The crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Synechococcus PCC 7942 in complex with NADP was determined at 2.5 Å resolution.
Keywords: glyceraldehyde-3-phosphate dehydrogenase, Synechococcus PCC 7942
Abstract
The crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase (NADP-GAPDH) from Synechococcus PCC 7942 (S. 7942) in complex with NADP was solved by molecular replacement and refined to an R factor of 19.1% and a free R factor of 24.0% at 2.5 Å resolution. The overall structure of NADP-GAPDH from S. 7942 was quite similar to those of other bacterial and eukaryotic GAPDHs. The nicotinamide ring of NADP, which is involved in the redox reaction, was oriented toward the catalytic site. The 2′-phosphate O atoms of NADP exhibited hydrogen bonds to the hydroxyl groups of Ser194 belonging to the S-loop and Thr37. These residues are therefore considered to be essential in the discrimination between NADP and NAD molecules. The C-terminal region was estimated to have an extremely flexible conformation and to play an important role in the formation of the supramolecular complex phosphoribulokinase (PRK)–regulatory peptide (CP12)–GAPDH, which regulates enzyme activities.
1. Introduction
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) catalyzes the reversible interconversion between 1,3-bisphosphoglycerate and d-glyceraldehyde 3-phosphate using either NAD(H) or NADP(H) as a coenzyme. NAD-GAPDH (EC 1.2.1.12) only catalyzes the glycolysis/glyconeogenesis of prokaryotes and eukaryotes in the presence of NAD(H). NADP-GAPDH (EC 1.2.1.13) utilizes both NADP(H) and NAD(H) with a preference for NADP(H) and functions in the second reaction of the photosynthetic carbon reduction (PCR) cycle of higher plants and algae (Wolosiuk & Buchanan, 1976 ▶). The activity of NADP-GAPDH is regulated in vivo by a system consisting of ferredoxin, thioredoxin f and ferredoxin/thioredoxin reductase (Buchanan, 1980 ▶; Cséke & Buchanan, 1986 ▶). This enzyme is also activated by addition of DTT, NADP(H) or ATP in vitro (Wolosiuk & Buchanan, 1976 ▶; Wirtz et al., 1982 ▶). In higher plants, NADP-GAPDH exists in the form of either a homotetramer A 4 consisting of a subunit A or a heterooligomer (A 2 B 2)n consisting of two subunits A and B in stoichiometric amounts (Cerff, 1978 ▶; Shih et al., 1991 ▶; Scagliarini et al., 1998 ▶). The subunits A (36 kDa) and B (39 kDa) of chloroplast GAPDHs are homologous and highly conserved, except for the C-terminal extension of the subunit B. The extended C-terminal region of subunit B, which contains two Cys residues, is thought to be involved in the regulation of GAPDH activity (Sparla et al., 2002 ▶, 2005 ▶).
The cyanobacterium Synechococcus PCC 7942 (S. 7942) possesses a unique homotetrameric NADP-GAPDH that is thought to work not only in the PCR cycle but also in glycolysis/gluconeogenesis. The activity of NADP-GAPDH is sixfold higher with NADPH than with NADH. Moreover, it is not greatly activated by light irradiation or by the addition of DTT, NADPH or ATP and is not regulated by a ferredoxin–thioredoxin system, thus making it different from those of higher plants (Tamoi et al., 1996 ▶, 1998 ▶). S. 7942 NADP-GAPDH has an amino-acid sequence that shares about 65–69% identity with subunits A and B of spinach and tobacco chloroplasts, with the differences in their primary structures mostly being found neighbouring the C-terminus. Of particular significance, S. 7942 GAPDH lacks the three thiol groups that might be involved in light/dark regulation of the chloroplast enzymes.
Crystal structures have been solved for cytosolic NAD-GAPDHs from a number of organisms including Escherichia coli (Duée et al., 1996 ▶), Bacillus stearothermophilus (Biesecker et al., 1977 ▶; Skarzynski et al., 1987 ▶), Trypanosoma cruzi (Vellieux et al., 1995 ▶; Souza et al., 1998 ▶), Leishmania mexicana (Kim et al., 1995 ▶), human muscle (Mercer et al., 1976 ▶) and lobster (Moras et al., 1975 ▶; Song et al., 1998 ▶). Crystal structures of NADP-GAPDHs from Methanothermus fervidus (Charron et al., 2000 ▶) and non-regulatory A 4 isoforms from spinach chloroplasts (Fermani et al., 2001 ▶; Falini et al., 2003 ▶; Sparla et al., 2004 ▶) have been reported. These GAPDH structures have contributed to the understanding of phosphate-recognition sites, which are important for the enzyme reaction. GAPDHs have two anion-binding sites, the Ps (substrate phosphate ion site) and the Pi (inorganic phosphate ion site) near the catalytic Cys residue (Moras et al., 1975 ▶). A new inorganic phosphate ion-binding site was recently reported in some GAPDH structures (Kim et al., 1995 ▶; Korndörfer et al., 1995 ▶; Song et al., 1999 ▶; Yun et al., 2000 ▶; Falini et al., 2003 ▶). This site is referred to as the ‘new Pi’ site to distinguish from ‘classical Pi’ sites.
The aim of the present study is to clarify the molecular mechanism of the novel NADP-GAPDH from S. 7942 by analyzing its crystal structure. A detailed study of the three-dimensional structure of the enzyme and comparison with that reported for the A 4 isoform of spinach chloroplasts in complex with NADP should provide information about the recognition mechanism of the two coenzymes NADP and NAD. Furthermore, structure analysis should help determine the activation mechanism produced by light irradiation. Here, we report the crystal structure of S. 7942 NADP-GAPDH in complex with NADP at 2.5 Å resolution.
2. Materials and methods
Expression and purification of recombinant NADP-GAPDH from S. 7942 was performed as reported previously (Tamoi et al., 1996 ▶). The initial crystallization condition was found using Crystal Screen 2 from Hampton Research (Nakamura et al., 2001 ▶). We slightly optimized the condition. Pure recombinant protein was concentrated to 3.5 mg ml−1 in 50 mM HEPES buffer containing 30 mM NaCl. Crystals of S. 7942 GAPDH were then grown at 293 K using the hanging-drop vapour-diffusion technique with initial conditions formed by mixing 2 µl protein solution with 1 µl reservoir solution consisting of 44% saturated ammonium sulfate, 0.1 M citrate buffer pH 5.2 and 0.2 M potassium sodium tartrate. Crystals complexed with NADP were produced by soaking the native crystals in solution containing 54% saturated ammonium sulfate, 0.1 M citrate buffer pH 5.2, 0.2 M potassium sodium tartrate and 40 mM NADP+.
Diffraction data from the crystals were collected at 100 K on an ADSC CCD detector using synchrotron radiation with a wavelength of 0.97 Å at the BL6A station of KEK-PF, Japan. The crystal-to-detector distance was 220 mm and 180 images were recorded at 1° intervals. For data collection at 100 K, the crystals were loop-mounted in cryoprotectant solution containing 70% saturated ammonium sulfate, 0.1 M citrate buffer pH 5.2, 0.2 M potassium sodium tartrate and 10%(v/v) glycerol. The intensity data were processed with MOSFLM (Steller et al., 1997 ▶) and scaled using SCALA from the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶). Unexpected diffraction overlap arising from high mosaisity of the crystals causes the 85.8% completeness. The crystals belong to the monoclinic space group C2, with unit-cell parameters a = 150.0, b = 79.6, c = 206.2 Å, β = 101.3°, and diffracted to 2.5 Å resolution.
The structure of NADP-GAPDH from S. 7942 was determined by the molecular-replacement method using EPMR (Kissinger et al., 1999 ▶). The dimeric structure of NAD-GAPDH from B. stearothermophilus (PDB code 1gd1), which exhibits 59.5% identity to S. 7942 NADP-GAPDH, was used as a search model. The solution revealed that the crystal contains one tetramer and one dimer as the asymmetric unit. Crystallographic refinement was performed by simulated-annealing, positional and individual B-factor refinements with CNS (Brünger et al., 1998 ▶), followed by manual rebuilding of the structure with O (Jones et al., 1991 ▶). Noncrystallographic symmetry (NCS) restraints were applied during refinement steps. After several rounds of refinement and model building, the R factor and free R factor were reduced to 25.6 and 28.9%, respectively. The F o − F c difference map showed clear electron density corresponding to the NADP molecule at the NADP-binding site, although density for the nicontinamide group was somewhat weak (Fig. 1 ▶). Two peaks exhibiting an electron density higher than 4σ were found close to the catalytic cavity in the map and were assigned as sulfate ions. After refinement including the NADP molecules and sulfate ions, water molecules were added to the model at locations with F o − F c densities higher than 3σ and hydrogen-bonding stereochemistry using the water-picking function of CNS without NCS restraints. The final R factor and free R factor were 19.1 and 24.0%, respectively. The stereochemistry of the final model was analyzed with PROCHECK (Laskowski et al., 1993 ▶). A Ramachandran plot showed 87.6% of the residues in the most favoured regions, with most of the remaining residues in additionally allowed regions. Only Val242 lies in a disallowed region. The electron density for this residue in each subunit clearly confirms its conformation. This structural feature is also present in other GAPDH structures (Skarzynski et al., 1987 ▶; Kim et al., 1995 ▶; Vellieux et al., 1995 ▶). The data-collection and final refinement statistics are summarized in Table 1 ▶.
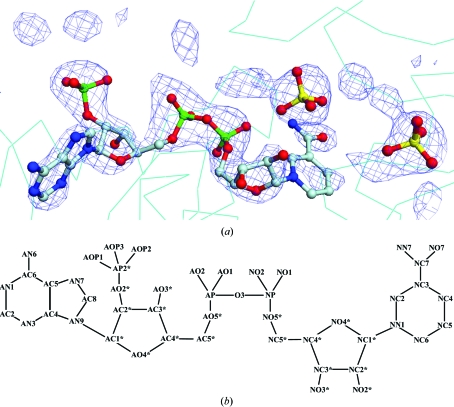
Figure 1.
(a) F o − F c map of NADP- and sulfate ion-binding sites contoured at 2.5σ. The NADP molecule and sulfate ions are shown as ball-and-stick models. This figure was prepared using DINO (http://www.dino3d.org). (b) Numbering scheme of the atoms in the NADP molecule using the general nomenclature (Carugo & Argos, 1997 ▶).
Table 1. Summary of data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell (2.6–2.5 Å).
| Data-collection statistics | |
| X-ray source | KEK-PF, BL6A |
| Wavelength (Å) | 0.97 |
| Temperature (K) | 100 |
| Resolution (Å) | 2.5 |
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 150.0, b = 79.6, c = 206.2, β = 101.3 |
| Rmerge† (%) | 6.7 (26.5) |
| I/σ(I) | 7.7 (2.8) |
| No. of observed reflections | 701767 |
| No. of unique reflections | 71057 |
| Completeness (%) | 85.8 (85.8) |
| Mosaicity (°) | 0.86 |
| Model details | |
| No. of protein residues | 2028 |
| No. of sulfate ions | 12 |
| No. of NADP molecules | 6 |
| No. of water molecules | 539 |
| Refinement statistics | |
| Resolution (Å) | 20–2.5 |
| R factor‡ (%) | 19.1 (26.8) |
| Rfree§ (%) | 24.0 (31.4) |
| R.m.s.d. in bond distance (Å) | 0.013 |
| R.m.s.d. in bond angle (°) | 1.7 |
| Average B values | |
| Overall (Å2) | 33.3 |
| NADP molecules (Å2) | 36.3 |
| Sulfate ions (Å2) | 31.3 |
| Water molecules (Å2) | 29.9 |
| Ramachandran plot | |
| Most favoured region (%) | 87.6 |
| Additionally allowed regions (%) | 11.4 |
| Generously allowed regions (%) | 0.7 |
| Disallowed regions (%) | 0.3 |
R
merge = 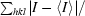
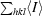 .
.
R factor = 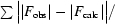
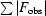 .
.
R free was calculated using 10% of the data.
3. Results and discussion
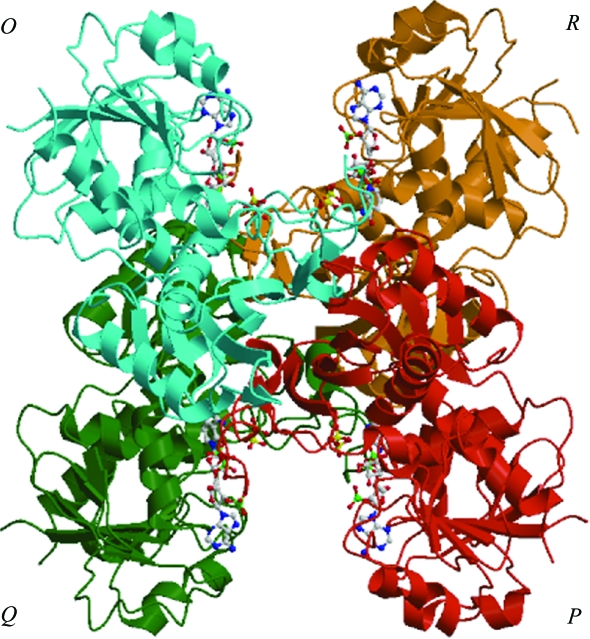
The final model comprises six monomers of S. 7942 GAPDH complexed with NADP and 539 water molecules in the asymmetric unit. The monomers, O, P, Q, R, O′ and P′, were named according to B. stearothermophilus GAPDH (Biesecker et al., 1977 ▶). Monomers O, P, Q and R form a tetramer, OPQR, as shown in Fig. 2 ▶. Monomers O′ and P′ are close to the cell origin and the crystallographic equivalents Q′ and R′ are created by a twofold symmetry operation to form the tetramer O′P′Q′R′. Although the two tetramers differ crystallographically, the difference in their quaternary structures cannot be regarded as significant.
Figure 2.
Overall view of the NADP-GAPDH tetramer from S. 7942 in a ribbon representation. NADP and sulfate ions are shown as ball-and-stick models. This figure was prepared using MOLSCRIPT (Kraulis, 1991 ▶) and Raster3D (Merritt & Murphy, 1994 ▶).
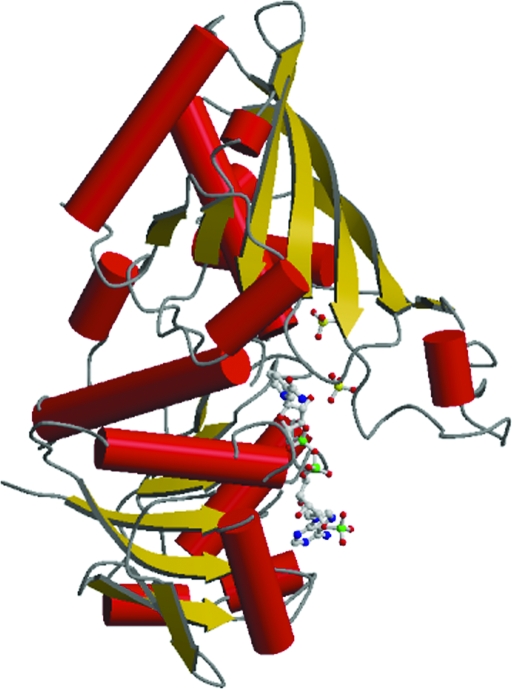
The structure of monomer O is shown in Fig. 3 ▶. In all six monomer structures, the 42 amino-acid residues from the C-terminus were disordered and did not show sufficient electron density for assignment, possibly because they are located on the surface of the tetramer. The six monomers present very similar conformations. The root-mean-square deviations (r.m.s.d.) calculated using LSQKAB with monomer O as a reference for superimposition of the P, Q, R, O′ and P′ Cα atoms are 0.157, 0.151, 0.165, 0.333 and 0.298 Å, respectively. The values are in the range 0.151–0.333 when calculated using other monomers as a reference. The monomer structures consist of NADP-binding (amino-acid residues 1–153 and 318–338) and catalytic (154–317) domains. The NADP-binding domain has a Rossmann fold (Rossmann et al., 1975 ▶) in which six β-strands form a parallel β-sheet and six α-helices lie on both sides of the sheet. The core sheet is completed by two extra β-strands. The catalytic domain is made up of an eight-stranded antiparallel β-sheet, seven α-helices and a loop structure (residues 183–208) called the S-loop (Skarzynski et al., 1987 ▶; Skarzynski & Wonacott, 1988 ▶). Overall, the topology of S. 7942 GAPDH is very similar to that of other known GAPDHs.
Figure 3.
Overall view of monomer O of NADP-GAPDH from S. 7942. NADP and sulfate ions are shown as ball-and-stick models. Helices are represented by crimson cylinders and β-strands are shown in gold. This figure was prepared using MOLSCRIPT (Kraulis, 1991 ▶) and Raster3D (Merritt & Murphy, 1994 ▶).
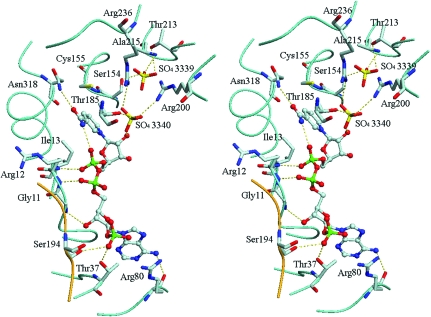
Each monomer contains one NADP molecule and two sulfate ions, as shown in Fig. 4 ▶. The NADP molecule has an extended conformation and is located in the coenzyme-binding site in each monomer, allowing additional interactions with residues of the S-loop in a neighbouring monomer (Table 2 ▶); for example, the NADP bound in the coenzyme site of monomer O forms additional interactions with the S-loop of monomer R. The adenine ring adopts an anti conformation, while the nicotinamide ring is in a syn conformation stabilized by an internal hydrogen bond between the NN7 atom of the nicotinamide and the NO1 atom of one of the phosphate groups, with an average distance of 2.96 Å between the two. The nicotinamide ring of NADP, which is involved in the redox reaction, is oriented toward the catalytic site. The distance between the NC5 atom of the nicotinamide ring and the Sγ atom of the catalytic Cys155 is 3.56 Å. One of the 2′-phosphate O atoms of NADP forms hydrogen bonds with the hydroxyl groups of Ser194 belonging to the S-loop and Thr37. The mode of interaction between the NADP molecule and protein is the same as that found in the crystal structure of A 4-GAPDH from spinach chloroplasts (Fermani et al., 2001 ▶). Of particular significance, hydrogen bonds between the 2′-phosphate O atoms of NADP and the hydroxyl groups of the residues Ser and Thr were also found in spinach A 4-GAPDH. Thus, it is suggested that these residues play an essential role in discrimination between NADP and NAD molecules because the two coenzymes only differ structurally in the phosphate group esterified at the 2′ position of adenosine ribose. This is supported by evidence that the affinity for NADPH is decreased and k cat (NADPH) is lowered in mutants of spinach A 4-GAPDH with the Ser of the S-loop and/or Thr replaced by Ala (Sparla et al., 2004 ▶).
Figure 4.
Stereo representation of the NADP- and sulfate-binding sites in monomer O. Hydrogen bonds are represented by dotted lines. C atoms are shown in grey, N atoms in blue, O atoms in red, S atoms in yellow and P atoms in green. A Cα trace of monomer O is shown in cyan and that of subunit R is shown in orange. This figure was prepared using DINO (http://www.dino3d.org).
Table 2. Short contacts (less than 3.4 Å) of NADP with the protein and sulfate ions.
| NADP | Distance (Å) | |
|---|---|---|
| AOP3 | Thr37 OG1 | 2.6 ± 0.1 |
| AOP3 | Ser194 OG† | 3.0 ± 0.4 |
| AN6 | Arg80 O | 3.3 ± 0.2 |
| AO3* | Gly11 N | 3.3 ± 0.2 |
| AO2 | Arg12 N | 2.8 ± 0.1 |
| NO2 | Ile13 N | 2.9 ± 0.2 |
| NO2* | SO4340 O4 | 2.8 ± 0.2 |
| NO7 | Asn318 ND2 | 3.0 ± 0.2 |
| NN7 | NADP NO1 | 3.0 ± 0.1 |
Belongs to the S-loop of the neighbouring subunit.
Two sulfate ions bind to the active site (Fig. 3 ▶). Sulfate ion 3339 occupies a classical Pi site, making hydrogen bonds with Thr213 Oγ1, Ser154 Oγ, Ala215 N and one water molecule. Sulfate ion 3340 occupies the Ps site, forming hydrogen bonds with the nicotinamide ribose O2′ of NADP, Thr185 Oγ, Arg200 Nη1 and one water molecule. In our structure, the conformational adjustment of the 220s loop that is necessary for formation of the ‘new Pi’ site (Kim et al., 1995 ▶) is not observed.
S. 7942 GAPDH is not greatly activated by light irradiation (Tamoi et al., 1996 ▶, 1998 ▶), although NADP-GAPDH in chloroplast stroma is activated by reduced thioredoxins in light. The regulatory isoform of the chloroplast enzyme is A 2 B 2-GAPDH, containing subunits A and B in stoichiometric amounts (Scheibe et al., 1996 ▶; Scagliarini et al., 1998 ▶; Sparla et al., 2002 ▶). Two extra Cys residues, Cys349 and Cys358, belonging to the extended C-terminal part of subunit B, are considered to be essential for the regulation of GAPDH activity (Sparla et al., 2005 ▶). These Cys residues are absent in S. 7942 GAPDH, although its subunit (41 kDa) has an extended C-terminus. Consequently, it seems likely that the light-activation property of S. 7942 GAPDH is related to the lack of Cys residues. The 42 amino-acid residues of the C-terminal region did not show sufficient electron density for assignment, indicating that the C-terminal region has an extremely flexible conformation. Recently, it has been reported that oligomerization of the regulatory peptide (CP12) with phosphoribulokinase (PRK) and GAPDH also regulates the activities of both enzymes in the cyanobacteria Synechococcus PCC7942 (Tamoi et al., 2005 ▶). Therefore, the C-terminal region of S. 7942 GAPDH might have to be flexible enough for the complicated formation of a supramolecular complex including GAPDH, CP12 and PRK.
Supplementary Material
PDB reference: NADP-GAPDH–NADP complex, 2d2i, r2d2isf
References
- Biesecker, G., Harris, J. I., Thierry, J. C., Walker, J. E. & Wonacott, A. J. (1977). Nature (London), 266, 328–333. [DOI] [PubMed] [Google Scholar]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Buchanan, B. B. (1980). Annu. Rev. Plant Physiol.31, 341–374.
- Carugo, O. & Argos, P. (1997). Proteins, 28, 10–28. [DOI] [PubMed] [Google Scholar]
- Cerff, R. (1978). Eur. J. Biochem.82, 45–53. [DOI] [PubMed] [Google Scholar]
- Charron, C., Talfournier, F., Isupov, M. N., Littlechild, J. A., Branlant, G., Vitoux, B. & Aubry, A. (2000). J. Mol. Biol.297, 481–500. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Cséke, C. & Buchanan, B. B. (1986). Biochim. Biophys. Acta, 853, 43–63.
- Duée, E., Olivier-Deyris, L., Fanchon, E., Corbier, C., Branlant, G. & Dideberg, O. (1996). J. Mol. Biol.257, 814–838. [DOI] [PubMed] [Google Scholar]
- Falini, G., Fermani, S., Ripamonti, A., Sabatino, P., Sparla, F., Pupillo, P. & Trost, P. (2003). Biochemistry, 42, 4631–4639. [DOI] [PubMed] [Google Scholar]
- Fermani, S., Ripamonti, A., Sabatino, P., Zanotti, G., Scagliarini, S., Sparla, F., Trost, P. & Pupillo, P. (2001). J. Mol. Biol.314, 527–542. [DOI] [PubMed] [Google Scholar]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kim, H., Feil, I. K., Verlinde, C. L., Petra, P. H. & Hol, W. G. (1995). Biochemistry, 34, 14975–14986. [DOI] [PubMed] [Google Scholar]
- Kissinger, C. R., Gehlhaar, D. K. & Fogel, D. B. (1999). Acta Cryst. D55, 484–491. [DOI] [PubMed] [Google Scholar]
- Korndörfer, I., Steipe, B., Huber, R., Tomschy, A. & Jaenicke, R. (1995). J. Mol. Biol.246, 511–522. [DOI] [PubMed] [Google Scholar]
- Kraulis, P. J. (1991). J. Appl. Cryst.24, 946–950. [Google Scholar]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–291. [Google Scholar]
- Mercer, W. D., Winn, S. I. & Watson, H. C. (1976). J. Mol. Biol.104, 277–283. [DOI] [PubMed] [Google Scholar]
- Merritt, E. A. & Murphy, M. E. P. (1994). Acta Cryst. D50, 869–873. [DOI] [PubMed] [Google Scholar]
- Moras, D., Olsen, K. W., Sabesan, M. N., Buehner, M., Ford, G. C. & Rossmann, M. G. (1975). J. Biol. Chem.250, 9137–9162. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., Tada, T., Wada, K., Kinoshita, T., Tamoi, M., Shigeoka, S. & Nishimura, K. (2001). Acta Cryst. D57, 879–881. [DOI] [PubMed] [Google Scholar]
- Rossmann, M. G., Liljas, A., Brändén, C. I. & Banaszak, L. J. (1975). The Enzymes, edited by P. D. Boyer, Vol. 11, pp. 61–102. New York: Academic Press.
- Scagliarini, S., Trost, P. & Pupillo, P. (1998). J. Exp. Bot.49, 1307–1315.
- Scheibe, R., Baalmann, E., Backhausen, J. E., Rak, C. & Vetter, S. (1996). Biochim. Biophys. Acta, 1296, 228–234. [DOI] [PubMed] [Google Scholar]
- Shih, M. C., Heinrich, P. & Goodman, H. M. (1991). Gene, 104, 133–138. [DOI] [PubMed] [Google Scholar]
- Skarzynski, T., Moody, P. C. & Wonacott, A. J. (1987). J. Mol. Biol.193, 171–187. [DOI] [PubMed] [Google Scholar]
- Skarzynski, T. & Wonacott, A. J. (1988). J. Mol. Biol.203, 1097–1118. [DOI] [PubMed] [Google Scholar]
- Song, S., Li, J. & Lin, Z. (1998). Acta Cryst. D54, 558–569. [DOI] [PubMed] [Google Scholar]
- Song, S. Y., Xu, Y. B., Lin, Z. J. & Tsou, C. L. (1999). J. Mol. Biol.287, 719–725. [DOI] [PubMed] [Google Scholar]
- Souza, D. H., Garratt, R. C., Araujo, A. P., Guimaraes, B. G., Jesus, W. D., Michels, P. A., Hannaert, V. & Oliva, G. (1998). FEBS Lett.424, 131–135. [DOI] [PubMed] [Google Scholar]
- Sparla, F., Fermani, S., Falini, G., Zaffagnini, M., Ripamonti, A., Sabatino, P., Pupillo, P. & Trost, P. (2004). J. Mol. Biol.340, 1025–1037. [DOI] [PubMed] [Google Scholar]
- Sparla, F., Pupillo, P. & Trost, P. (2002). J. Biol. Chem.277, 44946–44952. [DOI] [PubMed] [Google Scholar]
- Sparla, F., Zaffagnini, M., Wedel, N., Scheibe, R., Pupillo, P. & Trost, P. (2005). Plant Physiol.138, 2210–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steller, I., Bolotovsky, R. & Rossmann, M. G. (1997). J. Appl. Cryst.30, 1036–1040. [Google Scholar]
- Tamoi, M., Ishikawa, T., Takeda, T. & Shigeoka, S. (1996). Biochem. J.316, 685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamoi, M., Miyazaki, T., Fukamizo, T. & Shigeoka, S. (2005). Plant. J.42, 504–513. [DOI] [PubMed] [Google Scholar]
- Tamoi, M., Murakami, A., Takeda, T. & Shigeoka, S. (1998). Biosci. Biotechnol. Biochem.62, 374–376. [DOI] [PubMed]
- Vellieux, F. M., Hajdu, J. & Hol, W. G. (1995). Acta Cryst. D51, 575–589. [DOI] [PubMed] [Google Scholar]
- Wirtz, W., Stitt, M. & Heldt, H. W. (1982). FEBS Lett.142, 223–226.
- Wolosiuk, R. A. & Buchanan, B. B. (1976). J. Biol. Chem.251, 6456–6461. [PubMed] [Google Scholar]
- Yun, M., Park, C. G., Kim, J. Y. & Park, H. W. (2000). Biochemistry, 39, 10702–10710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: NADP-GAPDH–NADP complex, 2d2i, r2d2isf