The crystallization and preliminary X-ray crystallographic studies of the N-terminal domain of FadD28, a fatty-acyl AMP ligase from M. tuberculosis, are reported.
Keywords: FadD28, fatty-acyl AMP ligase (FAAL), phthiocerol dimycocerosate (PDIM), Mycobacterium tuberculosis
Abstract
FadD28 from Mycobacterium tuberculosis belongs to the fatty-acyl AMP ligase (FAAL) family of proteins. It is essential for the biosynthesis of a virulent phthiocerol dimycocerosate (PDIM) lipid that is only found in the cell wall of pathogenic mycobacteria. The N-terminal domain, comprising of the first 460 residues, was crystallized by the hanging-drop vapour-diffusion method at 295 K. The crystals belong to space group P212121, with unit-cell parameters a = 50.97, b = 60.74, c = 136.54 Å. The crystal structure of the N-terminal domain of FadD28 at 2.35 Å resolution has been solved using the MAD method.
1. Introduction
Mycobacterium tuberculosis is known to have the highest annual global mortality of all the bacterial pathogens. One-third of the world population has latent M. tuberculosis infection and 5–10% of them develop active tuberculosis at some time during their lives (World Health Organization, 2001 ▶). Although tuberculosis remains one of the leading causes of death worldwide, the mechanisms by which it establishes progressive disease are not well understood. Identification of the virulence factors of M. tuberculosis is of fundamental importance for the development of new vaccines and drugs against this pathogen. The genome sequence of the best characterized strain of M. tuberculosis, H37Rv, was made available in 1998. Analysis of the genome has revealed an array of fadD genes which activate long-chain fatty acids (Cole et al., 1998 ▶). After activation, these chains are transferred to the multifunctional polyketide synthases (PKSs) for further chain extension. In mycobacteria, PKSs in conjunction with fatty-acid synthases (FASs) have been implicated in generating diverse unusual lipids that constitute the complex cell wall of this organism (Brennan, 2003 ▶; Minnikin et al., 2002 ▶). We have undertaken a systematic study of the structure–function relationships of several PKSs and FASs from M. tuberculosis. One such newly identified PKS, which belongs to the chalcone synthase superfamily of proteins, is PKS18, which has an unusual specificity towards long-chain acyl CoA substrates (Saxena et al., 2003 ▶). We recently determined the crystal structure of PKS18 from M. tuberculosis, which revealed a novel tunnel through which metabolite diversity was generated (Sankaranarayanan et al., 2004 ▶). Our recent biochemical analysis of several FadD proteins showed that some of them activate fatty acids as acyl-adenylates and belongs to the fatty-acyl AMP ligase (FAAL) family, while the others activate them as acyl-CoAs and belong to the fatty-acyl CoA ligase (FACL) family (Trivedi et al., 2004 ▶). Modelling studies revealed that these proteins were comprised of large N-terminal and small C-terminal domains. FadD28, Rv2941, belongs to the FAAL family proteins and is essential for the biosynthesis of a virulent phthiocerol dimycocerosate (PDIM) lipid that is only found in the cell wall of slow-growing pathogenic mycobacteria (Azad et al., 1997 ▶; Cox et al., 1999 ▶; Camacho et al., 2001 ▶). Despite the presence of other functional FadD proteins and a plethora of acyl-CoAs in the bacteria, FadD28 knockout mutants of the mycobacterium lacked virulent PDIM lipid on the cell envelope (Azad et al., 1997 ▶; Cox et al., 1999 ▶; Camacho et al., 2001 ▶; Fitzmaurice & Kolattukudy, 1998 ▶).
Although the biochemical and gene-knockout studies have provided fundamental new insights into the functional importance of FadD28 protein, its structural and mechanistic aspects have not yet been investigated. Our efforts to crystallize the full-length 580-amino-acid protein were not successful. Therefore, we crystallized the N-terminal domain of FadD28 comprising of the first 460 residues (50.7 kDa). Although the enzymatic activity of the N-terminal domain of FadD28 was found to be compromised when compared with full-length protein owing to the absence of the C-terminal domain, all the residues determining the substrate specificity lie in N-terminal domain. Structural studies would further our understanding of the essential role played by FadD28 in the biosynthesis of the virulent PDIM lipid. We present here the expression, purification, crystallization and the preliminary X-ray crystallographic analysis of the FadD28 N-terminal domain from M. tuberculosis.
2. Materials and methods
2.1. Expression and purification
The gene for the FadD28 N-terminal domain from M. tuberculosis (consisting of the first 460 amino-acid residues) was cloned into the expression vector pET28c (Novagen). The recombinant protein contains an additional 20 amino acids including a His6 tag (MGSSHHHHHHSSGLVPRGSH) at the N-terminus. The recombinant plasmid was transformed and overexpressed in Escherichia coli BL21 (DE3) strain cells. The l-selenomethionine-derivative protein was produced using the feedback-inhibition method (Doublié, 1997 ▶). Transformed cells were grown in 1× M9 minimal medium pH 7.4 at 303 K containing 100 µg ml−1 kanamycin and l-selenomethionine at a concentration of 50 mg l−1. The culture was induced with 0.25 mM isopropyl β-d-thiogalactopyranoside (IPTG) at an OD of 0.6 and was grown for a further 10 h. The cells were harvested by centrifugation at 5000 rev min−1 for 30 min at 277 K. The pellet was resuspended in ice-cold lysis buffer containing 50 mM Tris–HCl buffer pH 8.0, 150 mM NaCl, 1 mM PMSF, 5 mM β-mercaptoethanol and 5 mM imidazole. Cells were lysed by sonication on ice and centrifuged at 20 000 rev min−1 for 30 min at 277 K to remove cell debris. The supernatant was applied onto an Ni–NTA column (Qiagen) pre-equilibrated with 50 mM Tris–HCl buffer pH 8.0, 150 mM NaCl, 5 mM β-mercaptoethanol and 5 mM imidazole. The target protein was eluted with buffer containing 50 mM Tris–HCl buffer pH 8.0, 5 mM β-mercaptoethanol and 50–100 mM imidazole. Fractions containing the protein were pooled and concentrated using an Amicon YM-30 membrane (Millipore) and loaded onto a HiLoad 16/60 Superdex-75 prep-grade gel-filtration column (Amersham Biosciences) pre-equilibrated with 20 mM Tris–HCl pH 8.0, 100 mM NaCl, 5 mM MgCl2, 5 mM β-mercaptoethanol and 1 mM EDTA. The purified protein was exchanged against a buffer containing 20 mM Tris–HCl pH 8.0, 20 mM NaCl, 5 mM MgCl2, 1 mM EDTA and 5 mM β-mercaptoethanol. The protein was concentrated to 10 mg ml−1 using an Amicon YM-30 membrane (Millipore) and stored at 277 K prior to crystallization.
2.2. Crystallization
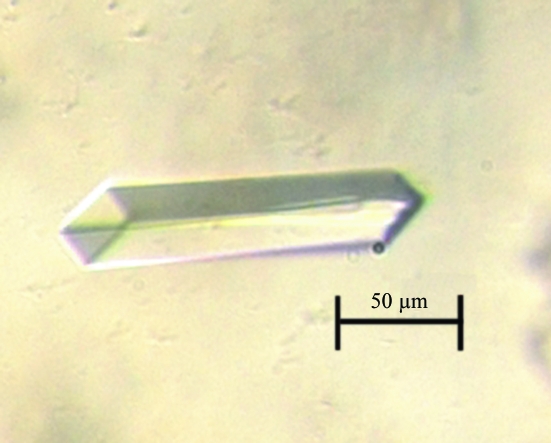
Crystallization was performed by the hanging-drop vapour-diffusion method at 277 and 295 K. Initial crystallization conditions were obtained using JBS (Jena Bioscience) screens. Further refinement of conditions at 295 K yielded well diffracting large rod-shaped crystals. Crystals grew to maximum dimensions of 0.15 × 0.03 × 0.02 mm over a period of 3 d (Fig. 1 ▶). A 3 µl droplet of 3 mg ml−1 protein solution mixed with the same amount of reservoir solution was equilibrated against 750 µl reservoir solution containing 20.3%(w/v) polyethylene glycol 3350, 0.1 M Na MES pH 6.0, 5 mM β-mercaptoethanol and 0.13 M lithium sulfate.
Figure 1.
Crystal of the FadD28 N-terminal domain from M. tuberculosis at 295 K.
2.3. Data collection and processing
At the in-house rotating-anode X-ray source, the crystals diffracted to 3.5 Å. The multiple-wavelength anomalous dispersion (MAD) data set was collected at the BW7A beamline, DESY synchrotron, EMBL, Hamburg, Germany using a MAR charged-coupled device detector. Prior to flash-freezing in a liquid-nitrogen stream at 100 K for data collection, crystals were soaked in mother liquor containing 15% glycerol for 90–120 s. Peak, inflection and remote data sets were collected at 0.9800, 0.9792 and 0.9776 Å, respectively. X-ray diffraction data sets were processed and analyzed using DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶).
3. Results
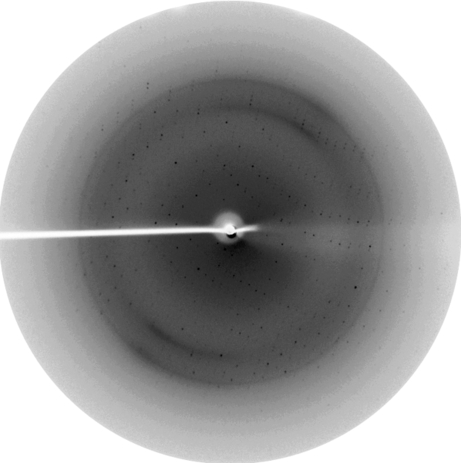
The protein was overexpressed in a soluble form in E. coli and purified to homogeneity using Ni–NTA and gel-filtration chromatographic techniques. Each litre of 1× M9 minimal medium yielded close to 10 mg of nearly 95% pure FadD28 N-terminal domain protein. The purified protein was crystallized by initially screening different conditions using Jena Bioscience screens and the best conditions were expanded using additives to obtain better crystals. Initial characterization was carried out at the in-house X-ray source (Fig. 2 ▶). A complete data set was collected at the BW7A beamline, Hamburg and the crystals diffracted to a resolution limit of 2.35 Å. Peak, inflection and remote data sets were collected with oscillations of 0.25° by rotating the crystal through an angle of 180, 180 and 162.5°, respectively. The X-ray diffraction data indicated that the crystals belong to space group P212121, with unit-cell parameters a = 50.97, b = 60.74, c = 136.54 Å, α = β = γ = 90°. The Matthews coefficient (V M = 2.0 Å3 Da−1) indicated the presence of one molecule in the asymmetric unit and a solvent content of about 37.8% (Matthews, 1968 ▶). The data-collection statistics are shown in Table 1 ▶. Calculation of the initial phases was performed with the program SOLVE with an overall experimental figure of merit of 0.49 at 2.35 Å resolution prior to density modification (Terwilliger & Berendzen, 1999 ▶). RESOLVE was used for density modification and automated model building and built around 240 residues out of the total 460 residues (Terwilliger, 2000 ▶, 2003 ▶).
Figure 2.
A diffraction image of a crystal of the N-terminal domain of FadD28 collected using the in-house X-ray system. The edge corresponds to 2.6 Å resolution.
Table 1. X-ray data-collection statistics.
Values in parentheses refer to the highest resolution shell.
| Inflection | Peak | Remote | |
|---|---|---|---|
| Wavelength (Å) | 0.9792 | 0.9800 | 0.9776 |
| Space group | P212121 | ||
| Resolution (Å) | 20.0–2.35 (2.39–2.35) | 20.0–2.35 (2.39–2.35) | 20.0–2.35 (2.39–2.35) |
| Unit-cell parameters (Å) | a = 50.97, b = 60.74, c = 136.54 | ||
| No. of observations | 124573 (4543) | 125245 (4570) | 113108 (4165) |
| Unique reflections | 17975 (826) | 18004 (831) | 18036 (850) |
| Completeness (%) | 99.3 (93.2) | 99.5 (93.8) | 99.7 (95.9) |
| Rmerge† (%) | 7.7 (41.4) | 9.3 (43.4) | 8.4 (42.7) |
| Average I/σ(I) | 23.13 (3.54) | 19.72 (3.44) | 21.87 (3.36) |
| Redundancy | 6.9 (5.5) | 7.0 (5.5) | 6.3 (4.9) |
| Mosaicity | 0.75 | 0.75 | 0.70 |
| Monomers per ASU | 1 | ||
R
merge = 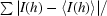
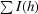 , where I(h) is the observed intensity and 〈I(h)〉 is the mean intensity of reflection h over all measurements of I(h).
, where I(h) is the observed intensity and 〈I(h)〉 is the mean intensity of reflection h over all measurements of I(h).
Acknowledgments
AG and ER thank the Council of Scientific and Industrial Research (CSIR), India for junior and senior research fellowships, respectively. RS is an International Senior Research Fellow (ISRF) of the Wellcome Trust UK in Biomedical Sciences in India. We also thank Dr Santosh Panjikar and the other staff of the synchrotron-radiation facility at EMBL, Hamburg for their help during data collection.
References
- Azad, A. K., Sirakova, T. D., Fernandes, N. D. & Kolattukudy, P. E. (1997). J. Biol. Chem.272, 16741–16745. [DOI] [PubMed] [Google Scholar]
- Brennan, P. J. (2003). Tuberculosis, 83, 91–97. [DOI] [PubMed] [Google Scholar]
- Camacho, L. R., Constant, P., Raynaud, C., Laneelle, M. A., Triccas, J. A., Gicquel, B., Daffe, M. & Guilhot, C. (2001). J. Biol. Chem.276, 19845–19854. [DOI] [PubMed] [Google Scholar]
- Cole, S. T. et al. (1998). Nature (London), 393, 537–544. [Google Scholar]
- Cox, J. S., Chen, B., McNeil, M. & Jacobs, W. R. Jr (1999). Nature (London), 402, 79–83. [DOI] [PubMed] [Google Scholar]
- Doublié, S. (1997). Methods Enzymol.276, 523–530. [PubMed] [Google Scholar]
- Fitzmaurice, A. M. & Kolattukudy, P. E. (1998). J. Biol. Chem.273, 8033–8039. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Minnikin, D. E., Kremer, L., Dover, L. G. & Besra, G. S. (2002). Chem. Biol.9, 545–553. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Sankaranarayanan, R., Saxena, P., Marathe, U. B., Gokhale, R. S., Shanmugam, V. M. & Rukmini, R. (2004). Nature Struct. Mol. Biol.11, 894–900. [DOI] [PubMed]
- Saxena, P., Yadav, G., Mohanty, D. & Gokhale, R. S. (2003). J. Biol. Chem.278, 44780–44790. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T. C. (2000). Acta Cryst. D56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T. C. (2003). Acta Cryst. D59, 38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi, O. A., Arora, P., Sridharan, V., Tickoo, R., Mohanty, D. & Gokhale, R. S. (2004). Nature (London), 428, 441–445. [DOI] [PubMed] [Google Scholar]
- World Health Organization (2001). WHO Report 2001. Global Tuberculosis Control. http://www.who.int/gtb/publications/glovrep01/.