Abstract
The human DEK protein has a long-standing association with carcinogenesis since the DEK gene was originally identified in the t(6:9) chromosomal translocation in a subtype of patients with acute myelogenous leukemia (AML). Recent studies have partly unveiled DEK's cellular functions including apoptosis inhibition in primary cells as well as cancer cells, determination of 3′ splice site of transcribed RNA, and suppression of transcription initiation by polymerase II. It has been previously shown that the N-terminal region of DEK, spanning residues 68–226, confers important in vitro and in vivo functions of DEK, which include double-stranded DNA (ds-DNA) binding, introduction of constrained positive supercoils into closed dsDNA, and apoptosis inhibition. In this paper, we describe the three-dimensional structure of the N-terminal domain of DEK (DEKntd) as determined using solution NMR. The C-terminal part of DEKntd, which contains a putative DNA-binding motif (SAF/SAP motif), folds into a helix-loop-helix structure. Interestingly, the N-terminal part of DEKntd shows a very similar structure to the C-terminal part, although the N-terminal and the C-terminal part differ distinctively in their amino acid sequences. As a consequence, the structure of DEKntd has a pseudo twofold plane symmetry. In addition, we tested dsDNA binding of DEKntd by monitoring changes of NMR chemical shifts upon addition of dsDNAs. We found that not only the C-terminal part containing the SAF/SAP motif but the N-terminal part is also involved in DEKntd's dsDNA binding. Our study illustrates a new structural variant and reveals novel dsDNA-binding properties for proteins containing the SAP/SAF motif.
Keywords: NMR, protein structure, SAF/SAP motif, DNA binding, AML, apoptosis inhibitor, chromatin-associated protein, DNA supercoiling
The 375-residue, 45 kDa DEK protein was first identified in patients with a subtype of AML as a fusion product in which its C-terminal 26 amino acids are replaced by the C-terminal two-thirds of the nucleoporin CAN (von Lindern et al. 1992). Overexpression of DEK has been found in a number of aggressive human tumors including acute myeloid leukemia (AML), bladder carcinoma, hepatocellular carcinoma, glioblastoma, and melanoma (Kondoh et al. 1999; Grottke et al. 2000; Kroes et al. 2000; Larramendy et al. 2002; Sanchez-Carbayo et al. 2003). Despite a strong association between DEK up-regulation and human carcinogenesis, intracellular function(s) of DEK have remained relatively elusive.
Recent studies revealed that DEK is down-regulated at both the message and protein levels during senescence of human papillomavirus (HPV)-positive HeLa cells as well as primary cells (Wise-Draper et al. 2005). Since overexpression of DEK resulted in partial inhibition of senescence in HeLa cells and extension of life span in primary cells, it was suggested that DEK is a senescence inhibitor (Wise-Draper et al. 2005). Further studies have shown that DEK depletion resulted in enhanced apoptosis in HeLa cells as well as in primary human keratinocytes, indicative of DEK's potential anti-apoptotic role (Wise-Draper et al. 2006). This anti-apoptotic function of DEK is coupled with p53 as DEK depletion elevated transcriptional activity of p53-regulated genes, and apoptosis was minimally induced by DEK depletion in SAOS-2 cells which do not express p53 (Wise-Draper et al. 2006). Furthermore, overexpression of amino acid residues 68–226 of DEK partially rescues cells from apoptosis induced by DEK depletion in HeLa cells (Wise-Draper et al. 2006).
DEK binds dsDNA and RNA, as 90% of DEK in HeLa cells is found associated with chromatin, whereas 10% is associated with RNA (Kappes et al. 2001). Since DEK prefers to bind structured DNA such as cruciform DNA and introduces supercoils into dsDNA in vitro, it was suggested that DEK might play a role in chromatin architecture (Waldmann et al. 2004). Two dsDNA-binding regions have been identified within DEK spanning amino acid residues 78–187 and 270–350 (Kappes et al. 2004). The N-terminal DNA-binding region (residues 78–187) includes a SAF/SAP DNA-binding motif spanning residues 149–183. These residues are indeed responsible for mediating DEK's dsDNA-binding activity in the N-terminal region of DEK (78–187), as shown by target bound assays (Bohm et al. 2005). The SAF/SAP motif was originally identified within sequences of chromatin-associated proteins such as scaffold attachment factor A and B (Aravind and Koonin 2000; Kipp et al. 2000). The SAF/SAP motif consists of 35 amino acid residues, of which 13 comprise the consensus sequence. DEK's SAF/SAP homologous region has a 69% similarity with this consensus sequence, which indicates that DEK is loosely related to other proteins within this family. Furthermore, the SAF/SAP motif containing the N-terminal region of DEK (residues 68–226), is sufficient for the introduction of constrained positive supercoils into closed circular dsDNA (Bohm et al. 2005).
Insights into a better understanding of DEK's cellular functions were provided by studying DEK's post-transcriptional modifications and cellular localizations. Acetylation of DEK, for example, changes its intracellular localization into interchromatin granule clusters that contain RNA processing factors and decreases its affinity for DNA elements within promoter regions (Cleary et al. 2005). Phosphorylation, another post-translational modification of DEK, peaks in the G1 phase and was shown to reduce DEKs overall DNA-binding affinity and enhance the multimerization of DEK molecules (Kappes et al. 2004). These observations are particularly interesting because DEK was found to be a member of a transcriptional repression complex with histone deacetylase II (Hollenbach et al. 2002), and more recently, because DEK has a role in determination of 3′ splice site by U2AF (Soares et al. 2006). Furthermore, a recent study uncovered a mechanism of transcriptional repression involving DEK (Faulkner et al. 2001; Gamble and Fisher 2007). In the proposed mechanism, histone chaperone SET protein dissociates DEK as well as another chromatin-associated protein PARP1 from chromatin, which allows assembly of the transcription pre-initiation complex on chromatin (Gamble and Fisher 2007).
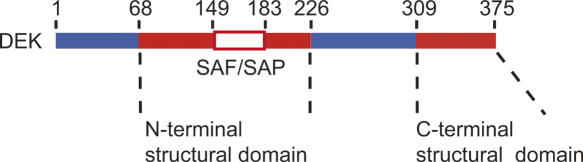
We have previously identified two structural regions within DEK, where the N-terminal region spans residues 68–226 and the C-terminal region spans residues 309–375 (Devany et al. 2005) (Fig. 1), and the three-dimensional structure of the C-terminal region of DEK has been reported. The C-terminal region of DEK has a three helix bundle structure similar to that of winged-helix DNA-binding proteins such as DP2 (Devany et al. 2004). In this report, we describe the three-dimensional structure of the N-terminal region of DEK and its dsDNA-binding properties.
Figure 1.
Schematic diagram of DEK. Two structural regions are colored orange, and the SAF/SAP motif (residues 149–183) is indicated by a red box.
Results
Assignments of NMR signals
We have previously found that residues 68–226 of DEK (DEK68–226) produce a soluble and structural protein (Devany et al. 2005). For the sequential assignments of NMR signals, fully 13C- and 15N-labeled DEK68–226 were produced, and two complementary pairs of three-dimensional triple resonance experiments, HNCA/HNCOCA and CBCANH/CBCACONH, were acquired. The sequential assignment of DEK68–226 was hindered by the homogenous and repetitive amino acid sequence (e.g., 27 lysines out of a total of 159 residues, and two KKK repeats). In order to facilitate the assignment process, DEK68–226 protein was prepared specifically labeled with 15N only on the lysine, arginine, leucine, valine, or isoleucine residues. HSQC spectra of each specifically labeled protein allowed the assignment of amino acid type to resonances.
Nearly complete backbone chemical shift assignments were obtained for the region from 78–226, whereas residues 68–77 are missing from 15N HSQC spectra. Protein sequence analysis (Edman degradation) confirmed that residues 68–77 are gradually degraded during purification, most likely due to co-purified proteases from host Escherichia coli cells. From these backbone chemical shifts it was possible to make a prediction of secondary structure elements within residues 78–226. A chemical shift analysis of the backbone Cα atoms indicated that residues from ∼78 through 193 exhibited well-defined structure, whereas Cα resonances from 192–226 exhibited chemical shifts indicative of a random coil sequence (chemical shifts have been deposited in the BioMagResBank [BMRB] with the entry number 6361).
In order to reduce the complexity of the spectra, a further truncated DEK construct, DEK78–208, was generated. Since residues 192–208 contain charged amino acid residues, these residues were retained in order to improve the solubility of the fragment. The heteronuclear single quantum coherence (HSQC) spectrum of DEK78–208 showed that most of the random coil residues appearing in the center of the spectrum of DEK68–226 had been removed in DEK78–208, and all of the structured residues retained the same chemical shifts (data not shown). CBCANH and CBCA(CO)NH (Grzesiek and Bax 1992a,b) spectra of DEK78–208 confirmed that the structured peaks retained the same chemical shifts, which indicated that the three-dimensional structures of DEK68–226 and DEK78–208 are the same. Therefore, we decided to use the DEK78–208 construct for structure determination. An HCCHTOCSY spectrum of 13C labeled DEK78–208 provided assignments of 85% of the side-chain 13C and 1H resonances (Devany and Matsuo 2005).
The three-dimensional structure of DEK78–208
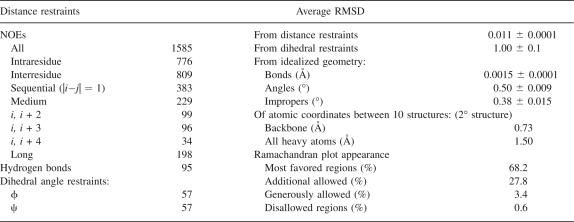
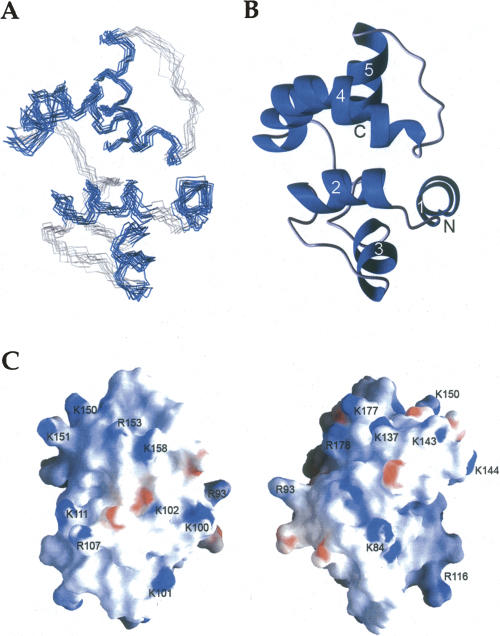
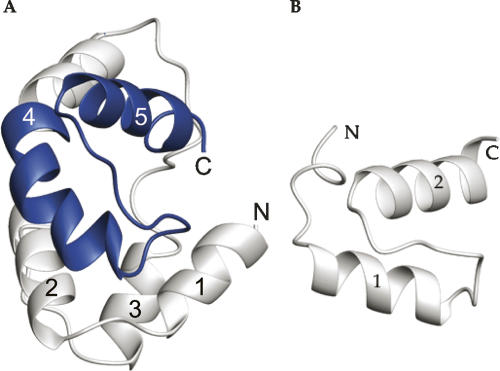
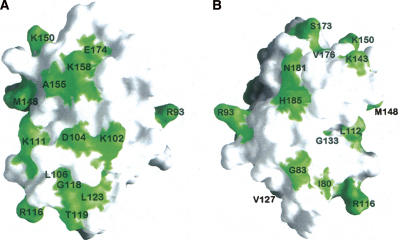
In each cycle of structure calculation using XPLOR-NIH (Schwieters et al. 2003), 80 starting structures were generated randomly and then refined to improve geometry. The 10 conformers with lowest total energy were selected and a summary of the structural statistics is shown in Table 1. Analysis of the dihedral angles using PROCHECK (Laskowski et al. 1996) revealed that >99% of the residues are within the most favorable and additional allowed regions. A best fit superposition of the backbone traces of the 10 lowest energy conformers is shown in Figure 2A, and the lowest energy structure is shown in ribbon diagrams with the same view in Figure 2B. The NMR structure is well-defined, with a root-mean-square deviation (RMSD) of 0.73 Å for the backbone heavy atoms and 1.5 Å for all of the heavy atoms of residues 78–191. Figure 2C shows the surface electrostatic potential of the DEK78–209 structure. DEK78–208 contains 23 lysines and arginines in total, and these residues are located mostly on the surface. These positive charges are scattered over the surface of the protein (Fig. 2C).
Table 1.
NMR structure parameters
Figure 2.
NMR solution structure of DEK78–208. (A) Superposition of the 10 lowest energy structures calculated of DEK78–208; helices are in blue, nonsecondary structured elements in gray. (B) Cartoon representation of a representative DEK78–208 structure. The N and C terminus are labeled N and C, respectively, and helices are numbered from the N-terminal. (C) Surface electrostatic potential of DEK78–208. (Left) Surface with the same orientation used for A and B. (Right) Molecule is 180° rotated.
The structure of DEK78–208 was determined and found to be a globular five-helix bundle. The five helices consist of amino acids 92–99(α1), 103–113(α2), 120–128(α3), 140–163(α4), and 172–182(α5). The SAP motif spans residues 149–183, corresponding to the C-terminal half of α4, an extended strand, and α5. The five-helix bundle adopts a unique topology of what is essentially two helix-extended loop-helix (HEH) (Aravind and Koonin 2000) subdomains. The first subdomain includes an N-terminal helix, α1, in addition to the helix-extended loop-helix motif, α2-loop-α3. The C-terminal subdomain includes the SAP HEH motif in α4 and α5. The two subdomains are then attached by a long linker loop. The C-terminus after residue 187 was lacking any nonsequential NOEs and their structure was not well-defined. Therefore, the structured portion of DEK68–226 shown in this work will now be referred to as DEK78–187.
dsDNA-binding property of DEKntd
To identify the nucleotide-binding interfaces of DEK68–226, 15N-labeled DEK68–226 was titrated with the three DNA molecules, short dsDNA (10 bp), long dsDNA (22 bp) and four-way junction DNA (4WJ-DNA). Unfortunately, DEK68–226 showed severe aggregation upon dsDNA binding, which resulted in poor NMR spectra (data not shown). It has been observed that DEK forms high molecular mass nucleoprotein complexes upon dsDNA binding using EMSA (Bohm et al. 2005). Therefore, we added a small protein tag, named B1 domain of streptococcal protein G (GB1), to DEK in order to increase solubility. Since residues 195–226 are not required for DEK's ds-DNA-binding and -supercoiling activity (Kappes et al. 2004), we deleted residues 196–226 and constructed the DEK68–194/GB1 fusion protein. We have tested the DNA-supercoiling property of DEK68–194/GB1, and confirmed that it can introduce positive supercoils into dsDNA as well as wild-type DEK (data not shown). The chemical shift perturbations were observed using 1H-15N HSQC spectra upon addition of dsDNA at various protein/dsDNA ratios. All of the NMR measurements were performed with the same buffer conditions used in the structure determination.
A binding preference of DEK for specific DNA sequences has yet to be definitively established. Following up on a previous report that DEK has a binding preference for supercoiled dsDNA and 4WJ-DNA (Waldmann et al. 2003), here we investigated DEK's specificity for DNA structure. We used four DNA oligomers with sequences reported to fold into a stable 4WJ complex (McKinney et al. 2003) to test binding of DEK68–194/GB1 to 4WJ-DNA molecules. As a control, we also used a sequence derived from the 4WJ oligomers to make a 22 bp linear ds DNA (referred to as DNA22). In addition, DEK was implied to have a transcription regulatory function through the Y-box sequence in Class II MHC gene promoters (Adams et al. 2003). Therefore, we used a 10 bp dsDNA containing the Y-box sequence to test whether DEK has a different DNA-binding mechanism for short dsDNA sequences. DEK68–194/GB1 was found to bind to all of these DNA molecules, and the results provide evidence that two distinct surfaces of DEK68–194 are involved in the dsDNA binding.
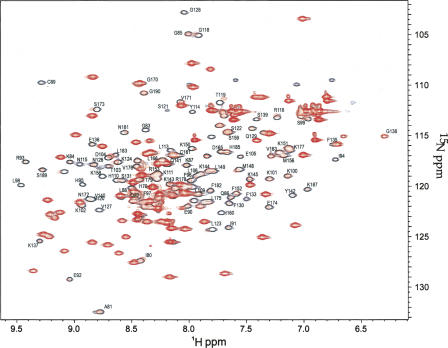
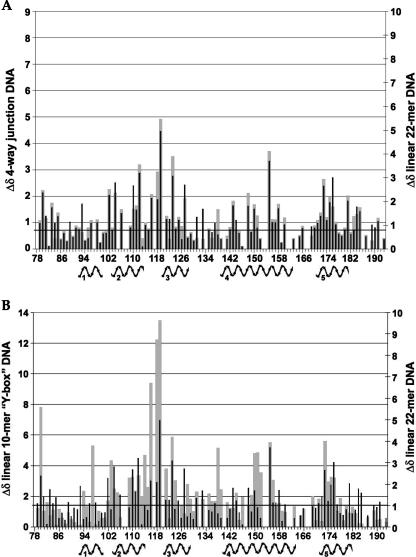
In Figure 3, the overlaid spectra of DEK68–194/GB1 in the absence and presence of the DNA22 at a DNA/protein ratio of 1:10 indicates residues that were perturbed by dsDNA binding shown as representative for dsDNA and 4WJ experiments (see Supplemental Fig. S1 for the titration spectra of Y-box DNA and 4WJ-DNA). The NMR signals of the DEK68–194/GB1 complex with each of the three DNA templates were assigned by following their changes upon increments of the DNA amount. Upon addition of DNA22, residues in the SAF/SAP motif were perturbed, particularly the helix α5 (S173, E174, L175, V176, F182, L183, M184, H185). In addition, residues in the N-terminal subdomain, particularly the extended loop between α2 and α3 (L112, Y114, R116, G118, T119, L123), were perturbed. Furthermore, residues surrounding exposed basic residues (R107, K111, K143, K144, K145, K150, K151, R153, and R168) were also perturbed. Similar experiments have been performed using Y-box or 4WJ-DNAs, and results are summarized as a comparison of the chemical shift indices between the different protein–dsDNA complexes (Fig. 4). Each index was calculated as Δδ = {(Δ1H)2 + {(Δ15N/5)2}1/2 in parts per million. Scales in the graphs were adjusted to fit the median values. The comparison of perturbations between the 4WJ-DNA (gray in Fig. 4A) and the DNA22 (black in Fig. 4A) indicates that they are binding in much the same manner, affecting the same residues to the same extent. Y-box DNA-binding (gray in Fig. 4B)-mediated chemical shift perturbations for the N-terminal subdomain are of much greater intensity than for the DNA22 (black in Fig. 4A,B). Conversely, for the binding of DNA22 or 4WJ-DNA, the C-terminal SAF/SAP motif shows a more significant level of perturbations throughout the full length of α5. This result indicates that a short dsDNA (10 bp) preferably bound the N-terminal subdomain that does not contain the SAF/SAP motif.
Figure 3.
DEK68–194/GB1 binds 22 mer dsDNA. Overlaid 1H-15N HSQC spectra of DEK68–194/GB1 in the absence (open black) and the presence (red) of DNA22 at a molar ratio of 1:10. Assignments are for the native residues of DEK.
Figure 4.
CSP comparison between different dsDNAs. Comparison of DEK68–194/GB1 backbone 1H and 15N chemical shift perturbation indices between complexes: (A) DNA22 vs. four-way junction. (B) DNA22 vs. Y-box DNA. The median line for the two Y-axes appears in bold.
Discussion
DEK78–187 shows a new variant structure of the SAF/SAP motif containing proteins
The structure of DEK78–187 has two subdomains facing each other with pseudo twofold planar symmetry, which provides a new variant structure of the SAF/SAP family proteins. The SAF/SAP motif is predicted by secondary structure algorithms and homology modeling to fold into two helices separated by an extended strand loop. It was suggested that the SAP domain was evolutionarily derived from the helix-extended loop-helix (HEH) fold (Aravind and Koonin 2000). The SAF/SAP motif of DEK has a unique fold among proteins containing this motif as the first helix of HEH is extended by three turns (Fig. 5). The 35-residue SAF/SAP motif of DEK runs within residues 149–183, and the two predicted helices would span residues 154–163 and 172–182. The calculated structure of DEK78–187 matches these predictions almost exactly, except the first helix here begins at residue 140. However, at residue N154 the helix has a sharp kink. N154 lacks an 1H-15N resonance in the 1H-15N HSQC, which indicates that this amide nitrogen is not in a canonical α-helix hydrogen bond and/or under influence of chemical exchange. Since residues 151–153 have negative deviations from random coil Cα chemical shift values, these residues also may not be in canonical α-helix, most likely a loop between two helices. Twenty-three nonsequential NOEs involving residues 151–154 indicated that these residues have stable structure. These NOE restraints and no hydrogen bond restraints involving residues 151–154 were used in the structure calculation. Interestingly, in all of the 10 low energy structures, residues 151–154 fall in the allowed region of the Ramachandran plot for right-handed α-helices, as are all of the residues in the α4. As a result, the extension of α4 forms an arm with a bent elbow to cradle the C-terminal helix.
Figure 5.
Comparison of the SAF/SAP HEH motifs of DEK and Ku70. The SAF/SAP helix-extended strand-helix (HEH) motif of DEK is in blue (A). Helix 4 is bent and extends from the SAF/SAP motif. The SAF/SAP motif located in the C terminus of Ku70 is shown in B (PDB ID: 1JEQ).
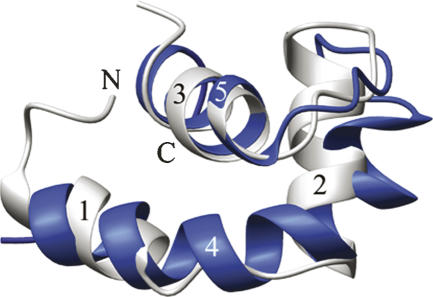
Curiously, the three helices, α1, α2, and α3, of the N-terminal subdomain have a remarkably similar topology to the SAF/SAP containing C-terminal subdomain (α4 and α5). The first two helices, α1 and α2, are connected by a short, four-amino acid linker loop that also allows them to form a crook to cradle the last α-helix of the subdomain. The subdomains are connected by a long, 11-amino acid linker oriented so that the bent arms face each other and run roughly parallel, forming a deep cleft between them that is lined with basic residues. The elbows in the arms are slightly offset between the two domains, but close enough so that the axes of the “cradled” helices, α3 and α5, appear to run in the same plane and form a “V,” with the apex opposite the elbow. Despite a lack of any sequence homology with the SAF/SAP motif, or any of the other identified HEH motifs found in rho-N or the lysyl tRNA synthetases, the N-terminal subdomain of DEK78–187 forms a helix-extended loop-helix with an amazingly similar topology to the C-terminal subdomain (Fig. 6). Overall, the arrangement of the helices is quite similar between the two subdomains, with a backbone RMSD of 2.0 Å.
Figure 6.
N- and C-terminal subdomains are structurally similar. SAF/SAP containing C-terminal helices in blue, helices of the N-terminal domain in gray.
We used the DALI server (Holm and Park 2000) to search the Protein Data Bank for structures with similar topology. The N- and C-terminal subdomains separately, as well as the entire structured region (residues 78–187) containing both subdomains were submitted as queries. No structural comparison with a Z-score >3.4 was returned as the result of structural similarity search, which indicates the unique topology of the DEK78–197 structure. It is not surprising that the DALI search discovered structural homologies with two of the three published SAP motif containing proteins, PIAS1 and Ku70, as well as the prototypical members of the HEH fold containing family of proteins, the RNA-binding domain of the transcriptional terminator rho and endonuclease VII from phage T4.
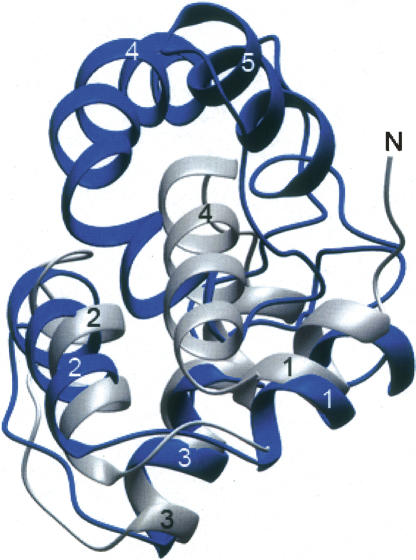
PIAS1 (PIAS, protein inhibitor of activated STAT) has been reported to function as an E3-type SUMO ligase for tumor suppressor p53 (Schmidt and Muller 2003). In addition to the N-terminal SAF/SAP motif (residues 11–45), PIAS1 contains a RING domain necessary for ligase function (Jackson 2001), a PINIT domain essential for nuclear retention (Duval et al. 2003), and a small C-terminal acidic domain. The SAP domain of PIAS1 binds dsDNA and may also contain a binding site for p53 (Okubo et al. 2004). The highest Z-score and lowest RMSD was discovered with the N-terminal SAF/SAP domain of PIAS1 superimposed on the N-terminal subdomain of DEK78–187 rather than with the SAF/SAP containing C-terminal subdomain. An overlay of the SAF/SAP domain of PIAS1 with the N-terminal subdomain of DEK78–187 is shown in Figure 7. This overlay shows a remarkable similarity in the length of the three helices, and their positions with respect to each other. This structural similarity between the N-terminal subdomain of DEK78–187 and the N-terminal SAF/SAP domain of PIAS1 implies the dsDNA-binding function of the N-terminal subdomain as discussed in the following section.
Figure 7.
Superimposed PIAS1 and the N-terminal subdomain of DEK78–187 PIAS1 and DEK78–187 are in gray and blue, respectively. Helices 1, 2, and 3 of the N-terminal SAF/SAP domain of PIAS1 are well superimposed with the N-terminal subdomain of DEK78–187 consisting of helices 1, 2, and 3.
dsDNA-binding and dsDNA-supercoiling mechanism of DEK
The SAP motif is a recently discovered putative DNA-binding motif identified in more than 44 proteins. Three SAP structures, besides that of DEK, have been published in the Protein Data Bank, but as yet, little information is available on the manner or mechanism of DNA binding. Direct evidence that the SAP motif binds DNA has come from the studies of SAF-A (Kipp et al. 2000), Ku70 (Walker et al. 2001; Zhang et al. 2001), and PIAS1 (Okubo et al. 2004), and indirect evidence exists for the spCCE1 SAP motif (Ahn and Whitby 2003) and DEK (Bohm et al. 2005). SAF-A was found to bind specifically to the minor groove of A-T-rich scaffold attachment regions (SAR) or matrix attachment regions (MAR) of DNA with what has been termed the “mass-binding mode.” In this model of DNA binding, high affinity for DNA can be generated from individual DNA-binding units with low affinity in isolation through protein–protein interactions that result in the formation of multimeric complexes (Kipp et al. 2000). The perturbations observed upon DNA binding of the C-terminal SAP domain of Ku70 suggested that the first helix and the extended loop are involved in DNA binding, and the investigators suggested that the α-helix is inserted into the major groove of the DNA (Walker et al. 2001; Zhang et al. 2001). Similarly, in the N-terminal SAP domain of PIAS1, perturbations upon DNA binding mapped to the first SAP helix and the beginning of the second SAP helix following the extended loop (Okubo et al. 2004). The SAP motif of spCCE1 has been shown to play an important role in the stable complex formation of spCCE1 with Holliday junction DNA (Ahn and Whitby 2003).
Consistent with previous observations that DEK binds to a four-way junction and dsDNA (Bohm et al. 2005), the chemical shift mapping experiments provide direct evidence that DEK68–194/GB1 binds to dsDNA (Figs. 3, 4). Figure 8 represents the surface of the DEK78–187 structure with those residues having a chemical shift perturbation upon binding DNA22 of Δδ >0.015 ppm. As indicated in Figure 4B, the N-terminal subdomain has more perturbed residues upon addition of dsDNA, which resulted in larger dsDNA-binding surfaces within the N-terminal subdomain than the C-terminal subdomain (Fig. 8). The SAF/SAP domain of PIAS1 uses residues R15, K30, and K34 for its dsDNA binding (Okubo et al. 2004). DEK's corresponding residues to R15, K30, and K34 of PIAS1 are R153, R168, and N172, respectively, and these residues did not show chemical shift perturbation stronger than the average perturbation value. Instead, S173, E174, and V176 located in the second SAP helix showed significant chemical shift perturbation (Fig. 4), which indicates that this helix is involved in dsDNA binding. It should be noted that hydrophobic residues that were perturbed upon addition of dsDNA, including L123, M148, and V176, may be involved in protein–protein interactions as DEK68–194/GB1 aggregated upon addition of dsDNA. Since aggregation was very minor with short dsDNA, 10 bp Y-box dsDNA, compared with larger dsDNAs including 4WJ or DNA22, it is plausible that protein–protein interactions become favorable when multiple molecules of DEK bind the same DNA molecule. This dsDNA-binding model is consistent with the mass-binding mode proposed for the interaction between the SAF-A protein and A-T-rich long DNA fragments (Kipp et al. 2000) in which the aggregation of protein that weakly binds dsDNA produce stronger dsDNA binding.
Figure 8.
Chemical shift perturbation upon protein/DNA complex formation mapped to the surface of DEK78–208. Resonance perturbations on the N-terminal HEH motif in green. (A) In the same orientation shown in Figure 2C (right side picture); (B) 180° opposite of A that has the same orientation of Figure2C (left).
It should be noted that we constructed 23 variants of DEK68–226 that contain single-alanine substitutions of lysine or arginine including K84, K87, R93, K100, K101, K102, R107, K111, R116, K124, K125, K137, K143, K144, K145, K150, K151, R153, K158, R168, K177, R178, and K187, and tested their dsDNA-binding and -supercoiling properties (see Supplemental Fig. S3). We found that all of our single-alanine substitution variants were capable of binding dsDNA and introduced supercoils into dsDNA as well as wild-type DEK68–226. Since the chemical shift perturbation indicated that the α5 helix (second helix in the SAF/SAP motif) is involved in the dsDNA binding, we made the K177A/R178A double-substitution variant, which removed all positive charges from the surface of the α5 helix. The K177A/R178A double-substitution variant bound dsDNA and introduced supercoils into dsDNA as well as wild-type DEK68–226 (data not shown). Since the chemical shift perturbation of the α5 helix is more significant when aggregation of the protein was observed upon addition of longer DNAs including 4WJ-DNA and DNA22, it was implied that this helix may be involved in protein–protein interaction upon dsDNA binding.
In summary, our current studies revealed that the N-terminal region of DEK has a unique three-dimensional structure that consists of two SAF/SAP folds, which provides a new structural variant for proteins containing the SAF/SAP motif. Each of these SAF/SAP fold domains interacts with dsDNA. Interestingly, the SAF/SAP amino acid sequence motif of DEK appeared to be involved in protein–protein interaction as well as the dsDNA interaction. Therefore, the N terminus of DEK may interact with two segments of DNA using its two distinctive dsDNA-binding domains, and may form nucleoprotein complexes through protein–protein interaction using its SAF/SAP motif, which would enable DEK to loop DNA, changing the writhe. One possibility how this could be accomplished is by a DNA-bending activity of DEK. As shown by Waldmann et al. (2003) and Kappes et al. (2004) full-length DEK and the SAP/SAF fragment (residues 78–187) were able to stimulate end-to-end joining of DNA molecules, but failed to bend 123 bp DNA fragments sufficiently for re-ligation in these assays. Another and more favorable possibility is a local change of twist by intercalation of multiple DEK molecules or by the individual molecule, changing the writhe in a closed circular DNA molecule. Indeed, electron microscopic analysis of linear 1000 bp fragments partly supported this idea as it revealed a 10% increase in length after DEK incubation, whereas the same fragment reconstituted with histones underwent a significant decrease in length (Waldmann et al. 2002). Further biochemical and structural studies are required to fully understand the mechanism by which DEK introduces constrained positive supercoils into closed dsDNA.
Materials and Methods
Protein expression and purification
All DEK constructs were cloned into the bacterial expression vector pET28a (Novagen) for expression and purification using a hexa-histidine tag (His6). DEK68–226 is C-terminally His6-taged, whereas DEK78–208 carries an N-terminal tag.
DNA fragments containing residues 68–226 of human DEK were ligated into the pET28a vector by using primers for PCR containing the NcoI and XhoI restriction enzyme sites to produce 5′ and 3′ cloning sites, respectively. For example, the protein encoded by the ensuing plasmid with the DEK68–226 insert had the amino acid sequence: (68MQVSSLQREPFTIAQGKGQKLCE-IERIHFFLSKKKTDELRNLHKLYNRPGTVSSLKKNVGQFSGFPFEKGSVQYKKKEEMLKKFRNAMLKSICEVLDLERSGVNSELVKRILNFLMHPKPSGKPLPKSKKTCSKGSKKERNSSGMARKAKRTCPEIL226)LEHHHHHH.
A DNA fragment encoding residues 78–208 of human DEK was ligated into the pET28a vector by using PCR primers containing the NdeI and Bpu1102I restriction enzyme sites. The protein encoded by the ensuing plasmid had the amino acid sequence: MGSSHHHHHHSSGLVPRGS(78FTIAQGKGQKLCEIERIHFFLSKKKTDELRNLHKLYNRPGTVSSLKKNVGQFSGFPFEKGSVQYKKKEEMLKKFRNAMLKSICEVLDLERSGVNSELVKRILNFLMHPKPSGKPLPKSKKTCSKGSKKER208).
For DNA binding experiments, amino acids 68–194 from human DEK were ligated into the bacterial expression vector pET30a (Novagen) as a fusion protein with the B1 domain of streptococcal protein G (GB1) with a His6-tag located at the C terminus (DEK68–194/GB1). DNA fragments containing residues 68–194 of human DEK and the immunoglobulin binding domain of streptococcal protein G (GB1) were first blunt-end ligated and then ligated into the pET30a vector by using primers for PCR containing the NcoI and XhoI restriction enzyme sites to produce 5′ and 3′ cloning sites, respectively. The protein sequence was (68MQVSSLQREPFTIAQGKGQKLCEIERIHFFLSKKKTDELRNLHKLYN-RPGTVSSLKKNVGQFSGFPFEKGSVQYKKKEEMLKKFRNAMLKSICEVLDLERSGVNSELVKRILNFLMHPKPSGKPLP194)MTYKLILNGKTLKGETTTEAVDAATAEKVFKQYANDNGVDGEWTYDDATKTFTVTELEHHHHHH.
The rare codon expressing Bl21(λDE3) RIL (Stratagene) E. coli host strain containing recombinant plasmid was grown at 37°C in LB medium for unlabeled protein. To produce uniformly 15N or 15N/13C labeled protein for NMR, E. coli were grown in M9 minimal medium supplemented with 1 g/L 15NH4Cl alone or with 2 g/L 13C glucose. The expression of the construct was induced by the addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside at an OD600 ≈ 0.6. After 16 h post-induction at 17°C, the cells were collected by centrifugation, and resuspended in 10 mL Y-PER lysis buffer (Promega) supplemented with protease inhibitors (EDTA-free Complete-mini, Roche). After 20 min of lysis the cell debris was pelleted at >35,000g, and the cleared lysate was applied to Ni-NTA beads (Qiagen) for 30 min and washed twice with 10 mL of 50 mM sodium phosphate, 300 mM sodium chloride, 10 mM imidazole, pH 8.0. DEK N-terminal domain constructs were then eluted with 250 mM imidazole and applied to a G-75 Sepharose size exclusion column (GE healthcare Amersham Biosciences).
NMR spectroscopy
NMR spectra were acquired at 25°C on Varian Inova 600 and 800 spectrometers equipped with an HCN triple-resonance probe and a triple-axis gradient unit. 1H, 15N, and 13C chemical shifts were referenced to sodium 4,4-dimethyl-4-silapentane sulfonate (DSS), and are quoted in parts per million (ppm). Backbone resonance assignment was achieved with NMR experiments using 15N single-labeled and 13C, 15N doubly labeled 0.35 mM DEK68–226 and DEK78–208 collected at 25°C in 50 mM sodium phosphate, 100 mM KCl, 20 mM DTT, 0.2% sodium azide, pH 6.8. NMR experiments for side chain resonance assignment and collection of restraints for structure determination were acquired on nonlabeled, 15N single-labeled or 13C, 15N doubly labeled 0.5 mM DEK78–208 with the same temperature and buffer conditions as above. Data were processed using NMRPipe (Delaglio et al. 1995) and analyzed with XEASY (Bartels et al. 1995) on Linux PCs. Spectral parameters are summarized in Table 2.
Table 2.
NMR experiment parameters
Sequential assignment was accomplished using conventional three-dimensional triple resonance experiments: HNCA, HN(CO)CA, CBCANH, CBCA(CO)NH, HNCO, and HN(CA)CO (Grzesiek and Bax 1992a,b; Powers et al. 1992; Matsuo et al. 1996).
Two- and three-dimensional TOCSY experiments (Marion et al. 1989) were used to identify the spin systems of the N-terminal domain of DEK.
The aromatic residue side chains of DEK78–208 were assigned using a 1H-1H 2D NOESY experiment and a 3D 13C-edited NOESY-HSQC experiment with 13C offset at 125 ppm.
Structural restraints
Three types of restraints were obtained for structural calculation of DEK78–208, including NOE distance restraints, hydrogen bond restraints, and dihedral angle restraints.
NOE distance restraints were collected from a 2D 1H-1H NOESY, and 3D 15N- and 13C-edited NOESY-HSQC spectra. Hydrogen bond restraints were added for residues in canonical secondary structure elements identified using Cα, Cβ, CO chemical shift deviation, and NOEs. For each hydrogen bond, two restraints were used: 1.8–2.5 Å for HN–O distances and 2.5–3.3 Å for N–O distances. Dihedral angle restraints were calculated by the program TALOS (Cornilescu et al. 1999) using the assigned backbone chemical shifts (HN, N, Cα, Cβ, and CO).
Structure calculation
Structure calculations were performed using XPLOR-NIH version 2.0.5 (Schwieters et al. 2003) on a Pentium4 3.0-GHz PC under the Linux OS. NMR structures were generated using the ab initio simulated annealing (SA) protocol starting from a template coordinate set (Brunger 1993). Final structures were selected under the criteria of no NOE violations >0.1 Å; no dihedral angle violations >5°; no angle, bond length, and improper angle deviation from the ideal geometry of >5°, 0.05 Å, and 5°, respectively, was allowed. The structures were then analyzed, displayed, and superimposed using INSIGHT, MOLMOL (Koradi et al. 1996), VMD-XPLOR (Schwieters and Clore 2001), GRASP (Nicholls et al. 1991), and SPDBV (Guex and Peitsch 1997).
DNA titration
Three DNA molecules were chosen to test the DNA-binding properties of DEK68–194/GB1: a 10 bp dsDNA, containing Y-box sequence, to which DEK has been reported to bind (Adams et al. 2003), a sequence reported to form a stable four-way junction complex (McKinney et al. 2003), and a 22 bp linear DNA sequence derived from the four-way junction sequence. The sequences of the eight DNA oligomers used to test DEK68–194/GB1 DNA binding were as follows: Y-box, (1) 5′ CTAATTGGCC 3′ and (2) 5′ GGCCAATTAG 3′; DNA22, (1) 5′ CCCTAGCAAGCCGAGCGGTGGG 3′, and (2) 5′ CCCACCGCTCGGCTTGCTAGGG 3′; and four-way junction, (1) 5′ CCCTAGCAAGCCGCTGCTACGG 3′, (2) 5′ CCGTAGCAGCGCGAGCGGTGGG 3′, (3) 5′ CCCACCGCTCGGCTCAACTGGG 3′, and (4) 5′ CCCAGTTGAGCGCTTGCTAGGG 3′.
Lyophilized DNA oligomers provided by the University of Minnesota Microchemical Facility were dissolved in annealing buffer (20 mM sodium phosphate, 50 mM sodium chloride, 10 mM EDTA, pH 6.5) at a concentration of 1 mM. DNAs were annealed by heating them to 90°C in a heat block and gently cooling to room temperature, then placing the solution on ice. The annealing buffer was exchanged with NMR buffer (50 mM sodium phosphate, 100 mM potassium chloride, 0.02% sodium azide, pH 6.8) by three repeated dilution and concentration steps in a Microcon 3 concentrator (Millipore). Successive additions of a 1 mM solution of the 10 bp, and 22 bp linear dsDNAs, and the 4WJ DNA were made to a 0.2 mM solution of 15N-labeled DEK68–194/GB1 (which was first used to acquire reference spectra). Following each addition, a one-dimensional 1H spectrum and a 2D [15N-1H] HSQC spectrum was recorded. The resulting data sets corresponded to the following approximate molar ratios of protein to DNA (P/D): for 10 bp dsDNA, 1:2 and 1:4; for 22 bp dsDNA, 40:1, 20:1, 10:1, 6.7:1, and 5:1; and for four-way junction DNA, 40:1, 20:1, and 10:1. Protein concentrations were derived from the A280 measured under denaturing conditions (6 M GuHCl, 20 mM sodium phosphate, pH 6.5), using a calculated extinction coefficient of 12,330 M−1 cm−1 (Gill and von Hippel 1989).
Data bank accession codes
The coordinates of the 10 energy-refined XPLOR conformers of DEK(78–208) have been deposited in the PDB with the ID code 2jx3. The NMR chemical shifts have been deposited in the BioMagResBank (BMRB) with the entry number 6361.
Electronic supplementary material
NMR spectra of the DNA titration of Y-box DNA and four-way junction DNA are supplied for publication in the Electronic Edition (Supplemental Fig. S1). A NMR spectrum showing aggregation of DEK upon addition of four-way junction DNA is provided as Supplemental Figure S2. Topology assays, aggregation assays and EMSAs of representative point mutants of DEK are provided in the Supplemental Figure S3.
Acknowledgments
NMR instrumentation was provided with funds from the NSF (BIR-961477), the University of Minnesota Medical School, and the Minnesota Medical Foundation. We thank Basic Science Computer Laboratory for providing us SGI workstations. This work is partially supported by a grant from Grant-in-Aid of Research, Artistry, and Scholarship from the Graduate School of University of Minnesota, and American Cancer Society Institutional grant (IRG-58-001-46-IRG14) to H.M. M.D. was partially supported by the National Institutes of Health predoctoral training grant. F.K. was supported by a grant from the Michigan Chapter of the Arthritis Foundation. D.M.M. was supported by grants from the NIH and a Clinical Scientist Award in Translational Research from the Burroughs Wellcome Fund. This work was carried out in part using computing resources at the University of Minnesota Supercomputing Institute.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Hiroshi Matsuo, Department of Biochemistry, Molecular Biology and Biophysics, University of Minnesota, 321 Church Street, S.E. Jackson Hall 6-155, Minneapolis, Minnesota 55455, USA; e-mail: matsu029@umn.edu; fax: (612) 625-2163.
Abbreviations: DEK68–226, the N-terminal structural domain of the human DEK protein, which has deletions of the N-terminal 67 amino acids and C-terminal 149 amino acids of the human DEK protein; DEK78–208, deletion of the N-terminal 77 amino acids and C-terminal 168 amino acids of the human DEK protein; SAF/SAP motif, nuclear scaffold attachment factor [SAF] and SAF, Acinus, PIAS [SAP] motif; AML, acute myelogenous leukemia; HSQC, heteronuclear single quantum coherence; NOE, nuclear Overhauser effect; RMSD, root-mean-square deviation.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073244108.
References
- Adams, B.S., Cha, H.C., Cleary, J., Haiying, T., Wang, H., Sitwala, K., Markovitz, D.M. DEK binding to class II MHC Y-box sequences is gene- and allele-specific. Arthritis Res. Ther. 2003;5:R226–R233. doi: 10.1186/ar774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, J.S., Whitby, M.C. The role of the SAP motif in promoting Holliday junction binding and resolution by SpCCE1. J. Biol. Chem. 2003;278:29121–29129. doi: 10.1074/jbc.M302314200. [DOI] [PubMed] [Google Scholar]
- Aravind, L., Koonin, E.V. SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- Bartels, C., Xia, T.-H., Billeter, M., Güntert, P., Wüthrich, K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- Bohm, F., Kappes, F., Scholten, I., Richter, N., Matsuo, H., Knippers, R., Waldmann, T. The SAF-box domain of chromatin protein DEK. Nucleic Acids Res. 2005;33:1101–1110. doi: 10.1093/nar/gki258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger, A.T. XPLOR version 3.1: A system for X-ray crystallography and NMR. Yale University Press; New Haven, CT: 1993. [Google Scholar]
- Cleary, J., Sitwala, K.V., Khodadoust, M.S., Kwok, R.P., Mor-Vaknin, N., Cebrat, M., Cole, P.A., Markovitz, D.M. p300/CBP-associated factor drives DEK into interchromatin granule clusters. J. Biol. Chem. 2005;280:31760–31767. doi: 10.1074/jbc.M500884200. [DOI] [PubMed] [Google Scholar]
- Cornilescu, G., Delaglio, F., Bax, A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Devany, M., Matsuo, H. NMR resonance assignments for the DNA-supercoiling domain of the human protein DEK. J. Biomol. NMR. 2005;31:65. doi: 10.1007/s10858-004-6889-5. [DOI] [PubMed] [Google Scholar]
- Devany, M., Kotharu, N.P., Matsuo, H. Solution NMR structure of the C-terminal domain of the human protein DEK. Protein Sci. 2004;13:2252–2259. doi: 10.1110/ps.04797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devany, M., Kotharu, N.P., Matsuo, H. Expression and isotopic labeling of structural domains of the human protein DEK. Protein Expr. Purif. 2005;40:244–247. doi: 10.1016/j.pep.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Duval, D., Duval, G., Kedinger, C., Poch, O., Boeuf, H. The “PINIT” motif, of a newly identified conserved domain of the PIAS protein family, is essential for nuclear retention of PIAS3L. FEBS Lett. 2003;554:111–118. doi: 10.1016/s0014-5793(03)01116-5. [DOI] [PubMed] [Google Scholar]
- Faulkner, N.E., Hilfinger, J.M., Markovitz, D.M. Protein phosphatase 2A activates the HIV-2 promoter through enhancer elements that include the pets site. J. Biol. Chem. 2001;276:25804–25812. doi: 10.1074/jbc.M006454200. [DOI] [PubMed] [Google Scholar]
- Gamble, M.J., Fisher, R.P. SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nat. Struct. Mol. Biol. 2007;14:545–555. doi: 10.1038/nsmb1248. [DOI] [PubMed] [Google Scholar]
- Gill, S.C., von Hippel, P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Grottke, C., Mantwill, K., Dietel, M., Schadendorf, D., Lage, H. Identification of differentially expressed genes in human melanoma cells with acquired resistance to various antineoplastic drugs. Int. J. Cancer. 2000;88:535–546. doi: 10.1002/1097-0215(20001115)88:4<535::aid-ijc4>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Grzesiek, S., Bax, A. Correlating backbone amide and side chain resonances in large proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 1992a;114:6291–6293. [Google Scholar]
- Grzesiek, S., Bax, A. An efficient experiment for sequential backbone assignment of medium sized isotopically enriched proteins. J. Magn. Reson. 1992b;99:201–207. [Google Scholar]
- Guex, N., Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Hollenbach, A.D., McPherson, C.J., Mientjes, E.J., Iyengar, R., Grosveld, G. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J. Cell Sci. 2002;115:3319–3330. doi: 10.1242/jcs.115.16.3319. [DOI] [PubMed] [Google Scholar]
- Holm, L., Park, J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- Jackson, P.K. A new RING for SUMO: Wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes & Dev. 2001;15:3053–3058. doi: 10.1101/gad.955501. [DOI] [PubMed] [Google Scholar]
- Kappes, F., Burger, K., Baack, M., Fackelmayer, F.O., Gruss, C. Subcellular localization of the human proto-oncogene protein DEK. J. Biol. Chem. 2001;276:26317–26323. doi: 10.1074/jbc.M100162200. [DOI] [PubMed] [Google Scholar]
- Kappes, F., Scholten, I., Richter, N., Gruss, C., Waldmann, T. Functional domains of the ubiquitous chromatin protein DEK. Mol. Cell. Biol. 2004;24:6000–6010. doi: 10.1128/MCB.24.13.6000-6010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipp, M., Gohring, F., Ostendorp, T., van Drunen, C.M., van Driel, R., Przybylski, M., Fackelmayer, F.O. SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol. Cell. Biol. 2000;20:7480–7489. doi: 10.1128/mcb.20.20.7480-7489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh, N., Wakatsuki, T., Ryo, A., Hada, A., Aihara, T., Horiuchi, S., Goseki, N., Matsubara, O., Takenaka, K., Shichita, M., et al. Identification and characterization of genes associated with human hepatocellular carcinogenesis. Cancer Res. 1999;59:4990–4996. [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., Wuthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Kroes, R.A., Jastrow, A., McLone, M.G., Yamamoto, H., Colley, P., Kersey, D.S., Yong, V.W., Mkrdichian, E., Cerullo, L., Leestma, J., et al. The identification of novel therapeutic targets for the treatment of malignant brain tumors. Cancer Lett. 2000;156:191–198. doi: 10.1016/s0304-3835(00)00462-6. [DOI] [PubMed] [Google Scholar]
- Larramendy, M.L., Niini, T., Elonen, E., Nagy, B., Ollila, J., Vihinen, M., Knuutila, S. Overexpression of translocation-associated fusion genes of FGFRI, MYC, NPMI, and DEK, but absence of the translocations in acute myeloid leukemia. A microarray analysis. Haematologica. 2002;87:569–577. [PubMed] [Google Scholar]
- Laskowski, R.A., Rullmannn, J.A., MacArthur, M.W., Kaptein, R., Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Marion, D., Driscoll, P.C., Kay, L.E., Wingfield, P.T., Bax, A., Gronenborn, A.M., Clore, G.M. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: Application to interleukin 1 beta. Biochemistry. 1989;28:6150–6156. doi: 10.1021/bi00441a004. [DOI] [PubMed] [Google Scholar]
- Matsuo, H., Kupce, E., Li, H., Wagner, G. Use of selective C alpha pulses for improvement of HN(CA)CO-D and HN(COCA)NH-D experiments. J. Magn. Reson. B. 1996;111:194–198. doi: 10.1006/jmrb.1996.0082. [DOI] [PubMed] [Google Scholar]
- McKinney, S.A., Declais, A.C., Lilley, D.M., Ha, T. Structural dynamics of individual Holliday junctions. Nat. Struct. Biol. 2003;10:93–97. doi: 10.1038/nsb883. [DOI] [PubMed] [Google Scholar]
- Nicholls, A., Sharp, K.A., Honig, B. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Okubo, S., Hara, F., Tsuchida, Y., Shimotakahara, S., Suzuki, S., Hatanaka, H., Yokoyama, S., Tanaka, H., Yasuda, H., Shindo, H. NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers. J. Biol. Chem. 2004;279:31455–31461. doi: 10.1074/jbc.M403561200. [DOI] [PubMed] [Google Scholar]
- Powers, R., Garrett, D.S., March, C.J., Frieden, E.A., Gronenborn, A.M., Clore, G.M. 1H, 15N, 13C, and 13CO assignments of human interleukin-4 using three-dimensional double- and triple-resonance heteronuclear magnetic resonance spectroscopy. Biochemistry. 1992;31:4334–4346. doi: 10.1021/bi00132a026. [DOI] [PubMed] [Google Scholar]
- Sanchez-Carbayo, M., Socci, N.D., Lozano, J.J., Li, W., Charytonowicz, E., Belbin, T.J., Prystowsky, M.B., Ortiz, A.R., Childs, G., Cordon-Cardo, C. Gene discovery in bladder cancer progression using cDNA microarrays. Am. J. Pathol. 2003;163:505–516. doi: 10.1016/S0002-9440(10)63679-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, D., Muller, S. PIAS/SUMO: New partners in transcriptional regulation. Cell. Mol. Life Sci. 2003;60:2561–2574. doi: 10.1007/s00018-003-3129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters, C.D., Clore, G.M. The VMD-XPLOR visualization package for NMR structure refinement. J. Magn. Reson. 2001;149:239–244. doi: 10.1006/jmre.2001.2300. [DOI] [PubMed] [Google Scholar]
- Schwieters, C.D., Kuszewski, J.J., Tjandra, N., Clore, G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Soares, L.M., Zanier, K., Mackereth, C., Sattler, M., Valcarcel, J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312:1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- von Lindern, M., Fornerod, M., Soekarman, N., van Baal, S., Jaegle, M., Hagemeijer, A., Bootsma, D., Grosveld, G. Translocation t(6;9) in acute non-lymphocytic leukaemia results in the formation of a DEK-CAN fusion gene. Baillieres Clin. Haematol. 1992;5:857–879. doi: 10.1016/s0950-3536(11)80049-1. [DOI] [PubMed] [Google Scholar]
- Waldmann, T., Eckerich, C., Baack, M., Gruss, C. The ubiquitous chromatin protein DEK alters the structure of DNA by introducing positive supercoils. J. Biol. Chem. 2002;277:24988–24994. doi: 10.1074/jbc.M204045200. [DOI] [PubMed] [Google Scholar]
- Waldmann, T., Baack, M., Richter, N., Gruss, C. Structure-specific binding of the proto-oncogene protein DEK to DNA. Nucleic Acids Res. 2003;31:7003–7010. doi: 10.1093/nar/gkg864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldmann, T., Scholten, I., Kappes, F., Hu, H.G., Knippers, R. The DEK protein—An abundant and ubiquitous constituent of mammalian chromatin. Gene. 2004;343:1–9. doi: 10.1016/j.gene.2004.08.029. [DOI] [PubMed] [Google Scholar]
- Walker, J.R., Corpina, R.A., Goldberg, J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- Wise-Draper, T.M., Allen, H.V., Thobe, M.N., Jones, E.E., Habash, K.B., Munger, K., Wells, S.I. The human DEK proto-oncogene is a senescence inhibitor and an up-regulated target of high-risk human papillomavirus E7. J. Virol. 2005;79:14309–14317. doi: 10.1128/JVI.79.22.14309-14317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise-Draper, T.M., Allen, H.V., Jones, E.E., Habash, K.B., Matsuo, H., Wells, S.I. The human DEK oncoprotein is an apoptosis inhibitor and interferes with p53 transcriptional activity. Mol. Cell. Biol. 2006;26:7506–7519. doi: 10.1128/MCB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Zhu, L., Lin, D., Chen, F., Chen, D.J., Chen, Y. The three-dimensional structure of the C-terminal DNA-binding domain of human Ku70. J. Biol. Chem. 2001;276:38231–38236. doi: 10.1074/jbc.M105238200. [DOI] [PubMed] [Google Scholar]