 DNA with probes corresponding to the NBS sequence derived from Hv1Lrr1 and Hv1Lrr2 suggested that several genes containing a NBS similar to Hv1Lrr1 and one gene containing a NBS similar to Hv1Lrr2 were present on the 160-kb fragment. Partial sequences of several genes (AF108011–AF108015) were obtained after isolation of clones either by screening the λ library with the Hv1Lrr1 NBS probe or by PCR amplification on YAC 102 DNA with primers encompassing this NBS region. Comparison at the nucleotide level of independent clones demonstrated the presence of at least five different genes containing NBS motives similar to Hv1Lrr1 in this region. These results show that a total of at least 11 genes are present in this barley genomic region of 160 kb corresponding to a gene density of at least 1 gene per 15 kb. Thus, the Lrk/Tak genes are located in a region of very high gene density on chromosome 1 in the Triticeae.
Lrk10-Related Genes Are Present on Homoeologous Chromosome Group 3 in the Triticeae and in the Homologous Chromosomes in Rice and Maize.
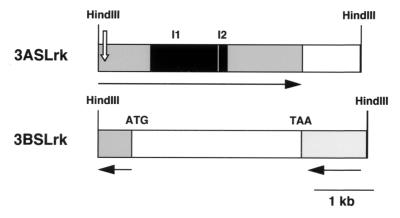
We have shown that Lrk10 belongs to a gene family that is located on group 1 chromosomes in wheat. Under high stringency, hybridization with the extracellular domain of Lrk10 detected only fragments located on chromosomes 1A, 1B, and 1D (12). However, when the kinase domain of Lrk10 was hybridized at high stringency or the extracellular domain at low stringency, we detected hybridizing fragments on chromosomes 3A and 3B (14). We isolated and characterized two wheat genomic sequences (3ASLrk and 3BSLrk) on group 3 chromosomes. Linkage analysis in wheat demonstrated that 3ASLrk and 3BSLrk were located on chromosomes 3A and 3B, respectively. Sequencing analysis showed that 3ASLrk contained a Lrk type of gene with the same intron/exon organization as Lrk10 (Fig. 2). When compared with the Lrk10 sequence, an insertion of 8 bp (AATAATAG), which could correspond to a transposon footprint, introduced a frameshift after amino acid 34. However, an in-frame ATG codon after the frameshift could be an alternative translation start site leading to a LRK protein without signal peptide. The 3BSLrk fragment contains, at one end, the last 220 aa of a Ser/Thr kinase domain of the TAK type and, at the other end, the first 130 aa of an extracellular domain of the LRK type (Fig. 2). This suggests that both Lrk and Tak types of genes are present on wheat chromosome group 3.
Figure 2.
Physical maps of two genomic fragments (3ASLrk and 3BSLrk) located in wheat chromosome group 3. The black boxes show the two introns (I1 and I2) of 3ASLrk. The solid arrows indicate the direction of the transcription. The open arrow indicates a frameshift in the ORF of the 3ASLrk sequence. ATG and TAA indicate the translation start site and the stop codon of the Lrk and Tak types of genes found on 3BSLrk, respectively.
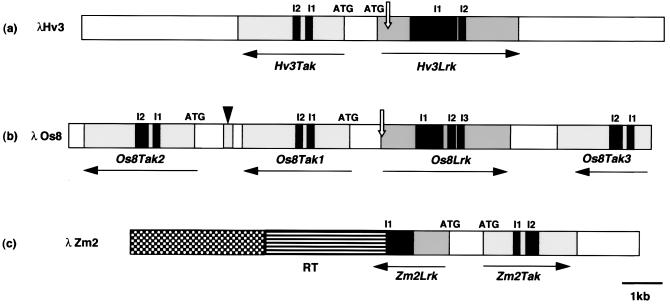
To compare the gene composition and organization at orthologous loci on the Triticeae chromosome group 3, the corresponding region on barley chromosome 3HS was analyzed. A gene of the Lrk type (Hv3Lrk) was found in opposite orientation to a gene of the Tak type (Hv3Tak) (Fig. 3a). A frameshift resulting from a deletion event occurred at the N-terminal sequence of Hv3Lrk at amino acid 35, i.e., at a very similar but not identical position as in the 3ASLrk gene in wheat. The intron/exon positions were identical to the Lrk10 type. In Hv3Tak, only the positions of exons 1 and 3 were well conserved.
Figure 3.
Physical maps of the genetic loci orthologous to wheat chromosome group 3 from barley (a), rice (b), and maize (c). The solid arrows start at the translation start site (ATG) and indicate the orientation of the transcription. The open arrows indicate the frameshift in the Hv3Lrk sequence and the point mutation at the Os8Lrk translation start site. The black boxes indicate the introns (I1, I2, and I3). The arrowhead indicates the 184-bp partial kinase sequence found between Os8Tak1 and Os8Tak2. The hatched box indicates the location of the retrotransposon (RT) homologous to colonist 1 in intron 1 of Zm2Lrk1. The dotted box indicates the sequence showing 70% homology with the maize 22-kDa zein cluster.
To study the conservation of the Lrk/Tak locus in other grass genomes, the orthologous loci were isolated from maize and rice. On a rice genomic fragment, (λOs8, 14,185 bp), three genes (Os8Tak1, Os8Tak2, and Os8Tak3) homologous to Tak10 were found as well as one gene (Os8Lrk) homologous to Lrk10 (Fig. 3b). The distance between the different genes did not exceed 2 kb. When compared with the sequence of Lrk10, a point mutation was detected at the translation start (AAG) site of Os8Lrk. Analysis of the three Os8Tak genes showed similar size and spacing for exons 1 and exon 3 compared with Tak10. A short, isolated sequence of 184 bp located between the Os8Tak1 and the Os8Tak2 genes was 92% identical to the kinase domain VII of Os8Tak2 between positions 3,334 and 3,518. This suggests that rearrangements occurred in this region. This very high gene density was confirmed by the partial analysis of other rice λ clones (data not shown). This result confirms the Southern hybridization data that indicated the presence of a large gene family of receptor-like kinase related to Lrk10 at the distal end of chromosome 1 in rice (14). Interestingly, this region has been shown to be syntenic to the chromosome group 3S of the Triticeae (17).
On the maize genomic fragment, two genes (Zm2Lrk and Zm2Tak) homologous to Lrk10 and Tak10 were found (Fig. 3c). A retrotransposon (RT) showing 98% identity at the nucleotide level with the maize RT colonist1 interrupted the sequence within the first intron of Zm2Lrk. Downstream of the retrotransposon, the sequence showed 70% homology with the noncoding region of the 22-kDa zein cluster described by Llaca and Messing (18). Genetic mapping of Zm2Lrk showed that two loci cosegregated on the short arm of chromosome 8. This region (bin 8.02) has been shown to be homologous to Triticeae chromosome group 3 (19). Thus, the Lrk- and Tak-related genes that were isolated here from maize and rice are located in chromosomal regions homologous to group 3 of the Triticeae. No orthologous genes of the group 1 loci in the Triticeae were found in maize or rice, either by genetic mapping or by gene isolation.
Comparison of the Orthologous Lrk/Tak Clusters Suggested Various Rearrangements During the Evolution in the Different Grass Genomes.
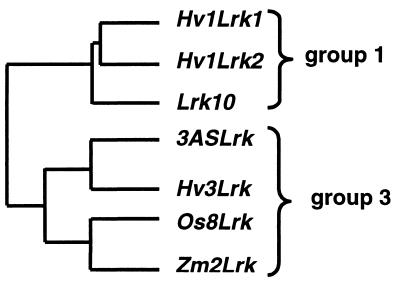
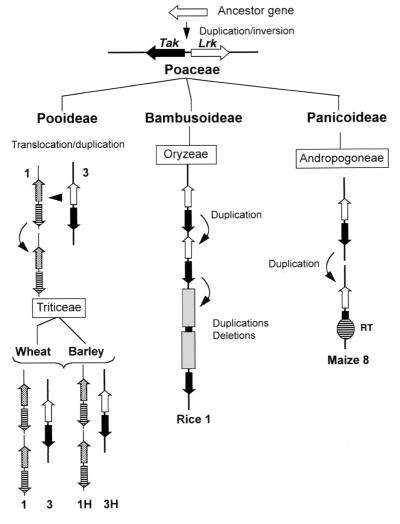
Comparison of the extracellular domains of the Lrk genes located on chromosome group 3 (3ASLrk) and group 1 (Lrk10) in wheat showed about 48% homology at the amino acid level. Similarly, in barley Hv3Lrk (on chromosome 3HS) and Hv1Lrk1/Hv1Lrk2 (on chromosome 1HS) are 50% homologous at the amino acid level. In contrast, there is about 72% homology between Lrk10 from wheat and Hv1Lrk1/Hv1Lrk2 from barley, which are located on chromosome 1AS and 1HS, respectively. Likewise, there is 78% homology between 3ASLrk from wheat and Hv3Lrk from barley. The same comparison was made with the Lrk genes of maize and rice, which are located on chromosomes corresponding to group 3 of the Triticeae. The best homology (59%) was found with the wheat and barley Lrk genes of group 3. Comparison of all the sequences resulted in two clearly distinct groups of genes corresponding to the two different chromosomal locations on Triticeae chromosome groups 1 and 3 (Fig. 4). These results confirmed the orthologous relationships established by cross-genome restriction fragment length polymorphism analysis between the different grass chromosomes at the sequence level. A model of the origin and the evolution of the Lrk/Tak clusters in the Poaceae family is given in Fig. 5. Lrk and Tak would have arisen from a common ancestor gene by duplication followed by an inversion on a chromosome homologous to the actual Triticeae group 3. The cluster then evolved by differential rearrangements in the different subfamilies. In the Pooideae subfamily, a duplication followed by a translocation occurred between segments of chromosomes 3 and 1 leading to the Lrk/Tak clusters found on the Triticeae chromosome groups 3 and 1. In maize, the detection of two loci on chromosome 8 also suggested duplication whereas the gene organization in rice indicated that several duplications and rearrangements occurred possibly because of unequal crossing over.
Figure 4.
Amino acid sequence comparison of Lrk gene products isolated from barley (Hv1Lrk1, Hv1Lrk2, and Hv3Lrk), wheat (Lrk10 and 3ASLrk), rice (Os8Lrk), and maize (Zm2Lrk). The extracellular domains of genes related to wheat gene Lrk10 showed two groups of homology corresponding to the Triticeae-homologous chromosome groups 1 and 3.
Figure 5.
Schematic representation of the putative evolution of the receptor-like kinase gene families Lrk and Tak in grass genomes. RT, retrotransposon; shaded boxes represent rearrangements such as deletions by unequal crossing over.
DISCUSSION
Organization and Gene Density at Receptor-Like Kinase Gene Clusters in Grass Genomes.
On Triticeae chromosome group 1, we identified and analyzed a high-gene-density region with four genes encoding receptor-like kinases (Lrk/Tak) and six or more genes encoding sequences that are characteristic for NBS and LRR. The molecular characterization of a gene-rich region at the distal end of chromosome group 1 confirms the conclusion of Gill et al. (9) from cytogenetic analysis in wheat. The authors identified small regions of high marker density interspersed by large chromosomal regions that were poor in markers. Two major marker clusters were defined on the short arm of chromosome group 1. One of them (FL 0.85) containing several genes of agronomic interest encompasses the genomic regions that were analyzed in this study. By sequencing wheat and barley fragments isolated from this region we found that the distance between the genes ranged from 450 bp to a maximum of 7 kb. Interestingly, Rahman et al. (10) found three genes within 16 kb encoding starch-branching enzymes I on chromosome 7DS of T. tauschii. However, these genes were tandemly repeated sequences. In the rice cluster on chromosome 1, the maximal distance found between the receptor-like kinase genes was 2 kb. The gene density observed around the Lrk10 gene and its homologues is considerably higher than what has been found in rice, maize, and sorghum at the Adh1/u22 and Sh2/A1 loci (5, 6, 20). At these loci, the genes were separated from each other by 20- to 140-kb regions of noncoding and/or repeated sequences. In barley, Panstruga et al. (8) found a gene density of one gene per 20 kb around the mlo gene. In this study, we found regions with a gene density of one gene every 4–5 kb similar to the gene density observed in A. thaliana (2). Bennetzen et al. (6) suggested that the length of the intergenic regions is correlated with the genome size, i.e., larger genomes would have accumulated noncoding sequences between the single-copy genes. Despite the genome size difference between rice (415 Mbp), barley (5,000 Mbp), and wheat (16,000 Mbp), the intergenic regions at the Lrk loci were of comparable length. In addition, we did not detect extensive repeated structures in the vicinity of the genes characterized in the different genomic regions. Interestingly, in rice, sorghum (5), and barley (8), the analysis of the intergenic regions also showed the presence of very few retroelements as compared with their abundance in maize (7). This might indicate that the dynamic structure of the maize genome with its active transposable element system is very distinct from the genomes of other grass species.
Genes and Gene Families at the Lrk/Tak Loci in Grass Genomes.
We analyzed and compared the gene composition at loci of the receptor-like kinase gene family Lrk (12) in wheat, barley, rice, and maize. In all cases, a second, distinct type of receptor-like kinase (Tak, Triticum aestivum kinase) was found in association with the Lrk type of genes. Moreover, the relative position and orientation of both genes were identical for all the species, i.e., the Lrk genes were always found in opposite orientation to the Tak genes. The physical distance between the genes varied between 450 bp and 1.2 kb, suggesting that both genes share a bidirectional promoter. The conservation of the gene organization of Lrk and Tak during the evolution without any obvious rearrangement raises the question of whether there is a selection pressure to maintain this organization as a functional unit. The clustering of different genes that are involved in specific signal transduction pathways already has been reported at several disease-resistance loci in plants. For example, the Ml-a12 gene and a modifier gene of its action are linked closely in barley (21). There is also a tight linkage between the Pto and Pfr genes in tomato (22). However, in the examples mentioned above, the genes were physically separated by some kilobases and encoded different protein types. To our knowledge, Lrk and Tak genes represent the first example of a tight association of receptor-like kinase genes in plants.
On the distal end of the Triticeae chromosome group 1, receptor-like kinase genes are found in the vicinity of genes that putatively encode proteins containing NBS and LRR. Moreover, the analysis of a large genomic region in barley suggested the presence of several NBS/LRR genes. Although we cannot ensure yet that they are all functional, the finding of a number of genes that possibly are involved in signal transduction in the vicinity of disease-resistance loci (Lr10/Pm3 in wheat; Ml-a alleles in barley) is highly interesting. In regard to the studies of disease-resistance gene loci, the type of genes (receptor-like kinase- and LRR-containing genes) that are associated here is intriguing. Indeed, two different types of genes, i.e., genes having a kinase domain and/or a leucine-rich repeat domain, have been found at disease-resistance loci (23), and the question of whether both components are necessary for resistance expression is still open (24). In the case of the resistance to Pseudomonas syringae in tomato, it is clear that both the Pto kinase and the Pfr NBS/LRR genes are necessary to express a specific resistance reaction (22).
Molecular Evolution at the Lrk/Tak Receptor-Like Kinase in Grass Genomes.
Lrk/Tak gene clusters were found on homologous chromosomes of the Triticeae group 3 in the four grass species examined whereas the locus on chromosome group 1 was found only in barley and wheat. This suggests that the emergence of the group 1 Lrk/Tak cluster occurred after the Triticeae speciation, about 10 millions years ago, relatively late in the evolution of the Poaceae family. Comparison of the gene organization and gene composition at both chromosomal locations between the different species showed that there is more homology between the genes located on homoeologous chromosomes in different plant species than between the genes located on different chromosomes within the same species. This confirms the hypothesis of a common ancestor with Lrk/Tak genes on chromosome group 3, which led, during the evolution, to the orthologous genes that were detected here on wheat chromosome group 3, barley chromosome 3H, rice chromosome 1, and maize chromosome 8. The conservation of several residues between the extracellular domain of LRK and TAK as well as the identical position of their intron suggested that both receptor-like kinase genes have arisen from a common ancestor gene. The evolutionary scheme we propose (Fig. 5) starts with the duplication of an ancestral, receptor-like kinase gene followed by an inversion in the ancestor species of the Poaceae family. Future work on the isolation of Lrk/Tak gene homologues from other monocot and dicot species should allow checking this hypothesis and should estimate at which time in the evolution this duplication occurred. If the Lrk and Tak genes then have descended side by side during the evolution, they could be defined as paralogs (25).
Duplications have been identified already between different chromosomes in grass species. Short fragments of a few centimorgans (26), large segments such as chromosome arms (27), or whole genomes such as in maize (28) were duplicated. The possible origin of the duplication of the Lrk/Tak cluster from chromosome group 3 to chromosome group 1 in the Triticeae species is highly interesting. On the circular alignment of the grass genome maps proposed by Devos and Gale (4), the distal ends of the short arms of Triticeae chromosome groups 1 and 3 face each other, similar to rice chromosomes 1 and 5 and the sorghum chromosomes III and V. In maize, the corresponding regions span chromosome 8. Interestingly, a duplication of a region spanning about 27 cM detected by Kishimoto et al. (29) between rice chromosomes 1 and 5 also was found in sorghum chromosomes III and V (30). Kishimoto et al. (29) suggested an ancestral relationship (homoeology) between the two chromosomes. In the case of the Lrk/Tak cluster we suggest that in the Poaceae ancestor, the cluster might have been duplicated on the ancestor chromosome group 3 (based on the Triticeae nomenclature). This situation remained in maize and in rice whereas in the Triticeae, a translocation occurred between chromosomes 3 and 1 (Fig. 5). During the evolution of rice, the cluster was duplicated several times and probably rearranged by unequal crossing over, as indicated by the presence of a partial kinase sequence between the Os8Tak1 and Os8Tak2 genes.
Genome conservation at the submegabase level is the basis for the use of model species in cross-genome map-based cloning strategies. The use of rice as a reference genome in the isolation of genes from barley (31, 32) and wheat (33) has shown the limits of colinearity with the presence of internal rearrangements within colinear regions. Together with the data obtained from the molecular analysis of large genomic regions in maize, rice, and sorghum, our results argue in favor of using a model species as related as possible to the species of interest. In this regard, barley represents a better reference genome for wheat than rice. Additional comparative studies at the molecular level between grass genomes will provide new insight into gene conservation and genome evolution, allowing to use the best model species for cross-genome map-based cloning.
Acknowledgments
We are very grateful to Dr. T. Musket (University of Missouri–Columbia) and Dr. A. Graner (IPK-Gatersleben, Germany) for their collaboration in linkage analysis in maize and barley, respectively. This project was supported by the Swiss Priority Programme Biotechnology Grant 5002-45033 and, within the European Union Biotechnology Project (The European Gramineae Mapping Programme), by the Bundesamt für Bildung und Wissenschaft, Grant BBW 97.0208.
ABBREVIATIONS
- NBS
nucleotide-binding site
- LRR
leucine-rich repeats
- YAC
yeast artificial chromosome
Footnotes
References
- 1.Rounsley S, Lin X, Ketchum K A. Curr Opin Plant Biol. 1998;1:136–141. doi: 10.1016/s1369-5266(98)80015-0. [DOI] [PubMed] [Google Scholar]
- 2.Bevan M, Bancroft I, Bent E, Love K, Goodman H, Dean C, Bergkam R, Dirkse W, van Steveran M, Stikema W, et al. Nature (London) 1998;391:485–488. doi: 10.1038/35140. [DOI] [PubMed] [Google Scholar]
- 3.Smith D B, Flavell R B. Chromosoma. 1975;50:223–242. [Google Scholar]
- 4.Devos K M, Gale M D. Plant Mol Biol. 1997;35:3–15. [PubMed] [Google Scholar]
- 5.Chen M, SanMiguel P, de Oliveira A C, Woo S-S, Zhang H, Wing R A, Bennetzen J L. Proc Natl Acad Sci USA. 1997;94:3431–3435. doi: 10.1073/pnas.94.7.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennetzen J L, SanMiguel P, Chen M, Tikhonov A, Francki M, Avramova Z. Proc Natl Acad Sci USA. 1998;95:1975–1978. doi: 10.1073/pnas.95.5.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.SanMiguel P, Tikhonov A, Young-Kwan J, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, et al. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 8.Panstruga R, Büschges R, Piffanelli P, Schulze-Lefert P. Nucleic Acids Res. 1998;26:1056–1062. doi: 10.1093/nar/26.4.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gill K S, Gill B S, Endo T R, Taylor T. Genetics. 1996;144:1883–1891. doi: 10.1093/genetics/144.4.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahman S, Abrahams S, Abbott D, Mukai Y, Samuel M, Morell M, Appels R. Genome. 1997;40:465–474. doi: 10.1139/g97-062. [DOI] [PubMed] [Google Scholar]
- 11.Feuillet C, Schachermayr G, Keller B. Plant J. 1997;11:45–52. doi: 10.1046/j.1365-313x.1997.11010045.x. [DOI] [PubMed] [Google Scholar]
- 12.Feuillet C, Reuzeau C, Kjellbom P, Keller B. Plant Mol Biol. 1998;37:943–953. doi: 10.1023/a:1006062016593. [DOI] [PubMed] [Google Scholar]
- 13.Whittaker P A, Rakesh A. In: Pulse Field Gel Electrophoresis, A Practical Approach. Monaco A P, editor. Oxford: IRL; 1995. pp. 69–94. [Google Scholar]
- 14.Gallego F, Feuillet C, Messmer M, Penger A, Graner A, Yano M, Sasaki M, Keller B. Genome. 1998;41:373–380. doi: 10.1139/g98-024. [DOI] [PubMed] [Google Scholar]
- 15.Grant M R, Godiart L, Straube E, Ashfield T, Lewald J, Sattler A, Innes R W, Dangl J L. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- 16.Leister D, Kurth J, Laurie D A, Yano M, Sasaki T, Devos K, Graner A, Schulze-Lefert P. Proc Natl Acad Sci USA. 1998;95:370–375. doi: 10.1073/pnas.95.1.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn S, Anderson J A, Sorrells M E, Tanksley S D. Mol Gen Genet. 1993;241:483–490. doi: 10.1007/BF00279889. [DOI] [PubMed] [Google Scholar]
- 18.Llaca V, Messing J. Plant J. 1998;15:211–220. doi: 10.1046/j.1365-313x.1998.00200.x. [DOI] [PubMed] [Google Scholar]
- 19.Van Deynze A E, Nelson J C, Yglesias E S, Harrington S E, Braga D P, McCouch S R, Sorrells M E. Mol Gen Genet. 1995;248:744–754. doi: 10.1007/BF02191715. [DOI] [PubMed] [Google Scholar]
- 20.Avramova Z, Tikhonov A, SanMiguel P, Jin Y K, Liu C, Woo S S, Wing R, Bennetzen J L. Plant J. 1996;10:1163–1168. doi: 10.1046/j.1365-313x.1996.10061163.x. [DOI] [PubMed] [Google Scholar]
- 21.Jorgensen J H. Genome. 1988;30:129–132. [Google Scholar]
- 22.Salmeron J M, Barker S J, Carland F M, Mehta A Y, Staskawicz B J. Plant Cell. 1994;6:511–520. doi: 10.1105/tpc.6.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammond-Kosak K E, Jones J D G. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- 24.Blumwald E, Aharon G S, Lam B C-H. Trends Plant Sci. 1998;3:342–346. [Google Scholar]
- 25.Huyen M J, Bork P. Proc Natl Acad Sci USA. 1998;95:5849–5856. doi: 10.1073/pnas.95.11.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.San-Alferez S, Richter T E, Hulbert S H, Bennetzen J L. Theor Appl Genet. 1995;91:25–32. doi: 10.1007/BF00220854. [DOI] [PubMed] [Google Scholar]
- 27.Moore G, Devos K M, Wang Z, Gale M D. Curr Biol. 1995;5:737–739. doi: 10.1016/s0960-9822(95)00148-5. [DOI] [PubMed] [Google Scholar]
- 28.White S, Doebley J. Trends Genet. 1998;14:327–332. doi: 10.1016/s0168-9525(98)01524-8. [DOI] [PubMed] [Google Scholar]
- 29.Kishimoto N, Higo H, Abe K, Arai S, Saito A, Higo K. Theor Appl Genet. 1994;88:722–726. doi: 10.1007/BF01253976. [DOI] [PubMed] [Google Scholar]
- 30.Devos K M, Wang Z M, Beales J, Sasaki T, Gale M D. Theor Appl Genet. 1998;96:63–68. [Google Scholar]
- 31.Killian A, Kudrna D A, Kleinhofs A, Yano M, Kurata N, Steffenson B, Sasaki T. Nucleic Acids Res. 1995;23:2729–2733. doi: 10.1093/nar/23.14.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunford R P, Kurata N, Laurie D A, Money T A, Minobe Y, Moore G. Nucleic Acids Res. 1995;23:2724–2728. doi: 10.1093/nar/23.14.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foote T, Roberts M, Kurata N, Sasaki T, Moore G. Genetics. 1997;147:801–807. doi: 10.1093/genetics/147.2.801. [DOI] [PMC free article] [PubMed] [Google Scholar]