Abstract
Background
Oil palm is the second largest source of edible oil which contributes to approximately 20% of the world's production of oils and fats. In order to understand the molecular biology involved in in vitro propagation, flowering, efficient utilization of nitrogen sources and root diseases, we have initiated an expressed sequence tag (EST) analysis on oil palm.
Results
In this study, six cDNA libraries from oil palm zygotic embryos, suspension cells, shoot apical meristems, young flowers, mature flowers and roots, were constructed. We have generated a total of 14537 expressed sequence tags (ESTs) from these libraries, from which 6464 tentative unique contigs (TUCs) and 2129 singletons were obtained. Approximately 6008 of these tentative unique genes (TUGs) have significant matches to the non-redundant protein database, from which 2361 were assigned to one or more Gene Ontology categories. Predominant transcripts and differentially expressed genes were identified in multiple oil palm tissues. Homologues of genes involved in many aspects of flower development were also identified among the EST collection, such as CONSTANS-like, AGAMOUS-like (AGL)2, AGL20, LFY-like, SQUAMOSA, SQUAMOSA binding protein (SBP) etc. Majority of them are the first representatives in oil palm, providing opportunities to explore the cause of epigenetic homeotic flowering abnormality in oil palm, given the importance of flowering in fruit production. The transcript levels of two flowering-related genes, EgSBP and EgSEP were analysed in the flower tissues of various developmental stages. Gene homologues for enzymes involved in oil biosynthesis, utilization of nitrogen sources, and scavenging of oxygen radicals, were also uncovered among the oil palm ESTs.
Conclusion
The EST sequences generated will allow comparative genomic studies between oil palm and other monocotyledonous and dicotyledonous plants, development of gene-targeted markers for the reference genetic map, design and fabrication of DNA array for future studies of oil palm. The outcomes of such studies will contribute to oil palm improvements through the establishment of breeding program using marker-assisted selection, development of diagnostic assays using gene targeted markers, and discovery of candidate genes related to important agronomic traits of oil palm.
Background
The oil palm (Elaeis guineensis Jacq.) is a perennial monocotyledonous plant which belongs to the family Arecaceae originating from West Africa. The fruit pulp and nut that provide palm and kernel oil, respectively; made oil palm a high yielding oil-producing crop [1]. At present, palm oil production is second only to that of soybean oil in terms of world vegetable oil production and the demand for palm oil is expected to increase in future. In order to meet the increasing demand for palm oil, an improvement in yield is required.
Clonal propagation of oil palm via tissue culture has been developed for mass propagation of elite planting materials. Although this approach has been widely used in the oil palm industries, the embryogenesis rate is low and a proportion of the tissue culture derived plants exhibited abnormalities. Therefore, it is important to understand the molecular events that happened during somatic embryogenesis and in vitro culture to improve the production scale and cost efficiency of the tissue culture process. In addition, the occurrence of abnormal fruit type known as mantled [2] has reduced the number of fertile fruits in palms propagated by tissue culture, thus resulting in loss of oil yield.
Root plays an important role in water and nutrient uptake from the soil. It also serves as an anchorage for plant and secretes root exudates with growth regulatory properties into the rhizosphere. Oil palm roots are usually infected by mycorrhizal fungus that assist the uptake of nutrients especially phosphate. Basal stem rot (BSR) caused by Ganoderma boniensis is a major disease in oil palm roots. The fungus attack the root of oil palm causing trunk rot. This disease remains to be the major constrain to sustainable palm oil production, causing significant yield losses either by direct loss of diseased palms or reduced yield of infected palms, in addition to requirement for earlier replanting [3]. Approximately 30–70% of oil palms are lost due to BSR by the end of each planting cycle, and the damage occurs increasingly early from one planting cycle to the next [4]. Understanding root physiology, diseases and symbiotic relationships will contribute towards the economical growth of healthy palms.
Single-pass sequencing of the 5' and/or 3' ends of randomly selected cDNA clones, is an effective approach to provide genetic information of an organism. These sequences can serve as markers or tags for transcripts, and have been used in the development of markers for reference genetic map and recovery of full-length cDNA and genomic sequences. Expressed sequence tags (ESTs) are also useful for the discovery of novel genes, investigation of genes of unknown function, comparative genomic study, and recognition of exon/intron boundaries. Currently, there are less than 3000 available oil palm sequences in the GenBank, and majority of these sequences are ESTs which had been reported by Jouannic et al. [5]. The lack of sequence information has limited the progress of gene discovery and characterisation, global transcript profiling, probe design for development of gene arrays, and generation of molecular markers for oil palm.
In this study, we have generated and analysed more than 14000 ESTs from oil palm zygotic embryos, suspension cells, shoot apical meristems, young and mature flowers, and roots. The availability of these EST sequences will allow comparative genomic studies between oil palm and other monocotyledonous and dicotyledonous plants, development of molecular markers for the establishment of reference genetic map, design and construction of cDNA microarray for global gene expression profiling.
Results
A total of six cDNA libraries were constructed from multiple oil palm tissues including zygotic embryos, suspension cell cultures, shoot apical meristems, young and mature flowers and roots; for the generation of ESTs from oil palm. The primary titer of all cDNA libraries used in this study consisted of at least 106 clones with more than 90% recombinant clones as revealed by X-Gal/IPTG screening (Table 1).
Table 1.
Summary of the oil palm cDNA libraries used in this study
| Library | Source of tissues | Titer of cDNA library (pfu/ml) | Percentage of recombinant clones (%) |
| Root | Root tissues of 3 month-old seedlings | 5.08 × 109 | 92.87 |
| Shoot apical meristem | Shoot apical meristem tissues of 6 month-old seedlings | 5.43 × 109 | 92.25 |
| Young flower | Male and female flowers of 4–6 cm | 6.4 × 109 | 90 |
| Mature flower | Female flowers of 26 cm | 3.19 × 109 | 97 |
| Suspension cell culture | Suspension cell culture | 1.3 × 1012 | 98 |
| Zygotic embryo | Zygotic embryo | 9.25 × 1010 | 90 |
In this study, two approaches were employed in the selection of cDNA clones for the generation of ESTs. In the first approach, cDNA clones were isolated randomly from each cDNA library after mass excision whereas 'cold' plaque screenings were performed in the second approach. In the latter approach, the cDNA library of suspension cell culture was hybridized with the first strand cDNA of suspension cell culture; whereas the young flower, mature flower and shoot apical meristem libraries were hybridized with the cDNA of young leaves and young or mature flowers or shoot apical meristems, respectively. Only cDNA clones that did not show positive signals in the respective hybridizations were selected for sequencing. This approach was aimed to increase the possibility of isolating rare sequences in respective cDNA libraries.
In total, 14537 ESTs from single-pass 5' sequencing of 16149 cDNA clones (GenBank: EL680967 – EL695503) passed the quality control for high confidence base call with an average read length of approximately 600 bp. Among these ESTs, 3772 were generated from 'cold' plaque screening whereas the remaining ESTs were generated from cDNA clones that were isolated randomly. The GC content of the EST sequences was approximately 48%. Approximately 56% of the sequences appeared twice or more times among the ESTs. The EST sequences comprised an estimated 2129 tentative unique contigs (TUCs, see additional file 1) and 6464 tentative unique sequences (TUSs) (Table 2).
Table 2.
Summary of ESTs from oil palm
| Number | % | |
| Total ESTs sequenced | 16149 | - |
| Number of EST sequences with readable sequence | 14537a | - |
| Redundant sequences | 8073 | - |
| Number of tentatively unique genes (TUGs) | 8593 | - |
| Number of unique sequences (TUSs, singletons) | 6464 | 75.2b |
| Number of tentatively unique contigs (TUCs) | 2129 | 24.8b |
| TUGs with significant matches | 6008 | 69.9b |
| TUGs with non-significant matches | 2585 | 30.1b |
aThis number consists of 4943 ESTs from root, 1854 from shoot apical meristem, 2623 from mature flower, 2807 from young flower, 1723 from suspension cell culture and 588 from zygotic embryo,
bPercentage of TUGs.
Among these sequences, approximately 70% had significant matches with sequences in the non-redundant protein database based on an E value cut off which was equal or less than 10-5, 20% with non-significant matches and 10% had no matches to sequences in the non-redundant protein database in GenBank. The percentages of oil palm sequences with significant matches varied from 63% in the root tissues to 78% in the oil palm suspension cell culture. Comparing the oil palm TUGs against the EST database using BLASTN demonstrated that the percentage of oil palm sequences that had significant matches was 39% based on an E value cut off which was equal or less than 10-5. Less than 4% of oil palm ESTs had no matches to the sequences in the EST database. These sequences may represent the novel sequences in oil palm.
The number of ESTs in TUCs ranged from 2 to 145 with more than 54% TUCs consisting of 2 ESTs, 18% with 3 ESTs and 25% with 4 – 12 ESTs. The top 20 most highly expressed genes (Table 3) accounted for 6 % of the sequence reads. The most widely expressed genes encoded for chaperonin 60 which was present in five cDNA libraries, whereas cyclophilin and glyceraldehyde 3-phosphate dehydrogenase were present in all six cDNA libraries. Among the predominant transcripts with more than 25 ESTs were glycine-rich RNA binding protein, alpha-tubulins, metallothionine-like proteins, PVR3-like protein, DNA J-like protein which were previously reported as transcripts that predominated the oil palm ESTs by Jouannic et al. [5]. However, other highly expressed proteins such as putative translation initiation factor and elongation factor were also identified and majority of them were involved in the housekeeping functions of cell. One of the highly expressed genes had no significant homology to the public database.
Table 3.
List of transcripts that predominate the oil palm TUCs
| Number of EST in each cDNA library | |||||||||||
| Contig ID | Putative identity | No of ESTs | Species | Accession number | E-value | Root | Shoot apical meristem | Young flower | Mature flower | Suspension cell culture | Zygotic embryo |
| Contig1765 | Metallothionein-like protein | 145 | Typha latifolia | gb|AAK28022.1 | 1E-60 | 50 | 8 | 35 | 39 | 13 | 0 |
| Contig962 | Glycine-rich RNA binding protein 2 | 59 | Pelargonium × hortorum | gb|AAB63581.1 | 8E-36 | 6 | 8 | 22 | 20 | 1 | 2 |
| Contig1185 | Putative translation initiation factor eIF-1A-like | 57 | Solanum tuberosum | gb|ABB55392.1 | 8E-51 | 21 | 3 | 16 | 17 | 0 | 0 |
| Contig1589 | Alpha-tubulin | 49 | Prunus dulcis | emb|CAA47635.1 | 0.0 | 5 | 6 | 9 | 28 | 1 | 0 |
| Contig447 | Cyclophilin | 43 | Ricinus communis | emb|CAC80550.1 | 6E-77 | 16 | 5 | 11 | 7 | 2 | 2 |
| Contig1818 | Type 2 metallothionein-like protein | 43 | Typha angustifolia | gb|ABQ14530.1 | 2E-10 | 41 | 0 | 0 | 0 | 2 | 0 |
| Contig1809 | Putative translation elongation factor eEF-1 beta' chain | 42 | Oryza sativa | dbj|BAC22427.2 | 3E-63 | 7 | 1 | 10 | 24 | 0 | 0 |
| Contig1384 | Translationally controlled tumor protein | 42 | Elaeis guineensis | gb|AAQ87663.1 | 7E-88 | 9 | 7 | 14 | 9 | 0 | 3 |
| Contig165 | Putative OsCTTP | 41 | Oryza sativa | dbj|BAD19560.1 | 5E-84 | 17 | 0 | 21 | 3 | 0 | 0 |
| Contig1595 | Type 2 metallothionein-like protein | 41 | Typha latifolia | gb|AAK28022.1 | 8E-11 | 2 | 6 | 12 | 21 | 0 | 0 |
| Contig2050 | hypothetical protein OsI_022576 | 37 | Oryza sativa | gb|EAZ01344.1 | 4E-45 | 17 | 1 | 4 | 15 | 0 | 0 |
| Contig1214 | PVR3-like protein | 36 | Ananas comosus | gb|AAM28295.1 | 2E-17 | 14 | 9 | 3 | 10 | 0 | 0 |
| Contig268 | DnaJ-like protein | 35 | Oryza sativa | dbj|BAD25681.1 | 3E-177 | 4 | 14 | 9 | 5 | 3 | 0 |
| Contig614 | Early-methionine-labelled polypeptide | 32 | Elaeis guineensis | gb|ABD66069.1 | 1E-43 | 0 | 0 | 0 | 0 | 0 | 32 |
| Contig2086 | Cationic peroxidase 2 | 30 | Glycine max | gb|AAC83463.1 | 1.E-155 | 3 | 0 | 5 | 10 | 12 | 0 |
| Contig80 | Glyceraldehyde 3-phosphate dehydrogenase | 29 | Magnolia quinquepeta | emb|CAA42905.1 | 2E-1744 | 2 | 5 | 4 | 4 | 12 | 2 |
| Contig1135 | Hypothetical protein CBG17156 | 29 | Caenorhabditis briggsae | emb|CAE70527.1 | 0.22 | 9 | 14 | 0 | 0 | 5 | 1 |
| Contig449 | T6D22.2 | 27 | Arabidopsis thaliana | gb|AAF79822.1 | 0.0 | 2 | 9 | 10 | 6 | 0 | 0 |
| Contig584 | Myo-inositol-1-phosphate synthase | 26 | Nicotiana tabacum | dbj|BAA95788.1 | 0.0 | 0 | 3 | 12 | 11 | 0 | 0 |
| Contig1454 | Alpha-tubulin | 26 | Prunus dulcis | emb|CAA47635.1 | 0.0 | 7 | 8 | 4 | 6 | 1 | 0 |
The most highly expressed genes in each cDNA library were listed in Table 4. The number of unique TUGs in each oil palm cDNA library that did not overlap with TUGs from other oil palm cDNA libraries were 2654, 940, 1193, 1094, 1027 and 299 for root, apical shoot meristem, young flower, mature flower, suspension cell culture and zygotic embryo, respectively. However, there were only 27 differentially expressed TUCs in multiple cDNA libraries according to the R statistic [6] with Bonferroni correction at the significance threshold of 2.35 × 10-5 (Table 5). Among these are a few TUCs that were predominant in individual cDNA libraries such as acidic class III chitinase, thaumatin-like protein 1 and 1-aminocyclopropane-1-carboxylic acid oxidase in the suspension cell culture; early methionine-labelled polypeptides, 7S globulin, dehydrin-like protein, embryogenesis abundant protein D-34 and chaperone in the zygotic embryo; and putative flavonol 3-sulfotransferase STF-1 in the young flowers. In addition, the results also demonstrated higher copy number of certain transcripts in both young and mature flower such as myo-inositol 1-phosphate synthase, alpha tubulin, translation elongation factor eEF-1, glycine-rich RNA binding protein and polyphenol oxidase. Both root and flower tissues (young and mature flowers) had high copy number of transcripts encoding for type 2 metallothionein-like proteins, however, the nucleotide sequences of these two contigs were different.
Table 4.
List of transcripts that predominate in different oil palm tissues
| Putative identity | No of ESTs | Species | Accession number | E-value |
| Root | ||||
| Type 2 metallothionein-like protein | 93 | Typha latifolia | gb|AAL09705.1 | 9E-10 |
| Sucrose synthase | 21 | Oncidium cv. 'Goldiana' | gb|AAM95943.1 | 0.0 |
| Putative translation initiation factor eIF-1A-like | 21 | Solanum tuberosum | gb|ABB55392.1 | 4E-51 |
| Putative OsCTTP | 17 | Oryza sativa | ref|XP_465531.1 | 3E-84 |
| OSJNBa0006M15.20 | 17 | Oryza sativa | ref|XP_472724.1 | 2E-45 |
| Cyclophilin | 16 | Ricinus communis | emb|CAC80550.1 | 2E-77 |
| Cytochrome b5 domain-containing protein-like | 15 | Oryza sativa | ref|XP_468235.1 | 4E-16 |
| PVR3-like protein | 14 | Ananas comosus | gb|AAM28295.1 | 6E-18 |
| Putative ubiquitin-conjugating enzyme | 11 | Oryza sativa | dbj|BAD34325.1 | 3E-79 |
| Shoot apical meristem | ||||
| rRNA intron-encoded homing endonuclease | 16 | Pan troglodytes | ref|XP_525925.1| | 8E-07 |
| DnaJ-like protein | 15 | Oryza sativa | emb|CAC39071.1 | E-178 |
| Hypothetical protein CBG17156 | 14 | Caenorhabditis briggsae | emb|CAE70527.1 | 0.13 |
| Putative fiber protein Fb2 | 10 | Oryza sativa | ref|XP_465147.1 | 1E-56 |
| Ethylene response factor | 9 | Manihot esculenta | gb|AAX84670.1 | 9E-73 |
| Hypothetical protein FG07171.1 | 9 | Gibberella zeae PH-1 | gb|EAA76630.1 | 6.8 |
| Type 2 metallothionein-like protein | 9 | Typha latifolia | gb|AAL09705.1 | 2E-20 |
| Annexin p33 | 9 | Zea mays | emb|CAA66900.2 | E-116 |
| Alpha-tubulin | 9 | Prunus dulcis | emb|CAA47635.1 | E-124 |
| Glycine-rich RNA binding protein 2 | 8 | Pelargonium × hortorum | gb|AAB63582.1 | 4E-36 |
| PVR3-like protein | 8 | Ananas comosus | gb|AAM28295.1 | 8E-18 |
| Drought-induced protein like | 8 | Arabidopsis thaliana | emb|CAB10370.1 | 4E-10 |
| Young flowers | ||||
| Cytoplasmic ribosomal protein S15a | 45 | Daucus carota | gb|AAK30203.1 | 4E-67 |
| Glycine-rich RNA binding protein 2 | 22 | Pelargonium × hortorum | gb|AAB63582.1 | 5E-36 |
| Putative OsCTTP | 21 | Oryza sativa | ref|XP_465531.1 | 3E-84 |
| Putative translation initiation factor eIF-1A-like | 17 | Solanum tuberosum | gb|ABB55392.1 | 4E-51 |
| Translationally controlled tumor protein | 14 | Elaeis guineensis | gb|AAQ87663.1 | 3E-88 |
| Putative STF-1 | 12 | Oryza sativa | dbj|BAD31135.1 | 9E-67 |
| Type 2 metallothionein-like protein | 12 | Typha latifolia | gb|AAL09705.1 | 6E-11 |
| Ribosomal protein L15 | 11 | Oryza sativa | ref|NP_909841.1 | 8E-92 |
| Polyphenol oxidase | 11 | Vitis vinifera | emb|CAA81798.1 | E-105 |
| Cyclophilin | 11 | Ricinus communis | emb|CAC80550.1 | 2E-77 |
| Mature flower | ||||
| Type 2 metallothionein-like protein | 60 | Typha latifolia | gb|AAL09705.1 | 8E-11 |
| Alpha-tubulin | 30 | Prunus dulcis | emb|CAA47635.1 | 0.0 |
| Putative translation elongation factor eEF-1 beta' chain | 24 | Oryza sativa | ref|NP_910927.2 | 5E-64 |
| Glycine-rich RNA binding protein 2 | 20 | Pelargonium × hortorum | gb|AAB63582.1 | 4E-36 |
| Putative translation initiation factor eIF-1A-like | 18 | Solanum tuberosum | gb|ABB55392.1 | 4E-51 |
| Similar to mucin 17 | 17 | Rattus norvegicus | ref|XP_578244.1 | 0.004 |
| OSJNBa0006M15.20 | 15 | Oryza sativa | ref|XP_472724.1 | 2E-45 |
| Cationic peroxidase 2 | 10 | Glycine max | gb|AAC83463.1 | E-155 |
| Suspension cell culture | ||||
| Lipid transfer protein homolog | 21 | Triticum aestivum | gb|AAB32995.1 | 3E-25 |
| Cationic peroxidase 2 | 12 | Glycine max | gb|AAC83463.1 | E-153 |
| Glyceraldehyde 3-phosphate dehydrogenase | 12 | Magnolia quinquepeta | emb|CAA42905.1 | E-174 |
| Hypothetical protein | 10 | Oryza sativa | dbj|BAD87021.1 | 7E-27 |
| Ubiquitin | 10 | Antirrhinum majus | emb|CAA48140.1 | E-121 |
| Non-symbiotic hemoglobin class 1 | 9 | Malus × domestica | gb|AAP57676.1 | 2E-67 |
| Putative beta-expansin | 9 | Triticum aestivum | dbj|BAD06319.1 | 2E-72 |
| TAPETUM DETERMINANT 1 | 9 | Arabidopsis thaliana | ref|NP_974612.1 | 4E-32 |
| 1-Aminocyclopropane-1-carboxylic acid oxidase | 9 | Elaeis guineensis | gb|AAP13098.1 | E-168 |
| Thaumatin-like protein precursor | 7 | Malus × domestica | gb|AAC36740.1 | 2E-82 |
| Zygotic embryo | ||||
| Early-methionine-labelled polypeptide | 46 | Secale cereale | emb|CAB88095.1 | 2E-35 |
| 7S globulin | 21 | Elaeis guineensis | gb|AAK28402.1 | 0.0 |
| Dehydrin-like protein | 10 | Elaeis guineensis | gb|AAF60172.1 | 1E-26 |
| Late embryogenesis abundant protein D-34 | 9 | Gossypium hirsutum | sp|P09444 | 4E-76 |
| Protein, small heat shock | 6 | Codonopsis lanceolata | gb|AAW02791.1 | 1E-23 |
| Seed maturation protein LEA 4 | 4 | Glycine tomentella | gb|AAG37452.1 | 6E-24 |
| Translationally controlled tumor protein | 4 | Elaeis guineensis | gb|AAQ87663.1 | 3E-88 |
| Ribosomal protein S27-like protein | 4 | Solanum tuberosum | gb|ABA40465.1 | 8E-35 |
| Class II metallothionein | 4 | Zea mays | emb|CAA84233.1 | 5E-17 |
Table 5.
Differentially expressed genes with the top hits (R values) in multiple cDNA libraries of oil palm
| Number of EST in each cDNA library | ||||||||||
| Contig ID | Description | E-value | Organism | Root | Shoot apical Meristem | Young Flower | Mature Flower | Suspension cell culture | Zygotic embryo | R value |
| Contig165 | Putative OsCTTP | 2.93E-84 | ref|XP_507481.1Oryza sativa | 17 | 0 | 21 | 3 | 0 | 0 | 0 |
| Contig1512 | Hypothetical protein XP_524016 | 1.00E-170 | ref|XP_524016Pan troglodytes | 0 | 0 | 0 | 20 | 1 | 0 | 0 |
| Contig584 | Myo-inositol-1-phosphate synthase | 1.00E-170 | dbj|BAA95788.1Nicotiana tabacum | 0 | 3 | 12 | 11 | 0 | 0 | 0 |
| Contig1589 | Alpha-tubulin | 1.00E-11 | emb|CAA47635.1Prunus dulcis | 5 | 6 | 9 | 28 | 1 | 0 | 0 |
| Contig1595 | Type 2 metallothionein-like protein | 1.00E-07 | gb|ABQ14530.1Typha angustifolia | 2 | 6 | 12 | 21 | 0 | 0 | 0 |
| Contig504 | Acidic class III chitinase OsChib3a | 1.00E-94 | NP_917360.1Oryza sativa | 0 | 0 | 0 | 0 | 13 | 0 | 0 |
| Contig874 | 1-Aminocyclopropane-1-carboxylic acid oxidase | 1.00E-168 | gb|AAP13098.1Elaeis guineensis | 0 | 0 | 0 | 0 | 9 | 0 | 0 |
| Contig1135 | Hypothetical protein | 0.130103 | emb|CAE70527.1Caenorhabditis briggsae | 9 | 14 | 0 | 0 | 5 | 1 | 0 |
| Contig1435 | rRNA intron-encoded homing endonuclease | 1.00E-08 | ref|XP_525925.1Pan troglodytes | 0 | 14 | 1 | 1 | 0 | 0 | 0 |
| Contig1809 | Translation elongation factor eEF-1 beta' chain | 1.00E-65 | ref|XP_506540.1Oryza sativa | 7 | 1 | 10 | 24 | 0 | 0 | 0 |
| Contig1818 | Type 2 metallothionein-like protein | 1.00E-10 | gb|AAL09705.1Typha latifolia | 41 | 0 | 0 | 0 | 2 | 0 | 0 |
| Contig614 | Early-methionine-labelled polypeptide | 1.00E-170 | emb|CAB88095.1Secale cereale | 0 | 0 | 0 | 0 | 0 | 32 | 0 |
| Contig611 | Early-methionine-labelled polypeptide | 1.00E-36 | emb|CAB88095.1Secale cereale | 0 | 0 | 0 | 0 | 0 | 12 | 0 |
| Contig634 | 7S globulin | 1.00E-25 | gb|AAK28402.1Elaeis guineensis | 0 | 0 | 0 | 0 | 0 | 21 | 0 |
| Contig1007 | Dehydrin-like protein | 1.00E-26 | gb|AAF60172.1Elaeis guineensis | 0 | 0 | 0 | 0 | 2 | 10 | 0 |
| Contig1024 | Embryogenesis abundant protein D-34 (LEA D-34) | 1.00E-76 | sp|P09444Gossypium hirsutum | 0 | 0 | 0 | 0 | 0 | 9 | 0 |
| Contig1097 | Chaperone | 5E-26 | gb|ABF61872.1Agave tequilana | 0 | 0 | 0 | 0 | 0 | 6 | 0 |
| Contig530 | Polyubiquitin | 3E-121 | gb|ABU40645.1Triticum aestivum | 2 | 0 | 0 | 0 | 10 | 0 | 0.000001 |
| Contig962 | Glycine-rich RNA binding protein 2 | 1.00E-36 | gb|AAB63582.1Pelargoniu × hortorum | 6 | 8 | 22 | 20 | 1 | 2 | 0.000001 |
| Contig775 | Polyphenol oxidase | 1.00E-105 | emb|CAA81798.1Vitis vinifera | 0 | 0 | 12 | 3 | 0 | 0 | 0.000002 |
| Contig701 | Unknown protein | 1.00E-38 | gb|ABF59206.1Arabidopsis thaliana | 0 | 0 | 2 | 0 | 9 | 0 | 0.000002 |
| Contig2113 | Putative STF-1 | 1.00E-72 | dbj|BAD31135.1Oryza sativa | 1 | 0 | 12 | 0 | 0 | 0 | 0.000002 |
| Contig2086 | Cationic peroxidase 2 | 1.00E-156 | gb|AAC83463.1Glycine max | 3 | 0 | 5 | 10 | 12 | 0 | 0.000004 |
| Contig546 | Non-symbiotic hemoglobin class 1 | 1.00E-67 | gb|AAP57676.1Malus × domestica | 3 | 0 | 0 | 0 | 9 | 0 | 0.000008 |
| Contig553 | OSJNBa0010H02.8 | 1.00E-73 | gb|AAK84683.1Oryza sativa | 1 | 0 | 0 | 0 | 8 | 0 | 0.000015 |
| Contig659 | Thaumatin-like protein 1 – apple tree | 1.00E-82 | gb|AAC36740.1Malus × domestica | 0 | 0 | 0 | 0 | 7 | 0 | 0.000016 |
More detailed functional annotation was performed by mapping tentative unique genes (TUGs) to the Gene Ontology Consortium structure which provides a structured and controlled vocabulary to describe gene products according to three ontologies: cellular components, biological processes and molecular functions. The GO classifications of TUGs from oil palm were summarized in Table 6, according to their involvement in various biological processes, molecular functions and cellular localization. In total, 2361 TUGs could be mapped to one or more ontologies, 2376 assignments were made to the category of molecular function (level 3), with approximately 42% in binding (including nucleotide, ion, nucleic acid, protein, and cofactor binding); 41% in catalytic activities (including transferase, hydrolase and oxidoreductase activities); 7% in structural constituents of ribosome; and 5% in transport activities (ion and carrier transport). Under the category of biological process, 4583 assignments (level 4) were made to cellular metabolism (23%), primary metabolism (21%), macromolecule metabolism (16%) and biosynthesis (9%). Finally, for cellular components, the vast majority of 1948 assignments (level 4) were assigned to the intracellular (40%) and intracellular parts (38%). The GO assignments of TUGs from each cDNA library were also summarized in Table 6. In general, the main GO categories assigned to TUGs from individual cDNA libraries were similar to that of the overall analysis mentioned above. The minor differences in percentage may be non-significant as only some of the TUGs were shown to be differentially expressed in multiple tissues (Table 5).
Table 6.
Gene Ontology (GO) classifications of TUGs from oil palm according to their involvement in biological processes, molecular functions and cellular localizations
| Percentage in each cDNA library (%) | |||||||
| Gene ontology (GO) classification | Root | Shoot apical meristem | Young flower | Mature flower | Suspension cell culture | Zygotic embryo | Overall |
| Biological processes | |||||||
| Biosynthesis | 9 | 10 | 9 | 9 | 7 | 13 | 9 |
| Catabolism | 2 | 1 | 2 | 2 | 4 | 3 | 2 |
| Cell organization and biogenesis | 3 | 3 | 4 | 4 | 3 | 3 | 3 |
| Cellular metabolism | 23 | 23 | 23 | 23 | 22 | 22 | 23 |
| Establishment of localization | 7 | 6 | 5 | 5 | 7 | 5 | 7 |
| Macrmolecule metabolism | 16 | 17 | 18 | 19 | 17 | 19 | 16 |
| Nitrogen compound metabolism | 2 | 2 | 1 | 1 | 2 | 1 | 2 |
| Primary metabolism | 21 | 22 | 22 | 22 | 21 | 22 | 21 |
| Protein localization | 1 | 1 | 1 | 1 | 1 | 3 | 1 |
| Regulation of cellular physiological process | 2 | 2 | 2 | 2 | 2 | 1 | 2 |
| Regulation of metabolism | 2 | 2 | 2 | 2 | 2 | < 1 | 2 |
| Response to chemical stimulus | 1 | 2 | 1 | 1 | 1 | 1 | 1 |
| Transport | 7 | 6 | 5 | 5 | 6 | 5 | 7 |
| Others | 4 | 3 | 5 | 4 | 5 | 2 | 4 |
| Molecular function | |||||||
| Antioxidant activity | 1 | 1 | 1 | 2 | 3 | 1 | 1 |
| Binding | |||||||
| Cofactor binding | 2 | 2 | 1 | 2 | 2 | 3 | 2 |
| Ion binding | 3 | 9 | 11 | 12 | 12 | 13 | 11 |
| Nucleic acid binding | 9 | 15 | 15 | 13 | 11 | 12 | 11 |
| Nucleotide binding | 11 | 15 | 15 | 12 | 9 | 11 | 13 |
| Protein binding | 6 | 6 | 8 | 5 | 6 | 4 | 5 |
| Catalytic activity | |||||||
| Hydrolase activity | 11 | 8 | 10 | 10 | 10 | 8 | 11 |
| Isomerase activity | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Ligase activity | 3 | 2 | 3 | 2 | 2 | 1 | 2 |
| Lyase activity | 3 | 2 | 1 | 2 | 3 | 3 | 3 |
| Oxidoreductase activity | 9 | 7 | 5 | 7 | 11 | 5 | 9 |
| Peroxidase activity | 1 | 1 | 1 | 1 | 2 | 0 | 1 |
| Transferase activity | 10 | 10 | 10 | 9 | 11 | 11 | 13 |
| Structural constituent of ribosome | 9 | 9 | 9 | 11 | 6 | 14 | 7 |
| Translation regulator activity | 1 | 3 | 3 | 2 | 1 | 4 | 2 |
| Transcription regulator activity | 1 | 2 | 1 | 1 | 2 | 0 | 1 |
| Transport | |||||||
| Carrier activity | 2 | 2 | 1 | 1 | 2 | 1 | 2 |
| Ion transport | 3 | 2 | 1 | 1 | 2 | 1 | 3 |
| Others | 13 | 2 | 2 | 5 | 3 | 7 | 1 |
| Cellular component | |||||||
| External encapsulating structure | < 1 | 1 | 1 | 1 | 1 | 0 | 1 |
| Intracellular | 39 | 43 | 42 | 43 | 40 | 39 | 40 |
| Intracellular part | 37 | 41 | 40 | 42 | 39 | 38 | 38 |
| Membrane | 12 | 8 | 9 | 7 | 11 | 11 | 11 |
| Membrane bound vesicle | 1 | 1 | 1 | 1 | 2 | 1 | 1 |
| Membrane part | 7 | 6 | 5 | 5 | 6 | 6 | 7 |
| Organelle inner membrane | 1 | < 1 | 1 | < 1 | < 1 | 1 | 1 |
| Others | 3 | < 1 | 1 | 1 | 1 | 4 | 1 |
Table 7 provides a list of TUGs from oil palm that are homologous to genes known to be involved in flower development. The ESTs related to oil palm flowering included CONSTANS-like protein, GIGANTEA-like protein, AGL20-like MADS box transcriptional factor, LFY-like protein, SQUAMOSA protein, SQUAMOSA promoter binding protein, AP domain containing proteins and aintegumenta-like protein. These proteins have been reported to be involved in flowering time, determination of floral identities and development of floral organs. Majority of these proteins were the first representatives of their gene families from oil palm. The results demonstrated that EST approach was successful in uncovering homologues of many (90 or more) putative floral regulatory genes, supporting the hypothesis that this regulatory pathway is largely conserved in angiosperms. Some of these gene families were not exclusively expressed in young and mature flowers, but were also expressed in other tissues. For example, AP2 domain containing protein was expressed in zygotic embryo and root tissues as well. However, 5 and 11 MADS box proteins were found in young and mature flowers respectively; 7 shoot-meristemless protein, 5 ZF-HD homeobox and 3 AGL20 in young flowers; and 10 knotted -like homeobox protein in both floral libraries. Functional elucidation of these proteins may shed light on the flowering process of oil palm. Two of these cDNA candidates, EgSBP (GenBank: EL682671) and EgSEP (GenBank: EL686357) isolated from young and mature flowers respectively; were further characterised by Northern analysis.
Table 7.
List of transcripts from oil palm that are homologous to genes involved in flower development and formation
| Number of ESTs in | |||||||
| Gene | Total EST | Root | Shoot apical meristem | Mature flower | Young flower | Suspension cell culture | Zygotic embryo |
| AGL2 | 8 | 0 | 0 | 8 | 0 | 0 | 0 |
| AGL20 | 3 | 0 | 0 | 0 | 3 | 0 | 0 |
| Putative argonaute protein | 4 | 0 | 2 | 0 | 0 | 2 | 0 |
| AP2 domain containing protein | 5 | 2 | 0 | 1 | 0 | 0 | 2 |
| CONSTANS-like protein 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| Early flowering protein 1 | 3 | 1 | 0 | 1 | 0 | 1 | 0 |
| Floral organ regulator 2 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| Flowering promoting factor-like 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Putative gigantea | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| Knotted1-like homeobox protein | 11 | 0 | 1 | 3 | 7 | 0 | 0 |
| LFY-like protein | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| Myb-like protein | 15 | 3 | 1 | 1 | 6 | 3 | 1 |
| NAC family protein | 11 | 2 | 3 | 0 | 0 | 6 | 0 |
| NAM-like protein | 2 | 1 | 0 | 0 | 1 | 0 | 0 |
| Putative GAMYB-binding protein | 4 | 1 | 3 | 0 | 0 | 0 | 0 |
| Putative LHY protein | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| Scarecrow-like protein | 5 | 1 | 2 | 0 | 0 | 2 | 0 |
| Shoot meristemless-like protein | 8 | 0 | 0 | 0 | 7 | 1 | 0 |
| SQUAMOSA protein | 7 | 2 | 0 | 3 | 2 | 0 | 0 |
| SQUAMOSA binding protein | 7 | 1 | 0 | 2 | 4 | 0 | 0 |
| STYLOSA protein | 2 | 1 | 0 | 0 | 0 | 1 | 0 |
| YABBY-like transcription factor | 5 | 3 | 1 | 1 | 0 | 0 | 0 |
| ZF-HD homeobox protein | 7 | 1 | 1 | 0 | 5 | 0 | 0 |
In order to study the temporal expression of EgSBP and EgSEP, blots were prepared from RNA extracted from different stages of normal and abnormal flower representing early, mid and late period of flowering. The transcripts of EgSBP were detected in normal male flower of 1.8 cm and above. Figure 1 also shows low transcript levels of EgSBP in the abnormal flower of 1.5 cm and the signal intensities increased gradually during the subsequent developmental stages until the size of the flower reached 11 cm. On the other hand, EgSEP was expressed during late floral development and no expression was detected in the shoot apex (Figure 2). In the abnormal female flower, it was first detected at very low levels in flowers of 18 cm and the expression gradually increased as flower development progressed. The expression was at its highest in flowers of 35 cm (Figure 2). In the normal male flower, EgSEP was first detected in flowers of 19 cm (data not shown).
Figure 1.
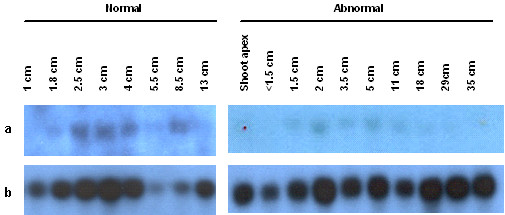
Transcriptional expression of EgSBP (a) in normal and abnormal male flowers of various sizes with cyclophilin (b) as a loading reference.
Figure 2.
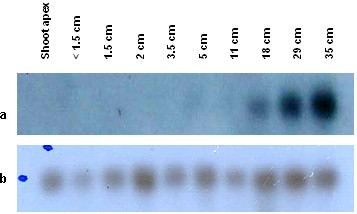
Transcriptional expression of EgSEP (a) in abnormal female flowers of various sizes with cyclophilin (b) as a loading reference.
In addition to the flowering related genes, we also surveyed a few well-characterised biological processes and metabolic pathways to determine the extent to which such pathways were represented within the TUGs of oil palm. Important enzymes that are involved in the oil biosynthesis were represented by ESTs encoding for acetyl-CoA carboxylase, malonyl-CoA: ACP transacylase, beta-ketoacyl-ACP synthase, beta-ketoacyl-ACP dehydratase, enoyl ACP reductase, enoyl ACP hydratase, palmitoyl protein thioesterase and desaturases such as steroyl-ACP desaturase, omega 3 desaturase and omega 6 desaturase. Besides, there were also ESTs encoding for enzymes in the ascorbate-glutathione cycle including ascorbate peroxidase, monohydroascorbate reductase, dehydroascorbate reductase, glutathione reductase, glutathione S-transferase, glutathione peroxidase, Cu/Zn-superoxide dismutase and catalase; and enzymes involved in nitrogen utilization such as nitrate reductase, nitrate transporter, glutamine synthase, glutamate synthase and asparagine synthase.
Discussion
In this study, we have analysed a complex data set of oil palm transcripts to gain insight into the gene content and to provide a preliminary assessment of the transcript and gene expression profile of this crop. The number of ESTs generated is more than 4.8 fold of the total number of entries for oil palm in GenBank. Despite the fact that more number of ESTs are required in order to have a good chance of finding any gene that is of interest, this may take probably several years before this number can be achieved. We have decided to report the existing number of ESTs as this sequence information is important to the science community since the oil palm genome has not been fully sequenced and the data is not currently available in the public databases.
Oil palm was represented by approximately 3000 sequences in the NCBI database prior to this study. Much of the previous EST sequencing in oil palm has focused on cDNAs derived from zygotic embryos, shoot apical meristems and flowers consistent with the overriding interest of the oil palm industry in yield improvement through clonal propagation of elite planting materials and reduction of losses attributed to abnormal epigenetic flowering. In this study, we have generated ESTs from oil palm tissues that were not covered by other research groups, most of the transcripts uncovered are the first representatives for oil palm especially those sequences from root and suspension cell culture. Eventhough the number of ESTs from flowers, shoot apical meristem and suspension cell culture was less, close to 50% of them were generated from 'cold' plaque screening. This approach increased the possibility of sequencing rare transcripts and tissue-specific ESTs. Overall, the GC content of the sequences is approximately 48 % which is close to the value reported by Jouannic et al. [5].
Since the oil palm materials used in this study were sampled from Dura × Pisifera hybrids developed by different companies and institution, there are possibilities that variations (including single nucleotide polymorphisms) may occur among the sequences encoding for the same transcript. Our preliminary screening of a few contigs assembled from high number of ESTs demonstrated some minor variations among the sequences. However, these sequences may have to be verified by resequencing to exclude any sequencing errors. In addition, we have also detected sequence variation in cDNAs encoding for type 2 metallothionein-like proteins in flower and root tissues, respectively. These EST sequences may serve as a source for tissue-specific markers or probes for the recovery of tissue-specific promoters.
In this study, we have surveyed several well-characterised biological processes and metabolic pathways to determine the extent to which such pathways were represented within the TUGs of oil palm. Our findings showed that the oil biosynthesis, ascorbate-glutathione cycle, nitrogen utilization and flowering pathways were well represented by ESTs encoding for some of the important proteins involved. We have also demonstrated that this EST data can be utilized not only for gene discovery but also for comparative analysis of gene expression within/between oil palm tissues. For example, methionine-labelled polypeptide (which also showed high identities to late embryogenesis abundant protein), 7S globulin, dehydrin-like protein and embryogenesis abundant protein D-34 that were predominant in zygotic embryos, were absent in suspension cell culture which was predominated by the transcripts of acidic class III chitinase and 1-aminocyclopropane-1-carboxylic acid oxidase. The differentially expressed genes in zygotic embryos indicated that they might be in a later developmental stage than the suspension cell culture although both tissues were embryogenic. These ESTs may have the potential to be developed as molecular markers for diagnostic assays, especially for tissue cultures. Besides, we also demonstrated that the transcripts encoding for polyphenol oxidase and flavonol 3-sulfotransferase were in high copy number in flowers. It is not surprising since both enzymes are involved in the flower coloration. In summary, our results showed that the differentially expressed transcripts reflected the physiological and developmental states of the oil palm tissues used for the construction of cDNA libraries. This information also provides a preliminary assessment of the gene expression profile of these genes in different oil palm tissues.
The oil palm homologue for SEPALLATA, EgSEP, was further characterized by northern analysis. SEPALLATA was shown to be required for petal, stamen and carpel identities thus necessary for the activities of the B and C function genes [7]. EgSEP is the most abundant transcription factor among the oil palm ESTs (7 clones) involved in flower development. It was expressed in normal male flowers with fully developed anthers and pollens (data not shown), and also in abnormal male flowers with fully developed supernumerary carpels [8]. However, EgSEP was not able to differentiate normal male flowers from abnormal male flowers with carpel-like structures. The results were consistent with the expression of oil palm SEPALLATA in both female and male flowers reported by Adam et al. [9].
EgSBP is a putative homologue for SQUAMOSA binding protein (SBP) which binds to the promoter of SQUAMOSA at unique sequence motif and involves in the control of early flower development [10]. EgSBP was first expressed in normal male oil palm flowers of 1.8 cm of which bracts have already developed around the rachillae. At this stage, the male rachillae can be differentiated from the female rachillae by the number and shape of the bracts. The expression of EgSBP increased in the subsequent flower developmental stages during the emergence of flower primordia (2 – 3 cm), emergence of male flowers in male rachillae (3 – 5 cm), and development of stamens (5 – 7 cm) and anther (7 – 9 cm). In abnormal flower, the development of supernumerary carpels occurs when the inflorescence is 5 – 7 cm [8]. Stamens of abnormal male flower develops into carpel-like structures at 7 cm and above. However, our results showed that the transcript profiles of EgSBP in normal and abnormal male flowers were similar (Figure 1). The onset of transcription of EgSBP is consistent with the findings of Cardon et al. [11] that some of the SBP genes were constitutively expressed during flower development. On the other hand, ESTs for SBP-like proteins were also identified in the root and mature flower tissues. It is not surprising as its target gene, SQUAMOSA, was also found to be expressed in vegetative tissue and did not have an expression pattern specific to either the male or female inflorescences [9].
Interestingly, the transcript for GIGANTEA – a circadian clock controlled gene that regulates photoperiodic flowering [12] was found in oil palm root instead of floral tissues. The role of this protein in root has not been characterised. On the other hand, the putative homologue for LHY which was proposed to function either within the circadian oscillator or in its output pathways, was found only in young flowers. In addition, ESTs for LFY-like, and CONSTANS-like, AGL20, SQUAMOSA proteins were also uncovered among the ESTs related to flowering. Among these sequences, only the cDNA sequence of SQUAMOSA have been isolated and examined in the vegetative and reproductive tissues of oil palm [9]. Further characterisation of these genes may enhance our understanding of the flowering pathway of oil palm which is largely unknown.
Conclusion
The EST sequences generated in this study will be a source of gene-targeted and tissue-specific markers. The data will also facilitate the fabrication of DNA array for future studies of oil palm. Transcript profiling through microarray could further contribute towards the understanding of fundamental aspects in oil palm biology such as transcriptional responses correlated with embryogenesis, abnormal flowering, diseases etc. The outcomes of such studies will contribute to oil palm improvements through the establishment of breeding program using marker-assisted selection, development of diagnostic assays using gene-targeted markers, recovery of genomic sequences and discovery of candidate genes related to important agronomic traits of oil palm.
Methods
Plant materials
The oil palm materials (Elaeis guineensis Jacq.) used in this study were summarized in Table 1. Oil palm suspension cell cultures and shoot apical meristems from 6-month old oil palm seedlings were kindly provided by Applied Agricultural Research Sdn Bhd., Sungai Buloh, Malaysia; whereas the root tissues were obtained from 3-month old oil palm seedlings from Guthrie Sdn Bhd., Seremban, Malaysia. The young flowers (4–6 cm), mature flowers (26 cm) and zygotic embryos used in this study were provided by Malaysian Palm Oil Board, Bangi, Malaysia. All of the above materials were sampled from Dura × Pisifera hybrids developed by respective institution and companies.
RNA extraction
RNA from oil palm shoot apical meristem tissues, young (male and female) and mature (female) flowers was extracted by using the method described by Rochester et al. [13], whereas RNA from oil palm suspension cells and zygotic embryos was extracted by using the method described by Schultz et al. [14]. CsCl gradient [15] was used to extract RNA from root tissues.
cDNA library construction
Poly A+ RNA isolation was performed using PolyA Tract Isolation System (Promega, USA) or μ MACS mRNA isolation kit (Miltenyi Biotec, Germany) according to the manufacturers' instructions. cDNA was prepared using a cDNA synthesis kit (Stratagene, USA) and directionally cloned into Uni-Zap vector (Stratagene, USA). The resulting primary cDNA library was amplified to more than 109 pfu/ml. Plasmids (pBluescript SK-) containing cDNA inserts were in vivo and mass-excised from phage stocks using ExAssist helper phage and propagated in Escherichia coli SOLR cells according to the manual provided by manufacturer.
'Cold' plaque screening
The cDNA library of suspension culture was screened with a 32P-labelled probe prepared from the first-strand cDNA of suspension culture. The young and mature female flower libraries were screened twice with 32P-labelled probes prepared from the cDNA of young leaves and young or mature flowers, respectively. The shoot apical meristem library was screened with cDNA from young leaves and shoot apical meristems, respectively. These probes were prepared from PCR amplified cDNA using SMART cDNA synthesis kit (Clontech, USA). Hybridization was performed in 5 × SSPE, 5 × Denhardt's solution, 0.5% (w/v) SDS, 100 μg/ml denatured herring sperm DNA at 60°C. Membranes were washed in 2 × SSC, 0.1% SDS at room temperature twice for 10 minutes each before the membranes were exposed to autoradiography films at -80°C. The 'cold' plaques that did not show any signals upon secondary screening were in vivo excised.
Nucleotide sequencing
The cDNA clones were cultured for plasmid DNA preparation manually [16] or by using Perfectprep® Plasmid 96 Vac kit (Eppendorf, Germany) according to the manual provided by the manufacturer. Automated cycle sequencing was performed by using T3 universal primer and BigDye Terminator (Applied Biosystems, USA) or ET Terminator (Amersham Pharmacia Bioscience, USA), and electrophoresed on DNA sequencer ABI 3730XL or ABI PRISM 377 (Applied Biosystems, USA) or Megabace™ 500 (Amersham Biosciences, USA).
Clustering analysis and annotation
Quality control of raw DNA sequences was performed by using Phred program [17] to remove sub-standard reads, followed by Lucy2 [18] to eliminate the vector and adapter sequences. Contig Assembly Program 3 (CAP3) was used to cluster the overlapping ESTs into contigs [19]. The edited EST was translated into six reading frames and compared with the non-redundant protein database at the National Center for Biotechnology Information (NCBI) using the default setting of BLASTX program [20]. BLASTN program was used to compare the nucleotide sequences with the sequences in the EST database at NCBI. BLASTX and BLASTN results with E-values equal or less than 10-5 were treated as 'significant matches', whereas ESTs with no hits or matches with E-values more than 10-5 to proteins in NCBI were classified as 'no significant matches'. The ESTs were mapped to Gene Ontology (GO) by using Blast2GO [21,22] and summarized according to their molecular functions, biological processes and cellular components. Differentially expressed transcripts in multiple oil palm cDNA libraries were detected by using R statistics [6] with Bonferonni correction at the significance threshold of 2.35 × 10-5 using the web tool IDEG6 [23,24].
Northern analyses
RNA blots were prepared from 10 μg total RNA from shoot apical meristems and flowers of different developmental stages. These blots were hybridised in 0.5 M SSPE pH 7.2, 1 mM EDTA, 7 % (w/v) SDS, 1 % (w/v) BSA and 100 μg/ml denatured herring sperm DNA and 32P-dCTP labelled probe at 60°C (for EgSBP) or 65°C (for EgSEP) for overnight. The membranes were washed in 40 mM sodium phosphate buffer, 5%(w/v) SDS for 10 min at 60°C (for EgSBP) or 65 °C (for EgSEP) twice, then followed by washing in 40 mM sodium phosphate buffer, 1 % (w/v) SDS at room temperature for 10 min. Subsequently, the membranes were exposed to autoradiography film for five days.
Authors' contributions
CLH, HK, SHT, SSRSA and MOA contributed to the conception and design of the study and coordinated the study. YYK, MCC, SST, WHN, KAL, YPL, SEO and JMT were involved in the generation of oil palm ESTs. CLH and WWL analysed the data. CLH drafted and revised the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Sequences of oil palm TUCs.
Acknowledgments
Acknowledgements
This work was financially supported by grants IRPA 01-02-04-0453, IRPA 01-02-04-0454, IRPA-PR 01-02-04-0386-PR0015/07-01 and IRPA 01-04-03-T0045-TC2 (under the Malaysian Palm Oil Board -Massachusettes Institute of Technology Biotechnology Partnership Programm) from the Ministry of Science, Technology and Innovation of Malaysia (MOSTI). We would like to thank the Director General of MPOB for granting us permission to publish this manuscript. We also gratefully acknowledge the contributions from Applied Agricultural Research Sdn Bhd., Sungai Buloh, Malaysia; Guthrie Sdn Bhd., Seremban, Malaysia; and Malaysian Palm Oil Board, Bangi, Malaysia. We thank the anonymous reviewers for constructive suggestions to improve this manuscript.
Contributor Information
Chai-Ling Ho, Email: clho@biotech.upm.edu.my.
Yen-Yen Kwan, Email: luvkeno@yahoo.com.
Mei-Chooi Choi, Email: mcchoi@its-interscience.com.
Sue-Sean Tee, Email: sueseant@gmail.com.
Wai-Har Ng, Email: waiharng@gmail.com.
Kok-Ang Lim, Email: limkokang@gmail.com.
Yang-Ping Lee, Email: leeyangping@yahoo.co.uk.
Siew-Eng Ooi, Email: oseng@mpob.gov.my.
Weng-Wah Lee, Email: leewengwah@gmail.com.
Jin-Ming Tee, Email: jmtee@niob.knaw.nl.
Siang-Hee Tan, Email: siangtsh@simedarby.com.
Harikrishna Kulaveerasingam, Email: harikrhk@simedarby.com.
Sharifah Shahrul Rabiah Syed Alwee, Email: shahrul.a@felda.net.my.
Meilina Ong Abdullah, Email: meilina@mpob.gov.my.
References
- Corley RHV, Tinker PB. The Oil Palm. 4. UK: Blackwell publishing; 2003. [Google Scholar]
- Corley RHV, Lee CH, Law LH, Wong CY. Abnormal development in oil palm clones. The Planter. 1986;62:233–240. [Google Scholar]
- Flood J, Keenan L, Wayne S, Hasan Y. Studies on oil palm trunks as sources of infection in the field. Mycopathologia. 2005;159:101–107. doi: 10.1007/s11046-004-4430-8. [DOI] [PubMed] [Google Scholar]
- Ariffin D, Idris AS, Gurmit S. Status of Ganoderma in oil palm. In: Flood J, Bridge PD, Holderness M, editor. Ganoderma diseases of perennial crops. UK: CABI Publishing; 2000. pp. 49–68. [Google Scholar]
- Jouannic S, Argout X, Lechauve F, Fizames C, Borgel A, Morcillo F, Aberlenc-Bertossi F, Duval Y, Tregear J. Analysis of expressed sequence tags from oil palm (Elaeis guineensis) FEBS Lett. 2005;579:2709–2714. doi: 10.1016/j.febslet.2005.03.093. [DOI] [PubMed] [Google Scholar]
- Stekel DJ, Git Y, Falciani F. The comparison of gene expression from multiple cDNA libraries. Genome Res. 2000;10:2055–2061. doi: 10.1101/gr.GR-1325RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- Zaidah R. MSc thesis. Universiti Putra Malaysia; 2001. The analysis of the tissue specific loacalization patterns of expression of several oil palm genes during floral development. [Google Scholar]
- Adam H, Jouannic S, Morcillo F, Richaud F, Duval Y, Tregear JW. MADS box genes in oil palm (Elaeis guineensis): Patterns in the evolution of the SQUAMOSA, DEFICIENS, GLOBOSA, AGAMOUS, and SEPALLATA subfamilies. J Mol Evol. 2006;62:15–31. doi: 10.1007/s00239-005-0333-7. [DOI] [PubMed] [Google Scholar]
- Klein J, Saedler H, Huijser P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA . Mol Gen Genet. 1996;250:7–16. doi: 10.1007/BF02191820. [DOI] [PubMed] [Google Scholar]
- Cardon G, Hohmann S, Klein J, Nettesheim K, Saedler H, Huijser P. Molecular characterization of the Arabidopsis SBP-box genes. Gene. 1999;237:91–104. doi: 10.1016/S0378-1119(99)00308-X. [DOI] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J. GIGANTEA : a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochester DE, Winter JA, Shah DM. The structure and expression of maize genes encoding the major heat shock protein, hsp70. EMBO J. 1986;5:451–458. doi: 10.1002/j.1460-2075.1986.tb04233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DJ, Craig R, Cox-Foster DL, Mumma RO, Medford JI. RNA isolation from recalcitrant plant tissue. Plant Mol Biol Reptr. 1994;12:310–316. [Google Scholar]
- Glišin V, Crkvenjakov R, Byus C. Ribonucleic acid isolation by cesium chloride centrifugation. Biochemistry. 1974;13:2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritisch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Ewing B, Hillier L, Wendhl MC, Green P. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Chou H, Holmes MH. DNA sequence quality trimming and vector removal. Bioinformatics. 2001;17:1093–1104. doi: 10.1093/bioinformatics/17.12.1093. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang I, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blast2Go http://www.Blast2GO.de
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- IDEG6 http://telethon.bio.unipd.it/bioinfo/IDEG6_form
- Romualdi C, Bortoluzzi S, d'Alessi F, Danieli GA. IDEG6: a web tool for detection of differentially expressed genes in multiple tag sampling experiments. Physiol Genomics. 2003;12:159–162. doi: 10.1152/physiolgenomics.00096.2002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of oil palm TUCs.


