The nucleocapsid protein of Bunyamwera virus, the prototypic member of the Bunyaviridae family of segmented negative-sense RNA viruses, has been expressed and crystallized. Complete X-ray diffraction data sets have been collected.
Keywords: Bunyamwera virus, nucleocapsid proteins
Abstract
Bunyamwera virus (BUNV) is the prototypic member of the Bunyaviridae family of segmented negative-sense RNA viruses. The BUNV nucleocapsid protein has been cloned and expressed in Escherichia coli. The purified protein has been crystallized and a complete data set has been collected to 3.3 Å resolution at a synchrotron source. Crystals of the nucleocapsid protein belong to space group C2, with unit-cell parameters a = 384.7, b = 89.8, c = 89.2 Å, β = 94.4°. Self-rotation function analysis of the X-ray diffraction data has provided insight into the oligomeric state of the protein as well as the orientation of the oligomers in the asymmetric unit. The structure determination of the protein is ongoing.
1. Introduction
Bunyamwera virus (BUNV) is the prototypic member of the Bunyaviridae family of segmented negative-sense RNA viruses. This family comprises the Orthobunyavirus, Hantavirus, Nairovirus, Phlebovirus and Tospovirus genera and several viruses within these groups are the causative agents of serious human disease (Elliott, 1997 ▶). In recognition of this fact, several bunyaviruses are classified by the Centers for Disease Control and Prevention (CDC) as priority A, B and C pathogens. Many features of BUNV molecular and cellular biology are common to other Bunyaviridae family members and as a result BUNV is recognized as a model to study the serious human pathogens within this family. The BUNV genome comprises three segments of negative-sense RNA, designated small (S), medium (M) and large (L). The coding potential of each segment is expressed as a single mRNA. The S segment encodes the nucleocapsid (N) and NSs proteins (Elliott, 1989b ▶; Fuller et al., 1983 ▶; Fuller & Bishop, 1982 ▶), the M segment encodes two envelope glycoproteins (Gn and Gc) and the NSm polypeptide (Elliott, 1985 ▶; Fuller & Bishop, 1982 ▶; Gentsch & Bishop, 1979 ▶) and the L segment encodes the BUNV RNA-dependent RNA polymerase (RdRp; Elliott, 1989a ▶). The BUNV RNA segments do not exist as naked RNAs, but rather as ribonucleoprotein (RNP) complexes in which the RNA is encapsidated with the N protein and associated with the BUNV RdRp. In this form, the N protein confers structural order to the RNA, which is required to allow access to the RNA-synthesis machinery. Furthermore, as all bunyaviruses lack a matrix protein, the RNP is also expected to directly interact with the viral glycoproteins and to allow packaging of the RNA segments into infectious particles.
RNP assembly implies that N proteins are able to interact homotypically and this activity has been best studied for members of the Hantavirus genus. The hantavirus N protein is proposed to form a trimeric structure as an intermediate in complete RNP formation, with adjacent N monomers interacting with each other at both ends (Kaukinen et al., 2004 ▶). It is proposed that association of pre-formed trimers encapsidate the growing RNA strand. In contrast, the N protein of Rift Valley Fever virus, a member of the Phlebovirus genus, has been reported to form homodimers both when harvested from RNPs and when expressed from recombinant sources (Le May et al., 2005 ▶). Finally, the N protein of BUNV has recently been shown to form high-order oligomers through the addition of one monomer at a time without passing through a lower-order intermediate (Leonard et al., 2005 ▶). The different preferred oligomeric states of orthobunyavirus, hantavirus and phlebovirus N proteins may reflect significant differences between these genera, suggesting that bunyaviruses may not utilize a common strategy of RNP formation.
We have begun the process of determining the three-dimensional structure of the BUNV N protein in order to gain insight into its oligomeric state and the mechanism of RNA binding. The N protein has been cloned and expressed in Escherichia coli. Expressed protein has been purified and crystallized.
2. Experimental procedures and results
2.1. Cloning, expression and purification
A cDNA containing the complete BUNV N-protein coding region was amplified by PCR using oligonucleotide primers BUN-Nde (5′-CAGTCATATGATTGAGTTGGAATTTCATGATGTCGC-3′) and BUN-Eco (5′-CTAAGAATTCTTACATGTTGATTCCGAATTTAGCAGG-3′) containing NdeI and EcoRI restriction enzyme recognition sites, respectively. The resulting cDNA fragment was digested with NdeI and EcoRI and ligated into the corresponding NdeI and EcoRI sites of expression plasmid pET28b(+).
The recombinant plasmid was transformed into E. coli BL21(DE3) strain and grown in Luria–Bertani broth (Fisher Scientific, Germany) with 25 mg ml−1 kanamycin at 310 K until the OD600 reached 0.7. Expression of the recombinant fusion protein was then induced with 1 mM isopropyl 1-thio-β-d-galactopyranoside (IPTG) for 4 d at 285 K. Cells were pelleted by centrifugation at 4629g for 20 min and frozen at 193 K for 4 h. Pellets were resuspended in binding buffer (20 mM Tris–HCl, 5 mM imidazole, 250 mM NaCl pH 7.9) and lysed using Benzonase Nuclease (Novagen, USA). The cell lysate was then disrupted by sonication on ice, after which debris was removed by centrifugation for 1 h at 8228g at 277 K. Unless described otherwise, all following steps were performed at 277 K. Ni–NTA Superflow (Qiagen, Germany) was added to the supernatant and the Ni–NTA-bound protein was washed five times in wash buffer (60 mM imidazole, 250 mM NaCl, 10 mM Tris pH 7.9). The bound protein was loaded onto a column and eluted using elution buffer (500 mM imidazole, 300 mM NaCl, 20 mM Tris pH 7.9). Fractions were then collected and checked by SDS–PAGE for size, purity and relative concentration. Selected fractions were then combined and dialyzed against 20 mM Tris–HCl, 150 mM NaCl pH 7.9. The N-terminal six-histidine tag was removed by thrombin cleavage according to the manufacturer’s instructions (Novagen), but with the omission of the thrombin-cleavage buffer. The cleaved protein was again dialyzed in 20 mM Tris–HCl, 150 mM NaCl pH 7.9 and cleavage was verified by SDS–PAGE analysis (Fig. 1 ▶). A sample of the purified protein was subjected to liquid-spray mass spectrometry. From this experiment, the molecular weight was determined to be 26 945 Da. The molecular-weight calculation aided in confirming the N-terminal amino-acid sequence of the protein, showing the existence of three additional N-terminal amino acids that remain from the thrombin-cleavage site (glycine, serine and histidine).
Figure 1.
The bacterially expressed BUNV N protein fused to an N-terminal His6 tag was analyzed using a Coomassie brilliant blue R-250 stained 15% polyacrylamide gel. BUNV N protein was analyzed before thrombin cleavage (lanes 1 and 2), after thrombin cleavage (lanes 4 and 5) and following partial thrombin cleavage (lane 3). Molecular-weight markers (kDa) are shown on the right.
2.2. Crystallization and data collection
Purified BUNV N protein was concentrated to 5 mg ml−1 and screened for crystallization conditions using the sitting-drop method with the aid of the Honeybee robotic system (Cartesian). Initial screens were performed with the commercially available sparse-matrix kits Crystal Screens I and II (Hampton Research) at 293 K. Crystallization hits were optimized. Ultimately, diffraction-quality crystals were grown in 10%(w/v) polyethylene glycol (PEG) 6000 containing 1.3–1.9 M sodium chloride buffered with 100 mM sodium citrate pH 5.7 at 293 K using the hanging-drop vapour-diffusion method (McPherson, 1982 ▶). In the final crystallization experiments, 2 µl protein solution was mixed with 2 µl precipitating agent. Crystals grown in higher concentrations of sodium chloride routinely appeared within 2 d and grew to maximal sizes in a week. Crystals grown in lower salt concentrations appeared in approximately three weeks and grew to larger sizes, with maximal crystal lengths of 0.5 mm (Fig. 2 ▶).
Figure 2.
Crystals of the N protein from BUNV. The maximum dimension of the crystal shown is 0.3 mm.
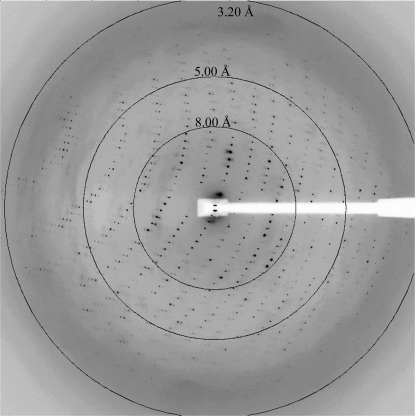
X-ray diffraction data were collected at the Advanced Photon Source SER-CAT beamline 22-BM (λ = 1.0 Å) on a MAR 225 CCD camera with a crystal-to-detector distance of 350 mm and 0.4° oscillations. Diffraction data were collected at 100 K. A single diffraction image is shown in Fig. 3 ▶. The optimal data-collection strategy was determined with the aid of the HKL2000 package (Otwinowski & Minor, 1997 ▶). All raw data images were processed, scaled and merged with DENZO and SCALEPACK from the HKL2000 package. Structure factors were calculated with TRUNCATE (French & Wilson, 1978 ▶). The self-rotation functions were calculated with the GLRF program (Tong & Rossmann, 1997 ▶). The Patterson integration radius was 40 Å and data in the resolution range 15.0–4.0 Å were used.
Figure 3.
A 0.4° oscillation X-ray diffraction image from a crystal of the N protein from BUNV. Resolution arcs are shown. Reflections are observed to beyond 3.2 Å.
The BUNV N crystals belong to the monoclinic system; the space group was identified as C2. The unit-cell parameters are a = 384.73, b = 89.89, c = 89.26 Å, β = 94.4°. Diffraction data statistics are summarized in Table 1 ▶. The large unit-cell volume suggests that multiple copies of the N protein should be present in the asymmetric unit. The Matthews coefficient (Matthews, 1968 ▶) was calculated and revealed that there are potentially eight copies of the N protein in the asymmetric unit, with a V M of 3.5 Å3 Da−1, corresponding to a solvent content of 65.1%.
Table 1. Crystal parameters, data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| X-ray source | APS SER-CAT 22-ID |
| Wavelength (Å) | 1.000 |
| Detector | MAR 225 |
| Space group | C2 |
| Unit-cell parameters | |
| a (Å) | 384.73 |
| b (Å) | 89.89 |
| c (Å) | 89.26 |
| β (°) | 94.4 |
| Resolution range (Å) | 30–3.30 (3.42–3.30) |
| No. of observations | 110618 |
| Unique reflections | 44239 |
| Mosaicity (°) | 0.79 |
| Completeness (%) | 97.4 (98.7) |
| Average 〈I/σ(I)〉 | 28.4 (2.9) |
| Rmerge (%) | 0.043 (0.307) |
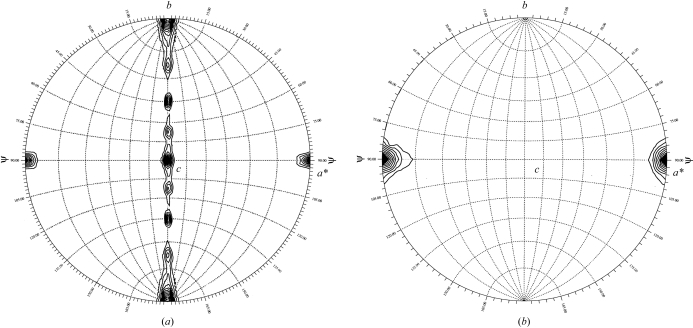
Given the large number of molecules in the asymmetric unit, self-rotation functions were calculated in order to search for non-crystallographic symmetry (NCS) elements. The results for the κ = 180° section of search space showed nine peaks (Fig. 4 ▶ a). Eight of these peaks were equally spaced by 22.5° around the crystallographic a axis. This is consistent with either two fourfold rotational symmetry axes separated by a 45° rotation with respect to each other along the direction of the a axis or an eightfold rotational symmetry axis. The fourfold rotational symmetry is confirmed by the appearance of a single peak (11σ) on the stereographic plot for κ = 90° (Fig. 4 ▶ b). A single peak suggestive of an eightfold is observed on the plot for κ = 45° (not shown), with a peak height of 2σ less than the height of the peak on the κ = 90° plot. Thus, the possibility of an eightfold rotational axis could not be eliminated.
Figure 4.
Self-rotation function calculated using the BUNV N protein crystals. Self-rotation searches with (a) κ = 180° and (b) κ = 90° were used to identify twofold and fourfold rotation angles, respectively. Contours for (a) start at 3 standard deviations (sd) with intervals of 1 sd and for (b) start at 6 sd with intervals of 1 sd. Calculations performed and plots produced using GLRF (Tong & Rossmann, 1997 ▶).
However, further inspection of the peaks on the κ = 180° plot might suggest the presence of multiple fourfolds. The eight peaks which are observed around the a axis on the κ = 180° plot could be separated into two sets of four peaks based on peak height. Peak set 1 [(0.0, 0.0), (90.0, 45.0), (90.0, 90.0), (90.0, 135.0)] has an average peak height of 7.5σ [this calculation excludes peak (0.0, 0.0) which has an elevated peak height owing to its coincidence with the crystallographic twofold], while peak set 2 [(90.0, 22.5), (90.0, 67.5), (90.0, 112.5), (90.0, 157.5)] has an average peak height of 5.1σ. The difference between the two sets of peaks could suggest that the each represents an independent fourfold rotational axis. This is further supported by preliminary analytical ultracentrifugation experiments, which show that the N protein at higher protein concentrations assembles into various aggregation states, each of which is a multiple of a tetramer (data to be published).
This is the first nucleocapsid protein from the Bunyaviridae family of segmented negative-sense viruses to be crystallized. Currently, there are no homologous structures that are suitable for use in molecular-replacement methods for the determination of initial phase information. We have successfully expressed and purified selenomethionine-substituted BUNV N protein in E. coli. Our present focus is the crystallization of the SeMet protein. This should allow structure determination by single or multiwavelength anomalous diffraction techniques in combination with real-space averaging and phase extension. With this structure, we hope to gain insight into RNA binding and RNP formation.
Acknowledgments
We thank the staff of the South East Regional Collaborative Access Team (SER-CAT) at the Advanced Photon Source for their assistance in data collection. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. W-31-109-Eng-38. We would like to thank David and Sylvia MacPherson (University of Alabama at Birmingham) for helpful discussions concerning N-protein purification, and Richard M. Elliott (University of St Andrews, Scotland) for supplying the cDNA encoding the BUNV N protein. This work is supported in part by grants to ML (NIH, AI050066) and JB (NIH, AI059174).
References
- Elliott, R. M. (1997). Mol. Med.3, 572–577. [PMC free article] [PubMed] [Google Scholar]
- Elliott, R. M. (1985). Virology, 143, 119–126. [DOI] [PubMed] [Google Scholar]
- Elliott, R. M. (1989a). Virology, 173, 426–436. [DOI] [PubMed] [Google Scholar]
- Elliott, R. M. (1989b). J. Gen. Virol.70, 1281–1285. [DOI] [PubMed] [Google Scholar]
- French, S. & Wilson, K. (1978). Acta Cryst. A34, 517–525. [Google Scholar]
- Fuller, F., Bhown, A. S. & Bishop, D. H. (1983). J. Gen. Virol.64, 1705–1714. [DOI] [PubMed] [Google Scholar]
- Fuller, F. & Bishop, D. H. (1982). J. Virol.41, 643–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch, J. R. & Bishop, D. L. (1979). J. Virol.30, 767–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaukinen, P., Kumar, V., Tulimaki, K., Engelhardt, P., Vaheri, A. & Plyusnin, A. (2004). J. Virol.78, 13669–13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le May, N., Gauliard, N., Billecocq, A. & Bouloy, M. (2005). J. Virol.79, 11974–11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard, V. H., Kohl, A., Osborne, J. C., McLees, A. & Elliott, R. M. (2005). J. Virol.79, 13166–13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson, A. (1982). Preparation and Analysis of Protein Crystals. New York: John Wiley.
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Tong, L. & Rossmann, M. G. (1997). Methods Enzymol.276, 594–611. [PubMed] [Google Scholar]