The crystal structures of two crystal forms of manganese superoxide dismutase (Mn-SOD) from the radiation-resistant bacterium D. radiodurans are reported and compared with the crystal structure of Mn-SOD from E. coli.
Keywords: manganese superoxide dismutase, Deinococcus radiodurans, radiation resistance
Abstract
The structure of the manganese superoxide dismutase (Mn-SOD; DR1279) from Deinococcus radiodurans has been determined in two different crystal forms. Both crystal forms are monoclinic with space group P21. Form I has unit-cell parameters a = 44.28, b = 83.21, c = 59.52 Å, β = 110.18° and contains a homodimer in the asymmetric unit, with structure refinement (R = 16.8%, R free = 23.6%) carried out using data to d min = 2.2 Å. Form II has unit-cell parameters a = 43.57, b = 87.10, c = 116.42 Å, β = 92.1° and an asymmetric unit containing two Mn-SOD homodimers; structure refinement was effected to a resolution of 2.0 Å (R = 17.2%, R free = 22.3%). The resulting structures are compared with that of Mn-SOD from Escherichia coli, with which they are shown to be essentially isostructural.
1. Introduction
Deinococcus radiodurans (DEIRA) is an aerobic, non-pathogenic, red-pigmented Gram-positive bacterium that exhibits significant resistance to ionizing and ultraviolet radiation, desiccation and the presence of reactive oxygen species (ROS; Minton, 1994 ▶; Mattimore & Battista, 1996 ▶; Battista, 1997 ▶). The capability of DEIRA to withstand acute radiation doses of up to 1500 krad is intriguing and the bacterium has been the subject of intensive investigation in order to try to understand how this is possible. Exposure to such high doses of ionizing radiation causes massive damage to genomic DNA and a phenomenal capability of DEIRA to repair such damage, perhaps aided by a unique packaging of DNA in its cells, is thought to be responsible for the bacterium’s ability to recover its viability (Minton, 1994 ▶; Makarova et al., 2001 ▶; Levin-Zaidman et al., 2003 ▶). However, it should be noted that the damaging effects of ionizing radiation are enhanced in the presence of oxygen, as ROS such as the hydroxyl radical (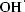 ), the superoxide radical (O
), the superoxide radical (O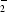 ), hydrogen peroxide (H2O2) and organic hydroperoxides which can also cause damage to cellular material are produced (Misra & Fridovich, 1976 ▶). Limiting the damage caused to DNA by ROS must therefore also be important to the ability of DEIRA to withstand the toxic effects of ionizing radiation.
), hydrogen peroxide (H2O2) and organic hydroperoxides which can also cause damage to cellular material are produced (Misra & Fridovich, 1976 ▶). Limiting the damage caused to DNA by ROS must therefore also be important to the ability of DEIRA to withstand the toxic effects of ionizing radiation.
As part of a structural genomics project focusing on the determination of the three-dimensional structures of gene products that may be important in contributing to the extremophile nature of DEIRA strain R1, we have targeted a number of enzymes involved in the neutralization of ROS. One such enzyme is the manganese-dependent superoxide dismutase (Mn-SOD; DR1279; EC 1.15.11), which catalyses the disproportionation of superoxide radicals to oxygen and hydrogen peroxide (Fridovitch, 1986 ▶).
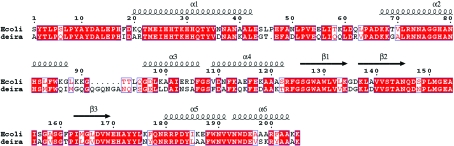
At first sight, Mn-SOD from DEIRA might seem to be a strange choice of target. After all, the three-dimensional structures and reaction mechanisms of Mn-SODs from other organisms (including that from Escherichia coli) have been extensively studied and characterized (Borgstahl et al., 2000 ▶; Edwards et al., 2001 ▶ and references therein). The amino-acid sequence of DEIRA Mn-SOD has (apart from a six-residue insert between residues 91 and 92) 57% identity (Fig. 1 ▶) with its homologue from E. coli and therefore its three-dimensional structure and mechanism of action must be very similar to those of E. coli Mn-SOD. However, Mn-SOD is highly (Chou & Tan, 1990 ▶) and apparently constitutively (Lipton et al., 2002 ▶) expressed by DEIRA and DEIRA mutants lacking Mn-SOD are much more sensitive to the effects of ionizing radiation than are both wild-type DEIRA or DEIRA mutants that do not express the catalase KatA (Markillie et al., 1999 ▶). This suggests that the neutralization of superoxide radicals is more important to the extremophile nature of DEIRA than the neutralization of hydrogen peroxide. Investigations of the structure of DEIRA Mn-SOD may provide structural reasons for the importance of Mn-SOD to the survival of DEIRA. We therefore now report the three-dimensional structure of Mn-SOD from DEIRA (Mn-SODDr) in two different crystal forms as determined using single-crystal X-ray diffraction techniques and, in particular, compare them with the crystal structure of the Mn-SOD from E. coli (Mn-SODEc; PDB code 1vew; Edwards et al., 1998 ▶).
Figure 1.
CLUSTAL W (Thompson et al., 1994 ▶) amino-acid sequence alignment of Mn-SODEc (top) and Mn-SODDr. Superimposed on these is a secondary-structure assignment as defined in Edwards et al. (1998 ▶). This figure was produced using ESPript (Gouet et al., 2003 ▶).
2. Materials and methods
2.1. Protein cloning, expression and purification
The Mn-SOD gene from D. radiodurans (DR1279) was amplified by Pfu DNA polymerase, subcloned into a Gateway pDest17 vector (Invitrogen) containing a 2.6 kDa N-terminal fusion tag that includes a hexahistidine region and transformed into BL21(DE3)pLysS E. coli cells (Novagen). One colony of bacteria was used to inoculate 10 ml of Luria–Bertani (LB) broth to which ampicillin (50 µg ml−1) and chloramphenicol (34 µg ml−1) had been added. An overnight culture was then grown at 310 K, shaking the flasks at ∼200 rev min−1. This was then centrifuged at 1500g for 10 min with the resulting pellet being resuspended in 10 ml 2×YT medium and used to inoculate 500 ml 2×YT medium supplemented with ampicillin (100 µg ml−1) and chloramphenicol (34 µg ml−1). This was incubated at 295 K until the cell density reached an OD600 of ∼0.6. The production of Mn-SODDr was induced with 1 mM isopropyl β-d-thiogalactopyranoside (IPTG) and cell growth continued for ∼16 h at 295 K. The cells were then harvested by centrifugation at 4500g for 20 min at 277 K. The pellet was resuspended in 10 ml of an ice-cold lysis buffer consisting of 100 mM Tris–HCl pH 7.5, 150 mM NaCl and 10 mM imidazole supplemented with 1 mM MgCl2, 10 µg ml−1 DNAseI, 100 µg ml−1 lysozyme and a Complete Mini EDTA-free cocktail of antiproteases (Roche). The cells were then sonicated on ice using five 30 s pulses and the cell debris was removed by centrifugation at 17 000g for 90 min at 277 K.
The resulting supernatant was applied onto a Hi-Trap column (Amersham Biosciences) previously loaded with 0.1 M nickel sulfate and pre-equilibrated with a buffer consisting of 20 mM Tris–HCl pH 7.5, 500 mM NaCl, 2 mM imidazole and 2 mM MnCl2 (buffer A). Mn-SODDr was eluted using a 50 ml linear gradient from buffer A to a buffer consisting of 20 mM Tris–HCl pH 7.5, 500 mM NaCl, 500 mM imidazole and 2 mM MnCl2 using a flow rate of 1 ml min−1. Mn-SODDr eluted at an imidazole concentration of ∼250 mM. The fractions containing Mn-SODDr were then pooled and dialysed against a buffer comprising 10 mM Tris–HCl pH 7.5 and 1 mM EGTA (buffer B) and applied onto a 1 ml Mono-Q ion-exchange chromatography column (Amersham Biosciences) previously equilibrated with buffer B. Mn-SODDr was eluted using a 30 ml linear gradient from buffer B to a buffer consisting of 20 mM Tris–HCl pH 7.5, 1 mM EGTA and 1 M NaCl. SDS–PAGE analysis (data not shown) confirmed that pure, tagged, recombinant Mn-SODDr of the correct molecular weight (231 amino acids, ∼26 kDa) had been obtained a with high degree of purity using the above protocol. For crystallization trials, protein stock solutions were prepared by pooling fractions containing the pure protein, dialysing them against a buffer consisting of 10 mM Tris–HCl pH 7.5, 20 mM NaCl and 1 mM MnCl2 and then concentrating to 10 mg ml−1. The stock solution used in the activity measurements described in §2.2 was prepared as described above except that the protein concentration was 20 mg ml−1.
2.2. Measurement of DEIRA Mn-SOD activity
The specific activity of Mn-SODDr was determined using an SOD Assay Kit-WST (Dojindo Molecular Technologies Inc., Maryland, USA) according to the manufacturer’s protocol (Dojindo Molecular Technologies Inc., 2005 ▶). WST-1 [2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt] is a soluble tetrazolium salt that produces a water-soluble formazan dye upon reduction with a superoxide anion. The rate of the reduction with 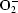 is linearly related to xanthine oxidase activity and is inhibited by SOD. Thus, the IC50 (50% inhibition activity of SOD or SOD-like materials) can be determined by a colorimetric method. Standard solutions of Mn-SODEc (Sigma–Aldrich Inc., MO, USA) were prepared at concentrations of 200, 100, 50, 20, 10, 5, 1, 0.1, 0.05, 0.01 and 0.001 U ml−1 in the SOD Assay Kit-WST dilution buffer and Mn-SODDr (20 mg ml−1) obtained as described above was diluted 100, 500 and 1000 times in the same buffer. 20 µl each of the solutions of Mn-SODEc, Mn-SODDr and three blank solutions (water blank 1, sample blank 2 and water blank 3) were transferred in triplets into microplate wells and 200 µl WST working solution and 20 µl enzyme working solution supplied by the manufacturer was added (except for blank 2 and 3, to which 20 µl dilution buffer was added). The reactions were incubated at 310 K for 20 min and absorbance was read at 405 nm in a V
max microplate reader (Molecular Devices Corp., CA, USA). The SOD activity (inhibition rate) was calculated using the equation
is linearly related to xanthine oxidase activity and is inhibited by SOD. Thus, the IC50 (50% inhibition activity of SOD or SOD-like materials) can be determined by a colorimetric method. Standard solutions of Mn-SODEc (Sigma–Aldrich Inc., MO, USA) were prepared at concentrations of 200, 100, 50, 20, 10, 5, 1, 0.1, 0.05, 0.01 and 0.001 U ml−1 in the SOD Assay Kit-WST dilution buffer and Mn-SODDr (20 mg ml−1) obtained as described above was diluted 100, 500 and 1000 times in the same buffer. 20 µl each of the solutions of Mn-SODEc, Mn-SODDr and three blank solutions (water blank 1, sample blank 2 and water blank 3) were transferred in triplets into microplate wells and 200 µl WST working solution and 20 µl enzyme working solution supplied by the manufacturer was added (except for blank 2 and 3, to which 20 µl dilution buffer was added). The reactions were incubated at 310 K for 20 min and absorbance was read at 405 nm in a V
max microplate reader (Molecular Devices Corp., CA, USA). The SOD activity (inhibition rate) was calculated using the equation
and plotted onto an inhibition curve. The amount of Mn-SODDr in the samples was determined from the curve in U ml−1 and used for calculations of the specific activity.
2.3. Crystallization, data collection and processing, structure solution and refinement
Crystals of Mn-SODDr appeared from several hanging-drop trials in which 2 µl protein stock solution and 2 µl reservoir solution were equilibrated at 293 K against 1 ml reservoir solution. Those most amenable to the collection of good-quality X-ray diffraction data were obtained using reservoir solutions consisting of either 10 mM Tris–HCl pH 8.5, 20 mM MnCl2, 30% PEG 4000 (crystal form I) or 10 mM Tris–HCl pH 8.5, 10% PEG 8000, 14% PEG 1500 and 10 mM MnCl2 (crystal form II). In both cases further cryoprotection of the crystals was not required and both were flash-frozen in a stream of gaseous nitrogen prior to data collection.
X-ray diffraction data from a single crystal of form I (Table 1 ▶) were collected at 100 K and λ = 0.933 Å on beamline ID14-EH1 of the ESRF, with the intensity data being indexed and integrated using the program MOSFLM, scaled and merged using SCALA and structure factors obtained using the program TRUNCATE. The indexing, integration, scaling and merging procedures clearly showed the crystals to be monoclinic, with unit-cell parameters a = 44.28, b = 83.21, c = 59.52 Å, β = 110.18°; examination of systematic absences in the intensity data indicated the space group to be P21. A calculated Matthews coefficient of 2.2 Å3 Da−1 and a theoretical solvent content of ∼43% are consistent with the asymmetric unit containing two Mn-SOD monomers.
Table 1. Summary of crystal parameters, data-collection and refinement statistics from crystal forms I and II of Mn-SODDr .
Values in parentheses are for the highest resolution shell.
| Crystal form I | Crystal form II | |
|---|---|---|
| Data collection and processing | ||
| Unit-cell parameters (Å, °) | a = 44.28, b = 83.21, c = 59.52, β = 110.18 | a = 43.57, b = 87.10, c = 116.42, β = 92.1 |
| Space group | P21 | P21 |
| Wavelength (Å) | 0.933 | 0.933 |
| Resolution range (Å) | 29.4–2.2 (2.32–2.2) | 41.2–2.0 (2.11–2.0) |
| No. of observations | 69134 (9955) | 452450 (26160) |
| No. of unique reflections | 20567 (2982) | 48281 (3564) |
| Completeness† (%) | 99.8 (99.8) | 82.2 (41.7) |
| Rsym (%) | 5.7 (16.5) | 7.5 (14.5) |
| 〈I/σ(I)〉 | 15.6 (6.0) | 31.8 (9.0) |
| Multiplicity | 3.4 (3.3) | 9.4 (7.3) |
| Final model composition | ||
| Amino-acid residues‡ | A1–A43, A45–A89, A96–A210, B1–B90, B96–B210 | A1–A92, A99–A210, B1–B43, B46–B91, B97–B210, C1–C91, C94–C210, D1–D91, D96–D210 |
| Protein atoms | 3248 | 6481 |
| Mn3+ ions | 2 | 4 |
| Water molecules | 366 | 567 |
| Refinement details | ||
| R/Rfree (%) | 16.8 (20.9)/23.6 (26.6) | 17.4 (18.4)/22.3 (23.1) |
| No. of reflections | 19552/996 | 45781/2483 |
| 〈B〉 (Å2) | 25.3 | 22.1 |
| E.s.u. (based on Rfree) (Å) | 0.23 | 0.19 |
| R.m.s. deviations from ideal geometry | ||
| Bond lengths (Å) | 0.012 | 0.012 |
| Bond angles (°) | 1.30 | 1.23 |
| Ramachandran plot | ||
| Favoured regions (%) | 94.8/93.9 | 96.1/92.6/93.9/94.0 |
| Allowed regions (%) | 3.4/5.0 | 2.8/6.3/3.9/4.0 |
| Generously allowed regions (%) | 1.7/1.1 | 1.1/1.1/1.7/1.1 |
| Disallowed regions (%) | 0.0/0.0 | 0.0/0.0/0.6 (Cys96)/0.0 |
For crystal form II the completeness in the resolution range 41.2–2.25 Å is 95.4%.
Some side chains are disordered and are not included in the final model. For crystal form I residues A126, B66 and B120 are truncated at Cδ, residues A40, A104, B40 and B43 are truncated at Cγ and residues A142 and B142 are truncated at Cβ. For crystal form II residues A142, A210, A53, A102, B40, B46, B53, B105, C91, C210, D139 and D158 are truncated at Cβ, while residues B23 and C94 are truncated at Sδ and Cγ, respectively.
X-ray diffraction data from a single crystal of form II (Table 1 ▶) were also collected at 100 K on beamline ID14-EH1, with the intensity data being treated in the same way as for form I. The crystals of form II are also monoclinic, with space group P21 and unit-cell parameters a = 43.57, b = 87.10, c = 116.42 Å, β = 92.1°. For this crystal form a calculated Matthews coefficient of 2.3 Å3 Da−1 and a theoretical solvent content of ∼47% are consistent with the asymmetric unit containing four Mn-SOD monomers.
Structure solution for crystal form I was carried out using the molecular-replacement (MR) technique as implemented in the program CNS (Brünger et al., 1998 ▶). The search model was a monomer from the crystal structure of Mn-SODEc (PDB code 1vew; Edwards et al., 1998 ▶) stripped of all solvent molecules and metal ions which was modified such that the residues in two loop regions (44–46, 92–95; Mn-SODEc numbering) were removed and amino-acid residues larger in the sequence of Mn-SODDr than in that of Mn-SODEc (see Fig. 1 ▶ for the sequence alignment) were truncated to alanine. The MR protocol used data in the resolution range 20.0–4.0 Å, resulted in Patterson and packing correlation coefficients of 58.0 and 61.0%, respectively, and confirmed that form I contains two molecules arranged as a homodimer in the asymmetric unit. The MR solution was then subjected to a round of rigid-body refinement using data in the range 40.0–3.5 Å, yielding R = 34.3%, R free = 34.6%. An initial round of simulated-annealing-based model refinement was then carried out using CNS. This was followed by a refinement protocol using the program REFMAC5 (Winn et al., 2001 ▶ and references therein) interspersed with rounds of manual rebuilding using the program QUANTA (Accelrys Inc.; http://www.accelrys.com), during which the model of Mn-SODDr was improved and water molecules and Mn3+ ions included.
Structure solution for crystal form II was also carried out using the MR technique, this time using the program AMoRe (Navaza, 1994 ▶). The search model was a monomer from the partially refined crystal form I structure of Mn-SODDr stripped of all solvent molecules and Mn3+ ions. The MR protocol was carried out using data in the resolution range 15–3 Å, resulted in R = 35.2%, CC(Fobs, Fcalc) = 66.8% and clearly showed that form II contains four molecules arranged in two homodimers in the asymmetric unit. Model refinement was carried out using the program REFMAC5 interspersed with rounds of manual rebuilding using the program O (Jones et al., 1991 ▶), during which the model was improved and water molecules and Mn3+ ions were included. Details concerning diffraction data quality and showing the results of both refinement protocols are shown in Table 1 ▶. The programs MOSFLM, SCALA, TRUNCATE, REFMAC5 and AMoRe are from the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶).
3. Results and discussion
The composition of the final refined models for both crystal forms of Mn-SODDr are summarized in Table 1 ▶. Briefly, crystal form I contains a homodimer in the asymmetric unit and the model consists of amino-acid residues A1–A43, A45–A89, A96–A210, B1–B90, B96–B210, two Mn3+ ions and 366 solvent molecules. Crystal form II contains two homodimers in the asymmetric unit, with the final refined model consisting of residues A1–A92, A99–A210, B1–B43, B46–B91, B97–B210, C1–C91, C94–C210, D1–D91, D96–D210, four Mn3+ ions and 567 solvent molecules. For both crystal forms the residue-numbering scheme refers to the sequence of DR1279 as deposited in the UniProtKB/Swiss-Prot database (http://www.expasy.org; primary accession No. Q9RUV2) and the final models described ignore residues from the purification tag modelled into electron density.
The three-dimensional structures of all the monomers found in both crystal forms are extremely similar, yielding r.m.s. deviations in Cα positions of 0.26, 0.30, 0.30, 0.31 and 0.32 Å when monomer A of crystal form I is superimposed on monomer B of crystal form I and monomers A, B, C and D of crystal form II, respectively.
During the course of this work, coordinates for an independently determined structure of DR1279 in crystal form II described above were deposited in the Protein Data Bank (PDB code 1y67; S. Chan, S. Tanaka, M. R. Sawaya & L. J. Perry, unpublished work). Superposing the structure of monomer A in crystals forms I and II as determined in this work with the four SOD monomers found the asymmetric unit of form II as determined by Chan and coworkers results in r.m.s. deviations in Cα positions of 0.26, 0.28, 0.38 and 0.32 Å (crystal form I) and 0.35, 0.25, 0.26 and 0.32 Å (crystal form II), respectively. Similarly, superimposing the AB homodimer in crystal forms I and II with the AB homodimer found in the structure deposited in the PDB results in r.m.s. deviations in Cα positions of 0.30 and 0.48 Å, respectively. Thus, the structures determined in this work and that determined by Chan and coworkers are essentially identical. However, it is intriguing to note that in the structure of DR1279 as determined by Chan and coworkers the metal ions bound to the protein are assigned as Fe3+ rather than Mn3+. This is despite the fact that DR1279 is clearly annotated as an Mn-SOD in the UniProtKB/Swiss-Prot database (see http://ca.expasy.org/uniprot/Q9RUV2) and that in the sequence of DR1279 the residues proposed as being the most likely candidates as markers for distinguishing between Mn-SODs and Fe-SODs (Parker & Blake, 1988 ▶) clearly indicate DR1279 to be a Mn-SOD. Moreover, the wavelength (1.1 Å) at which Chan and coworkers collected diffraction data for their structure solution does not allow one to distinguish between Fe3+ and Mn3+.
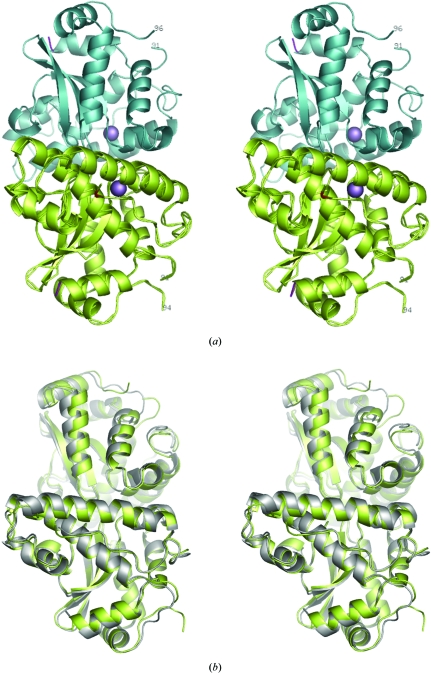
Nevertheless, in all cases the three-dimensional structures of the Mn-SODDr monomers (Fig. 2 ▶ a) are very similar to those reported in crystal structures of Mn-SODEc (Edwards et al., 1998 ▶, 2001 ▶; Borgstahl et al., 2000 ▶). Similarly, the structures of the physiologically relevant Mn-SODDr dimers (Fig. 2 ▶ b) in the crystal structures reported here and by Chan and coworkers are essentially isostructural to those found for Mn-SODEc and other Mn-SODs. The r.m.s. deviations between around 400 common Cα positions from superpositions of the Mn-SODEc dimer and those found in crystal forms I and II of Mn-SODDr average around 0.75 Å.
Figure 2.
(a) Stereoview of a ribbon representation of the structure of a physiologically relevant Mn-SODDr dimer. The two monomers making up the dimer are coloured lime-green (with Cα also shown) and cyan, respectively, with the positions of the Mg3+ ions shown in magenta. The C-terminus of each monomer is coloured red and the N-terminus is blue. Residues forming part of the six-residue QGQNGA insert are numbered to show the likely position of disordered residues with respect to the rest of the protein. (b) A superposition of the CD dimer found in crystal form II of Mn-SODDr and the dimer found in the crystal structure of Mn-SODEc. This shows that both monomers and dimers of the two Mn-SODs are essentially isostructural. The figure was produced using PyMOL (DeLano, 2002 ▶).
The metal-ion coordination sphere, which is absolutely conserved in all Mn-SODs, consists of the side chains of the amino-acid residues His26, His80, Asp172 and His176, which along with a conserved water molecule are arranged in a trigonal bipyramidal geometry around the metal ion. As in the structures of other Mn-SODs, the active site lies at the bottom of a funnel with access to the catalytic centre being guarded by several so-called ‘gateway residues’ (see Fig. 1 ▶; Edwards et al., 2001 ▶). These residues are very highly conserved throughout Mn-SOD sequences. In Mn-SODDr these residues are His30, Tyr34, Gln151, Glu175′ and Tyr179′ (the ′ denotes that the residues are from the second monomer of the homodimer). Both these amino-acid residues and their structural arrangement are absolutely conserved between Mn-SODDr and Mn-SODEc.
Based upon the above comparison of the crystal structures of Mn-SODDr and Mn-SODEc, one can infer that their tertiary and quaternary structures are extremely similar and that the geometric and electrostatic properties of their catalytic centres are essentially identical. One might thus expect the activities of the two enzymes to be similar. However, in our hands (see §2.2), the measured activities of 1250 and 2740 U mg−1 for Mn-SODDr and Mn-SODEc indicate that the activity of Mn-SODDr is rather lower than that of its E. coli counterpart. Thus, the importance of Mn-SOD to the radiation resistance of DEIRA cannot be attributed to a higher activity of Mn-SODDr with respect to that of Mn-SODEc.
It has been shown that in E. coli Mn-SOD preferentially protects DNA from damage caused by the presence of ROS (Hopkin et al., 1992 ▶). This, coupled with later discovery that E. coli Mn-SOD binds in a non-specific manner to DNA (Steinman et al., 1994 ▶), led to the suggestion that in E. coli Mn-SOD acts as a ‘tethered antioxidant’ which by binding to DNA concentrates its activity on scavenging superoxide radicals found close to DNA, thus preferentially ensuring its protection. The determination of the crystal structure of Mn-SODEc allowed the construction of a plausible model of how Mn-SODs and DNA might interact (see Figs. 6 and 7 of Edwards et al., 1998 ▶). In this model, DNA binds in a positively charged semi-circular groove formed at the monomer–monomer interface of the Mn-SOD homodimer, with amino-acid side chains from two loops on the surface of each monomer (formed by residues 61–67 and 152–158, respectively) providing anchoring points for the DNA. Both the positive charge of the hemihedral groove and the amino acids in the DNA-anchoring loops are highly conserved between the structures of Mn-SODDr and Mn-SODEc. Therefore, it may well be that in DEIRA Mn-SOD plays the same role as it is proposed to play in E. coli.
As mentioned in §1, the only really significant difference between the amino-acid sequences of Mn-SODDr and Mn-SODEc is a six-residue insert of sequence QGQNGA between residues 91 and 92 (E. coli numbering; Fig. 1 ▶). In the crystal structures of Mn-SODDr, this region of the sequence forms part of a solvent-accessible loop joining the helices α1 and α2. However, in the structures reported here and in that deposited in the PDB by Chan and coworkers, the residues making up the insert are disordered (Fig. 2 ▶) and are in a region of structure that is unlikely to affect either the active site or (potential) DNA binding by Mn-SODDr. However, they clearly point away from the body of the Mn-SODDr structure. Thus, although there is currently no evidence to support such a hypothesis, it may be that the residues in this insert provide a means by which Mn-SODDr can interact with other proteins and thus act as a ‘tethered antioxidant’ protecting not only DNA but also proteins that are crucial to the radiation resistance of DEIRA.
Supplementary Material
PDB reference: manganese superoxide dismutase, crystal form I, 2ce4, r2ce4sf
PDB reference: crystal form II, 2cdy, 2cdysf
Acknowledgments
The authors would like to thank the staff of Protein’eXpert SA, Grenoble, France for carrying out initial cloning and expression tests on DR1279.
References
- Battista, J. R. (1997). Annu. Rev. Microbiol.51, 203–224. [DOI] [PubMed] [Google Scholar]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Borgstahl, G. E. O., Pokross, M., Chehab, R., Sekher, A. & Snell, E. H. (2000). J. Mol. Biol.296, 951–959. [DOI] [PubMed] [Google Scholar]
- Chou, F. I. & Tan, S. T. (1990). J. Bacteriol.171, 2029–2035. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- DeLano, W. L. (2002). The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA, USA. http://www.pymol.org.
- Dojindo Molecular Technologies Inc. (2005). SOD Assay Kit-WST Technical Manual. http://www.dojindo.com/newimages/SODKitTechnicalInformation.pdf.
- Edwards, R. A., Baker, H. M., Whittaker, M. M., Whittaker, J. W., Jameson, G. B. & Baker, E. N. (1998). J. Biol. Inorg. Chem.3, 161–171.
- Edwards, R. A., Whittaker, M. M., Whittaker, J. W., Baker, E. N. & Jameson, G. B. (2001). Biochemistry, 40, 4622–4632. [DOI] [PubMed] [Google Scholar]
- Fridovitch, I. (1986). Arch. Biochem. Biophys.247, 1–11. [DOI] [PubMed] [Google Scholar]
- Gouet, P., Robert, X. & Courcelle, E. (2003). Nucleic Acids Res.31, 3320–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkin, K. A., Papazia, M. A. & Steinman, H. M. (1992). J. Biol. Chem.267, 24253–24258. [PubMed] [Google Scholar]
- Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed] [Google Scholar]
- Levin-Zaidman, S., Englander, J., Shimoni, E., Sharma, A. K., Minton, K. W. & Minsky, A. (2003). Science, 299, 254–256. [DOI] [PubMed] [Google Scholar]
- Lipton, M. S. et al. (2002). Proc. Natl Acad. Sci. USA, 99, 11049–11054. [Google Scholar]
- Makarova, K. S., Aravind, L., Wolf, Y. I., Tatusov, R. L., Minton, K.W., Koonin, E. V. & Daly, M. J. (2001). Microbiol. Mol. Biol. Rev.65, 44–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markillie, M. N., Varnum, S. M., Hradeky, P. & Wong, K.-K. (1999). J. Bacteriol.181, 666–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattimore, V. & Battista, J. R. (1996). J. Bacteriol.178, 633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton, K. W. (1994). Mol. Microbiol.13, 9–15. [DOI] [PubMed] [Google Scholar]
- Misra, H. P. & Fridovich, I. (1976). Arch. Biochem. Biophys.176, 577–581. [DOI] [PubMed] [Google Scholar]
- Navaza, J. (1994). Acta Cryst. A50, 157–163. [Google Scholar]
- Parker, M. W. & Blake, C. C. F. (1988). FEBS Lett.229, 377–382. [DOI] [PubMed] [Google Scholar]
- Steinman, H. M., Weinstein, L. & Brenowitz, M. J. (1994). J. Biol. Chem.269, 28629–28634. [PubMed] [Google Scholar]
- Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994). Nucleic Acids Res.22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001). Acta Cryst. D57, 122–133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: manganese superoxide dismutase, crystal form I, 2ce4, r2ce4sf
PDB reference: crystal form II, 2cdy, 2cdysf