Cytochrome P460 from N. europaea, a novel mono-heme protein containing an unusual lysine cross-link to the porphyrin ring, has been recombinantly expressed and purified from E. coli and crystallized. The crystals belong to the trigonal space group P31/221, with unit-cell parameters a = b = 53.3, c = 127.1 Å, one monomer in the asymmetric unit and diffract to 1.7 Å on a Cu Kα rotating-anode X-ray source.
Keywords: cytochrome P460, cross-linked heme, nitrification
Abstract
Cytochrome P460 from Nitrosomonas europaea, a novel mono-heme protein containing an unusual cross-link between a conserved lysine and the porphyrin ring, has been recombinantly expressed and purified from Escherichia coli. The protein crystallizes readily and diffraction to 1.7 Å has been obtained in-house. The crystals belong to the trigonal space group P31/221, with unit-cell parameters a = b = 53.3, c = 127.1 Å, and contain one monomer in the asymmetric unit.
1. Introduction
Nitrosomonas europaea is an aerobic chemolithotrophic ammonia-oxidizing Gram-negative bacterium belonging to the class β-proteobacteria in which ammonia is oxidized as the sole energy source and assimilated CO2 serves as the carbon source (Chain et al., 2003 ▶; Hooper et al., 2004 ▶). Ammonia- and nitrite-oxidizing bacteria constitute the nitrifying bacteria and are major biological participants in the global nitrogen cycle. Bacterial nitrification is a key process in the loss of fertilizer nitrogen from soils and in wastewater treatment and may be the major biological source of the greenhouse gases NO and N2O (Lipschultz et al., 1981 ▶; Anderson et al., 1993 ▶; Conrad, 1996 ▶; Colliver & Stephenson, 2000 ▶; Pérez et al., 2001 ▶; Wrage et al., 2001 ▶; Schmidt et al., 2003 ▶; Egli et al., 2003 ▶; Pynaert et al., 2004 ▶).
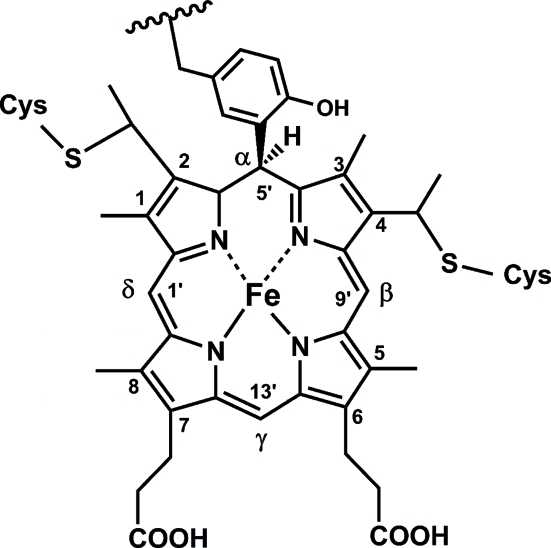
Much of the ammonia-oxidation pathway in Nitrosomonas has been elucidated and involves a series of membrane-bound and periplasmic metalloenzymes and electron-transfer proteins (Arciero & Hooper, 1998 ▶; Whittaker et al., 2000 ▶; Hooper et al., 2004 ▶). The second step, hydroxylamine oxidation to nitrite, is responsible for the net gain in reductive equivalents required for oxidative phosphorylation and other metabolic processes. This step is catalyzed by hydroxylamine oxidoreductase (HAO; EC 1.7.3.4), a multi-heme protein containing seven c-type hemes and a catalytic heme P460 with unusual spectral properties (named after the broad Soret band in the reduced protein, centered at 460 nm; Hooper et al., 1978 ▶, 1983 ▶; Hooper & Terry, 1977 ▶; Lipscomb et al., 1982 ▶; Andersson et al., 1984 ▶; Arciero & Hooper, 1993 ▶; Hendrich et al., 1994 ▶, 2001 ▶, 2002 ▶). Biochemical studies indicated the presence of a novel cross-link between a tyrosine side chain of HAO and heme P460 and this was confirmed by the crystal structure of HAO (Fig. 1 ▶) (Arciero et al., 1993 ▶; Igarashi et al., 1997 ▶).
Figure 1.
Schematic of the heme-protein cross-link observed in HAO.
Cytochrome P460 is a small (18.8 kDa) periplasmic mono-heme protein found in Nitrosomonas and Methylococcus capsulatus (Erickson & Hooper, 1972 ▶; Zahn et al., 1994 ▶). It has very similar electronic absorption, resonance Raman and Mössbauer features, ligand binding and hydrogen peroxide sensitivity as the heme P460 of HAO (Erickson & Hooper, 1972 ▶; Miller et al., 1984 ▶; Andersson et al., 1984 ▶, 1991 ▶; Numata et al., 1990 ▶), although there is no sequence similarity between the two proteins (Bergmann & Hooper, 1994 ▶).
It has been shown to bind hydroxylamine and hydrazine and is able to catalyze the oxidation of hydroxylamine to nitrite, although at much lower rates than HAO (Erickson & Hooper, 1972 ▶; Numata et al., 1990 ▶; Zahn et al., 1994 ▶). Furthermore, biochemical characterization of the N. europea cytochrome P460 has indicated the existence of a similar heme-protein cross-link to that observed in HAO, which involves a lysine residue that is conserved in the 21 homologous sequences currently identified as putative cytochrome P460s (Arciero & Hooper, 1997 ▶; Bergmann & Hooper, 2003 ▶).
Since detailed studies of the catalytic mechanism of HAO have been hampered by the presence of the seven non-catalytic hemes in each subunit, cytochrome P460 is an attractive and tractable model system for studies of the dehydrogenation of hydroxylamine by these cross-linked heme enzymes. It also provides a means to investigate the biogenesis and chemical properties of this unusual class of protein-bound cross-linked heme cofactors.
2. Experimental
2.1. Expression and purification
The mature coding sequence for cytochrome P460 (NCBI Accession No. NP_840112) was amplified by PCR using two primer-adapters (5′-CAGTATGCCATGG CAGGTGTGGCAGAGTTTAAC and 5′-TTAATAAGCTT CTTGGGCGCGACTACACCGAA; NcoI and HindIII sites are in italics and recognition sequences are in bold). Restriction digestion and ligation into plasmid pET20b(+) (Novagen) gave the expression vector pETP460His, a construct featuring the pelB signal sequence, mature cytochrome P460 coding sequence and a C-terminal six-His tag. The sequence of the His-tagged mature recombinant cytochrome P460 (rP460His) differs from that of the native protein in having an additional N-terminal Met residue and C-terminal KLAAALEHHHHHH residues. DNA sequencing confirmed the desired product. Vent DNA polymerase, restriction enzymes and T4 DNA ligase were supplied by New England Biolabs.
The expression plasmid and pEC86 containing the cytochrome c maturation genes ccmABCDEFGH (Arslan et al., 1998 ▶) were co-tranformed into E. coli strain BL21(DE3) with selection on LB plates supplemented with 100 µg ml−1 ampicillin and 34 µg ml−1 chloramphenicol. A single colony was used to inoculate 3 ml LB liquid medium and grown to late log phase at 310 K. This starter culture was used to inoculate 50 ml LB medium (0.5% inoculum), which was grown to late log phase and subsequently used to inoculate four Fernbach culture flasks, each containing 1 l Terrific Broth medium, 0.4% glycerol. The litre-volume cultures were grown at 310 K until the OD600 was ∼0.6; the temperature was then decreased to 303 K and expression induced by addition of IPTG to 0.4 mM. Liquid media contained 100 µg ml−1 carbenicillin and 34 µg ml−1 chloramphenicol.
Cells were harvested by centrifugation 10–12 h post-induction, yielding a green-colored cell paste. The periplasmic fraction was prepared as in Ausubel et al. (1992 ▶) and loaded directly onto a 4.5 × 8 cm DEAE Sepharose Fast Flow (Sigma–Aldrich) column equilibrated in 10 mM Tris–HCl pH 8.0. A five column-volume linear gradient to 10 mM Tris–HCl, 1 M NaCl pH 8.0 eluted the bound protein. The rP460His is readily monitored by its characteristic visible absorption spectrum and green color. This anion-exchange column served as an initial purification step and, more importantly, quickly concentrated the dilute protein. The eluted protein was loaded directly onto a 2.6 × 7 cm column of Talon Superflow Metal Affinity Resin (BD Biosciences). The manufacturer’s instructions were followed for washing and elution of the bound rP460His.
At this point the protein appeared to be pure, with a single band on SDS–PAGE stained with Coomassie Blue; the ratio of the ferric Soret band (436 nm) to the absorbance at 280 nm was ∼1.3. However, this measure of hemoprotein purity could be improved to ∼1.5 following an additional ion-exchange step and gel filtration. Anion exchange was carried out using a 10 ml Source 15Q Tricorn column (Amersham Biosciences) and a ten column-volume gradient from 20 mM HEPES pH 7.0 to 20 mM HEPES, 500 mM NaCl pH 7.0. Gel filtration used a 1.6 × 60 cm Toyopearl HW-55S (Tosoh Biosciences) column equilibrated in 20 mM HEPES pH 7.0.
2.2. Crystallization
Initial crystallization trials were carried out at 293 K using Hampton Research Crystal Screens I and II and the sodium/potassium phosphate Quik Screen. The recombinant protein was concentrated with Amicon Ultra Centrifugal Filter Devices (Millipore) to ∼300 µM as determined by the absorption of the ferric Soret band at 436 nm (∊ = 52 mM −1 cm−1; Numata et al., 1990 ▶). Crystals were grown by hanging-drop vapor diffusion in Nextal Crystallization Tools using equal amounts (2 µl) of protein solution and reservoir solution. Small crystals were obtained using the Quik Screen (1.8 M phosphate pH 5.6). This condition was optimized by screening pH and protein and phosphate concentrations. Large crystals suitable for X-ray studies appeared in 5 d by sitting-drop vapor diffusion in VDX plates (Hampton Research).
2.3. Characterization
2.3.1. Spectrophotometry
Crystals of rP460His were washed in fresh mother liquor and dissolved in 20 mM HEPES pH 7 for spectral studies. Fresh rP460His was prepared in the same buffer. In addition, this protein was prepared in 2.2 M phosphate pH 5.4 for comparison. Spectra were measured using a Cary50 spectrophotometer (Varian).
2.3.2. Mass spectrometry
10 µl of 300 µM rP460His was desalted with a C4 ZipTip (Millipore) as per the manufacturer’s instructions, mixed with an equal volume of matrix solution (saturated sinapinic acid in 75% acetonitrile, 0.1% trifluoroacetic acid) and dried onto a target. Mass spectra were acquired on a Biflex III MALDI–TOF mass spectrometer (Bruker Daltonics) operating in linear mode and calibrated using horse heart cytochrome c (MW 12 384 Da; Sigma–Aldrich).
2.4. X-ray data collection
Data collection was carried out in-house on a Rigaku/MSC MicroMax-007 rotating-anode X-ray generator (40 kW, 20 mA) with copper anode, VariMax Confocal MaxFlux optics and a R-AXIS IV++ detector (Rigaku MSC). Cryogenic temperatures (100 K) were maintained using an X-stream cryojet (Rigaku/MSC). For data collection at cryogenic temperatures, the crystal was soaked for 1 min in a cryoprotectant solution containing 8 µl mother liquor and 2 µl 100% glycerol. The crystal was then mounted in a nylon loop and frozen directly in the cryostream. Diffraction data were processed and scaled using HKL2000 (Otwinowski & Minor, 1997 ▶). Further data analysis was carried out using the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶).
3. Results and discussion
3.1. Expression and purification
Cytochrome P460 from N. europaea has been overexpressed in an E. coli BL21(DE3) cell line co-transformed with the expression plasmid and plasmid pEC86 containing the genes required for the incorporation of the heme cofactor in c-type cytochromes (Arslan et al., 1998 ▶). Initial expression gave more than 10 mg recombinant protein per litre; however, purification proved difficult. Therefore, a C-terminal six-His tag was included in the expression construct (calculated molecular weight 21 113.6 Da). Although this allowed rapid purification, it also decreased the recombinant protein expression to <5 mg l−1. Immobilized metal-affinity purification yielded high-purity protein as indicated by a single band on SDS–PAGE. An indicator of hemoprotein purity is the ratio of the absorbance of the Soret band to the 280 nm band. This A 436/A 280 ratio for the rP460His was improved from 1.3 to 1.5 by subsequent anion-exchange and gel-filtration chromatography. The values for cytochrome P460 purified from the natural source are reported as 0.1–0.9 (Erickson & Hooper, 1972 ▶; Miller et al., 1984 ▶; Numata et al., 1990 ▶). It is likely that these additional chromatography steps remove a population of rP460His lacking the correctly cross-linked cofactor.
3.2. Crystallization
After three weeks, small single crystals were obtained in 1.8 M sodium/potassium phosphate pH 5.6 at 293 K. Further screening yielded optimized conditions in which reproducible larger crystals appeared over 5 d. The final optimal conditions were 600 µM protein, 2.2 M sodium/potassium phosphate pH 5.2 or 2.4 M sodium/potassium phosphate pH 5.4 at 293 K. The crystals are reddish–brown in color with hexagonal bipyramidal morphology. A crystal grown in 2.4 M sodium/potassium phosphate pH 5.4 was used for X-ray data collection.
3.3. Characterization
3.3.1. Spectrophotometry
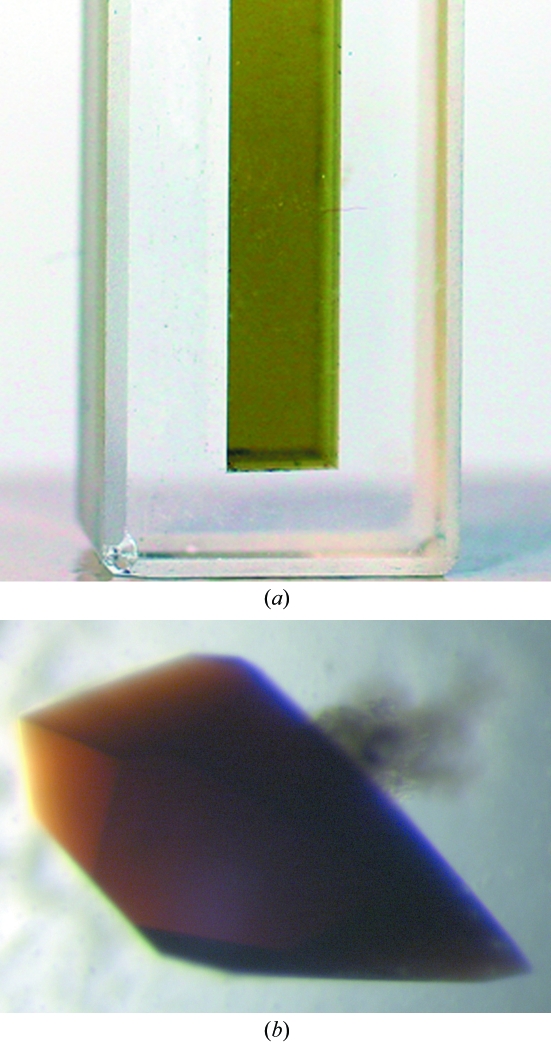
As purified, rP460His has a green color owing to the 436 nm Soret absorption peak in the ferric protein. However, the crystals visually appear deep red–brown in color (Fig. 2 ▶). In order to confirm that there has been no significant alteration in the chromophore during crystallization, crystals were dissolved and the solution spectrum compared with that of the purified protein. The two spectra are identical. In addition, pure uncrystallized protein transferred into mother liquor exhibits an essentially identical spectrum. This suggests that the anisotropic ordered packing of the chromophore in the crystals is responsible for the red visual appearance, rather than any intrinsic change in the chromophore. UV–visible single-crystal spectrophotometry was attempted, but the absorbance was so strong that even small crystals did not allow the transmission of any light to the detector.
Figure 2.
(a) rP460His solution. (b) Crystals of rP460His (0.8 × 0.4 × 0.4 mm) grown by sitting-drop vapor diffusion.
3.3.2. Mass spectrometry
The calculated monomer weight of rP460His (21 113.6 Da) was confirmed by MALDI–TOF mass spectrometry, which yielded an observed weight of 21 112 Da.
3.4. Preliminary X-ray diffraction studies
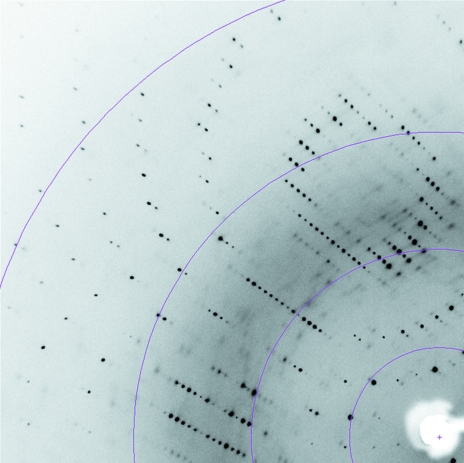
Crystals of rP460His diffract to 1.7 Å on an in-house Cu Kα rotating-anode X-ray source. Data-collection statistics are summarized in Table 1 ▶. A diffraction image is shown in Fig. 3 ▶. The crystals belong to the trigonal space group P31/221, with unit-cell parameters a = b = 53.3, c = 127.1 Å.
Table 1. Data-collection statistics.
Values in parentheses refer to the last resolution shell.
| Space group | P31/221 |
| Unit-cell parameters | |
| a = b (Å) | 53.3 |
| c (Å) | 127.1 |
| Wavelength (Å) | 1.5418 |
| Resolution range (Å) | 50–1.69 (1.75–1.69) |
| No. of unique reflections | 20852 (522) |
| Completeness (%) | 86.0 (22.0) |
| Redundancy | 8.8 (2.2) |
| Rmerge† | 0.048 (0.177) |
| Mean I/σ(I) | 20.6 (2.5) |
R
merge = 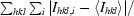
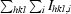 , where I is the observed intensity and 〈I〉 is the average intensity for multiple measurements.
, where I is the observed intensity and 〈I〉 is the average intensity for multiple measurements.
Figure 3.
Part of an X-ray diffraction image from rP460His crystals. Colored arcs show resolution (from left to right, arcs are at 2.0, 2.7, 4 and 8 Å).
Calculation of the Matthews coefficient suggests that a monomer is present in the asymmetric unit with a solvent content of 51.1% (Matthews, 1968 ▶). The data are of good quality, with an R merge of 4.8% and an estimated mosaicity after scaling of 0.4°. The average B factor derived from a Wilson plot is 25.3 Å2 (French & Wilson, 1978 ▶).
Acknowledgments
This work was carried out in part using computing resources in the Basic Sciences Computing Laboratory of the University of Minnesota Supercomputing Institute and we thank Patton Fast for his support. Funds for this research were supplied by The University of Minnesota, NSF (MCB 0093447 to ABH), DOE (DE-FG02-95ER20191 A009 to ABH) and NIH (GM-66569 to CMW).
References
- Anderson, I. C., Poth, M., Homstead, J. & Burdige, D. (1993). Appl. Environ. Microbiol.59, 3525–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, K. K., Babcock, G. T. & Hooper, A. B. (1991). Biochem. Biophys. Res. Commun.174, 358–363. [DOI] [PubMed] [Google Scholar]
- Andersson, K. K., Kent, T. A., Lipscomb, J. D., Hooper, A. B. & Munck, E. (1984). J. Biol. Chem.259, 6833–6840. [PubMed] [Google Scholar]
- Arciero, D. M. & Hooper, A. B. (1993). J. Biol. Chem.268, 14645–14654. [PubMed] [Google Scholar]
- Arciero, D. M. & Hooper, A. B. (1997). FEBS Lett.410, 457–460. [DOI] [PubMed] [Google Scholar]
- Arciero, D. M. & Hooper, A. B. (1998). Biochem. Soc. Trans.26, 385–389. [DOI] [PubMed] [Google Scholar]
- Arciero, D. M., Hooper, A. B., Cai, M. & Timkovich, R. (1993). Biochemistry, 32, 9370–9378. [DOI] [PubMed] [Google Scholar]
- Arslan, E., Schulz, H., Zufferey, R., Kunzler, P. & Thöny-Meyer, L. (1998). Biochem. Biophys. Res. Commun.251, 744–747. [DOI] [PubMed] [Google Scholar]
- Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (1992). Short Protocols in Molecular Biology. New York: John Wiley & Sons.
- Bergmann, D. J. & Hooper, A. B. (1994). FEBS Lett.353, 324–326. [DOI] [PubMed] [Google Scholar]
- Bergmann, D. J. & Hooper, A. B. (2003). Eur. J. Biochem.270, 1935–1941. [DOI] [PubMed] [Google Scholar]
- Chain, P., Lamerdin, J., Larimer, F., Regala, W., Lao, V., Land, M., Hauser, L., Hooper, A. B., Klotz, M., Norton, J., Sayavedra-Soto, L., Arciero, D., Hommes, N., Whittaker, M. & Arp, D. (2003). J. Bacteriol.185, 2759–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Colliver, B. B. & Stephenson, T. (2000). Biotechnol. Adv.18, 219–232. [DOI] [PubMed] [Google Scholar]
- Conrad, R. (1996). Microbiol. Rev.60, 609–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli, K., Bosshard, F., Werlen, C., Lais, P., Siegrist, H., Zehnder, A. J. & Van der Meer, J. R. (2003). Microb. Ecol.45, 419–432. [DOI] [PubMed] [Google Scholar]
- Erickson, R. H. & Hooper, A. B. (1972). Biochim. Biophys. Acta, 275, 231–244. [DOI] [PubMed] [Google Scholar]
- French, S. & Wilson, K. S. (1978). Acta Cryst. A34, 517–525. [Google Scholar]
- Hendrich, M. P., Logan, M., Andersson, K. K., Arciero, D. M., Lipscomb, J. D. & Hooper, A. B. (1994). J. Am. Chem. Soc.116, 11961–11968.
- Hendrich, M. P., Petasis, D., Arciero, D. M. & Hooper, A. B. (2001). J. Am. Chem. Soc.123, 2997–3005. [DOI] [PubMed] [Google Scholar]
- Hendrich, M. P., Upadhyay, A. K., Riga, J., Arciero, D. M. & Hooper, A. B. (2002). Biochemistry, 41, 4603–4611. [DOI] [PubMed] [Google Scholar]
- Hooper, A. B., Arciero, D., Bergmann, D. & Hendrich, M. P. (2004). Respiration in Archaea and Bacteria, edited by D. Zannoni, pp. 121–147. Berlin: Springer.
- Hooper, A. B., Debey, P., Andersson, K. K. & Balny, C. (1983). Eur. J. Biochem.134, 83–87. [DOI] [PubMed] [Google Scholar]
- Hooper, A. B., Maxwell, P. C. & Terry, K. R. (1978). Biochemistry, 17, 2984–2989. [DOI] [PubMed] [Google Scholar]
- Hooper, A. B. & Terry, K. R. (1977). Biochemistry, 16, 455–459. [DOI] [PubMed] [Google Scholar]
- Igarashi, N., Moriyama, H., Fujiwara, T., Fukumori, Y. & Tanaka, N. (1997). Nature Struct. Biol.4, 276–284. [DOI] [PubMed] [Google Scholar]
- Lipschultz, F., Zafiriou, O. C., Wofsy, S. C., McElroy, M. B., Valois, F. W. & Watson, S. W. (1981). Nature (London), 294, 641–643.
- Lipscomb, J. D., Andersson, K. K., Munck, E., Kent, T. A. & Hooper, A. B. (1982). Biochemistry, 21, 3973–3976. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Miller, D. A., Wood, P. M. & Nicholas, D. J. D. (1984). J. Gen. Microbiol.130, 3049–3054.
- Numata, M., Saito, T., Yamazaki, T., Fukumori, Y. & Yamanaka, T. (1990). J. Biochem.108, 1016–1021. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Pérez, T., Trumbore, S. E., Tyler, S. C., Matson, P. A., Ortiz-Monasterio, I., Rahn, T. & Griffith, D. W. T. (2001). J. Geophys. Res.106, 9869–9878.
- Pynaert, K., Smets, B. F., Beheydt, D. & Verstraete, W. (2004). Environ. Sci. Technol.38, 1228–1235. [DOI] [PubMed] [Google Scholar]
- Schmidt, I., Sliekers, O., Schmid, M., Bock, E., Fuerst, J., Kuenen, J. G., Jetten, M. S. & Strous, M. (2003). FEMS Microbiol. Rev.27, 481–492. [DOI] [PubMed] [Google Scholar]
- Whittaker, M., Bergmann, D., Arciero, D. & Hooper, A. B. (2000). Biochim. Biophys. Acta, 1459, 346–355. [DOI] [PubMed] [Google Scholar]
- Wrage, N., Velthof, G. L., van Beusichem, M. L. & Oenema, O. (2001). Soil Biol. Biochem.33, 1723–1732.
- Zahn, J., Duncan, C. & DiSpirito, A. A. (1994). J. Bacteriol.176, 5879–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]