Abstract
The Escherichia coli inner membrane enzyme DsbB catalyzes disulfide bond formation in periplasmic proteins, by transferring electrons to ubiquinone from DsbA, which in turn directly oxidizes cysteines in substrate proteins. We have previously shown that DsbB can be prepared in a state that gives highly resolved magic-angle spinning (MAS) NMR spectra. Here we report sequential 13C and 15N chemical shift assignments for the majority of the residues in the transmembrane helices, achieved by three-dimensional (3D) correlation experiments on a uniformly 13C, 15N-labeled sample at 750-MHz 1H frequency. We also present a four-dimensional (4D) correlation spectrum, which confirms assignments in some highly congested regions of the 3D spectra. Overall, our results show the potential to assign larger membrane proteins using 3D and 4D correlation experiments and form the basis of further structural and dynamical studies of DsbB by MAS NMR.
Keywords: solid-state NMR, membrane protein, chemical shift assignment, magic-angle spinning, disulfide bond formation
Membrane proteins are estimated to constitute 30%–40% of the proteins encoded by the human genome (Liu et al. 2002), and the majority of the drugs on the market are targeted to membrane proteins (Lundstrom 2005). Despite their high abundance and importance, they are underrepresented in current protein structure databases. In recent years, structural biology tools including both X-ray crystallography (Byrne and Iwata 2002) and solution NMR (Pervushin et al. 1997; Fernandez and Wider 2003; Tamm and Liang 2006) have been increasingly applied to membrane proteins (Torres et al. 2003), resulting in more than 100 unique high-resolution structures. Both structure and dynamics can be examined by NMR, and recent advances in solid-state NMR methods (McDermott 2004) are especially beneficial for membrane proteins since samples can be prepared in either native-like lipid environments or chemically defined lipid bilayers without using detergents. Another advantage of SSNMR is that line widths are not correlated to the size of the protein.
SSNMR techniques capable of determining the relative orientation of transmembrane helices from macroscopically aligned samples have been well established (Opella and Marassi 2004). However, the spectral quality depends on the degree of alignment. On the other hand, magic-angle spinning (MAS) experiments do not require sample alignment but require signals to be assigned in a site-specific fashion that is analogous to solution NMR. Due to the smaller chemical shift range within transmembrane helices (relative to β-strand secondary structures), this task can be time-consuming and technically challenging. Moreover, few membrane proteins have solution NMR assignments available, and so the SSNMR assignments must be performed de novo. So far, nearly complete or partial assignments have been reported for only a few membrane proteins, compared with more than a dozen soluble proteins (with molecular weight ranging from 5 to 20 kDa) that have been completely assigned (McDermott 2004; Pintacuda et al. 2007). In these few studies, partially selective labeling schemes have been employed to simplify spectra (Egorova-Zachernyuk et al. 2001; Gammeren et al. 2005; Etzkorn et al. 2007) except for a small single transmembrane protein (Andronesi et al. 2005). Here we show that the majority of the signals from the transmembrane helices of a 20-kDa membrane protein, DsbB, can be assigned using one uniformly 13C, 15N-labeled sample and three-dimensional (3D) correlation spectroscopy.
DsbB is an integral membrane enzyme located on the inner membrane of Escherichia coli. Together with soluble protein DsbA, it catalyzes disulfide bond formation in many periplasmic proteins and facilitates their folding (Kadokura et al. 2003). DsbB contains four transmembrane helices and binds DsbA via one of two periplasmic loops. There is so far one published high-resolution structural study of DsbB (Inaba et al. 2006), in which DsbB was co-crystallized with DsbA in a covalently linked complex. Efforts to crystallize DsbB in the absence of DsbA were not successful. In the present study, we use MAS NMR techniques to study the C41S mutant of DsbB, which represents a transient intermediate state in the proposed reaction pathways (Inaba et al. 2005). Besides reporting the signal assignment of transmembrane helices, we demonstrate that four-dimensional (4D) correlation spectra can be acquired on the same sample with good sensitivity, illustrating the potential to assign membrane proteins of larger size.
Results and Discussion
Four 3D chemical shift correlation experiments (NCACX, NCOCX, CAN(CO)CX, and CON(CA)CX) were acquired at 750 MHz 1H frequency for signal assignment. Three-dimensional NCACX and NCOCX experiments form the basis of most previous chemical shift assignment work on solid proteins by MAS NMR (McDermott 2004). However, these two experiments are not sufficient for uniformly labeled polytopic membrane proteins, which typically contain a large number of hydrophobic residues in helical conformation. Within these amino acid types, both 13C and 15N chemical shifts are highly degenerate, though to less extent for 15N. In a previous study, we have reported the method to prepare DsbB samples that give narrow line widths (∼0.5 ppm for 13C and ∼0.7 ppm for 15N at 750 MHz 1H frequency) and partial assignment of signals from NCOCX, NCACX, and CANCO spectra (Li et al. 2007). The same method was first employed to prepare a 144-kDa membrane protein, cytochrome bo3 oxidase, for MAS NMR studies (Frericks et al. 2006). In this study, we have employed a revised sample preparation to obtain protein pellets with less bulk lipids, which enabled us to pack a larger quantity of protein into the rotor. The enhanced sample sensitivity allows for two additional experiments, CAN(CO)CX and CON(CA)CX, to be performed, which provide unambiguous connectivity information.
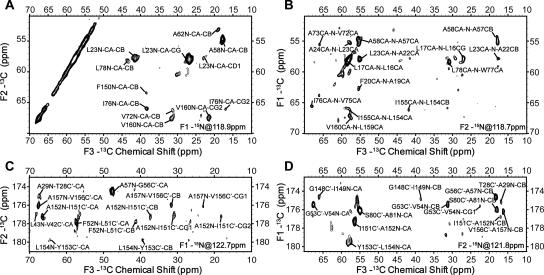
For all 3D experiments, relatively short 13C-13C mixing times were chosen to give only intraresidue correlations. Figure 1 shows representative two-dimensional (2D) planes from these 3D spectra. Among these experiments, CAN(CO)CX and CON(CA)CX 3D are more technically challenging due to the fact that three cross-polarization steps, two of which involve 15N-13C transfers, must be performed with high stability and transfer efficiency (Franks et al. 2007). However, they provide a few advantages over NCOCX and NCACX 3D with long 13C-13C mixing times, which can also provide a similar type of connectivity information. First, the correlation pattern is more predictable since long mixing times often result in cross-peaks between spins that are not close in the primary sequence. Second, the number of cross-peaks is approximately the same as in NCOCX and NCACX with short mixing times rather than being significantly increased. This presents a significant advantage to the time-consuming stages of data analysis, which are not yet sufficiently automated for SSNMR. Third, the CAN(CO)CX and CON(CA)CX experiments provide unambiguous interresidue 13C-13C correlations and therefore eliminate the need to infer the types (intra- or interresidue) of correlations from peak intensity, which can be significantly affected not only by the internuclear distances but also by molecular dynamics.
Figure 1.
Representative 2D planes from 3D (A) NCACX, (B) CAN(CO)CX, (C) NCOCX, and (D) CON(CA)CX chemical shift correlation spectra acquired on a [U-13C,15N] DsbB C41S sample (∼1 μmol) at 750-MHz 1H frequency, 12.5-kHz MAS frequency, and 223 K; 75-kHz 1H TPPM decoupling (6.5 μs, 9°) was used during indirect evolution periods and acquisition period, and 90-kHz CW decoupling was used during 15N-13C and 13C-15N SPECIFIC CP periods. Additional acquisition and all processing parameters for each spectrum are included in the Electronic supplementary material.
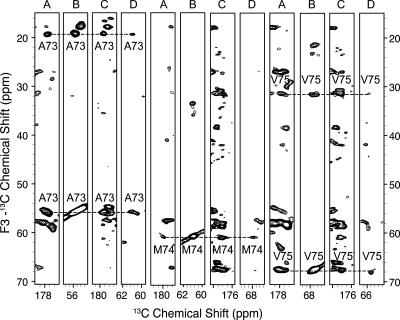
Similar to solution NMR protocols, the sequential assignment process involves spin system identification, the linking of spin systems, amino acid type identification, and mapping to the amino acid sequence. Because amide proton chemical shifts are generally not observed in SSNMR (with notable exceptions) (Paulson et al. 2003; Hologne et al. 2006; Zhou et al. 2007a,b), more accurate identification of amino acid types is required to efficiently link spin systems and map them back to the sequence. The iterative assignment process proceeds in stages. In the first stage, CON(CA)CX and NCACX spectra are combined to identify all spins from the i residue and C′[i − 1]. Similarly, NCOCX and CAN(CO)CX spectra can be combined to identify all spins from the i − 1 residue, Cα[i] and N[i], forming another group of spin systems. Subsequently, these two groups of spin systems are compared to link i and i − 1 residues. The results from this linking step are highly reliable since three backbone chemical shifts (N[i], C′[i − 1], Cα[i]) are matched and the overall degeneracy for this particular set of chemical shifts is quite low. The second stage of the linking process involves taking two residue pairs to form a triplet by matching the chemical shifts of all 13C spins from the same residue. False matching can occur at this stage due to the high 13C chemical shift degeneracy within residues of the same type and secondary structure. Therefore, at this stage, accurate determination of amino acid types is essential to eliminate the triplets that are not present in the amino acid sequence. For DsbB, this can be done for a large percentage of residue pairs since the three most abundant amino acid types (Leu, Ala, and Val) are among those that can be unambiguously determined from Cα, Cβ, and Cγ chemical shifts. This is expected to be generally the case for the transmembrane regions of other membrane proteins since they are always featured by high abundance in hydrophobic residues, among which most of the nonaromatic types can be easily determined. Figure 2 shows a strip plot of 3D spectra that contain cross-peaks from the triplet A73-V75. If a single amino acid type cannot be assigned, a generic type consisting of several types of amino acids with similar chemical shift ranges could be assigned and sequence information can be used in a similar way. Linking triplets and mapping them back to the sequence is straightforward and therefore is not discussed here.
Figure 2.
A strip plot of residues A73–V75. The plot consists of strips from four 3D spectra: (A) CON(CA)CX, (B) NCACX, (C) NCOCX, and (D) CAN(CO)CX.
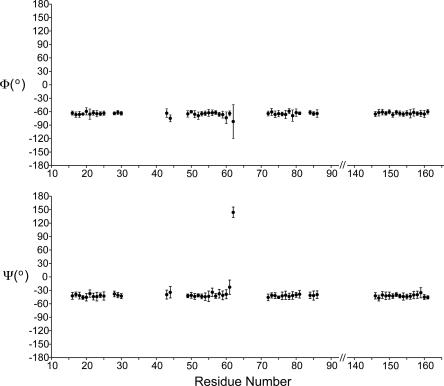
By following the protocol described above, we were able to assign 90% of the signals that were detected in at least one of the 3D spectra. The remaining 10% of signals were not assigned due to either very high degeneracy in the chemical shifts of many Leu residues or signals missing in one or two 3D spectra. The assignment table is included in the Supplementary material and has been submitted to the BMRB as entry 15,546 (Seavey et al. 1991). Figure 3 shows the backbone torsion angles of all assigned residues determined from assigned chemical shifts by the program TALOS (Cornilescu et al. 1999). All assigned residues have helical secondary structure except for G61 and A62. These two residues are located at the end of the second transmembrane helix and the beginning of the short intracellular loop. Other signals in this loop are missing, and similarly, we find no long stretches of amino acids consistent with the periplasmic region. Since essentially all of the signals observed in the spectra have been assigned at least by amino acid type, we conclude that the signals from two periplasmic regions must be below the detection limit in the 3D spectra. The missing signals can be explained by either low CP efficiency caused by motion or inhomogeneous line broadening caused by multiple conformations in slow chemical exchange. If the former is the case, the overall 13H-13C CP enhancement factor determined from 13C one-dimensional (1D) experiments with and without CP should be significantly lower than that for a completely rigid protein since ∼90 out of 176 residues are located outside of the transmembrane helices. However, the (1H-13C) CP enhancement factor remains ∼2.0 throughout the temperature range from 223 K to 283 K, which is close to the values previously detected for completely rigid proteins (Franks et al. 2005); likewise the 15N-13C transfers proceed with high efficiency (∼50% for N-C′ and ∼40% for N-CA), which is only possible if the majority of the protein backbone is rigid. In addition, at 283 K we did not detect backbone 13C signals with significantly longer T2 values than 15 ms, suggesting that the periplasmic regions are not mobile even when the actual sample temperature is close to room temperature. Therefore, we conclude that static disorder is the reason why periplasmic signals are too broad to be detected in the 3D spectra yet contribute to the overall integrated intensity of 1D CP-MAS spectra. In the X-ray structure (Inaba et al. 2006), the second periplasmic region contains an additional helix, which was not observed in our spectra. This we attribute to the likelihood that these residues are more disordered when DsbB is not in complex with DsbA. This structural difference is consistent with biochemical studies (Kadokura and Beckwith 2002; Inaba et al. 2005) that suggest the intrinsic flexibility of the second periplasmic region of DsbB is likely to be important for its function.
Figure 3.
Backbone torsion angles of assigned residues in DsbB C41S determined by TALOS analysis.
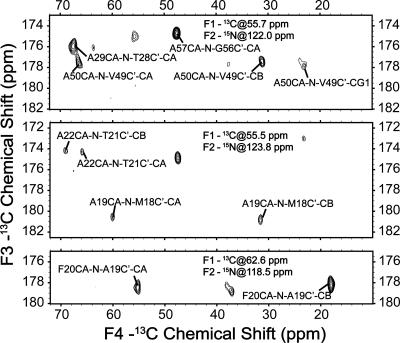
To evaluate the potential to assign larger membrane proteins, such as receptors and channels with seven or more transmembrane helices (von Heijne 2006), we acquired a 4D CANCOCX spectrum on the same DsbB sample. The polarization transfer pathway is the same as the 3D CAN(CO)CX experiment, but in addition, an evolution period is incorporated while the polarization resides on the CO spin (Franks et al. 2007). Figure 4 shows representative 2D planes from this 4D spectrum with assignments labeled. Many strong cross-peaks can be detected with chemical shifts in each dimension matching very well with the 3D data. Since 4D experiments are often associated with long sampling time, the choice of acquisition and sampling parameters can affect spectral quality significantly. A detailed investigation of how 4D experiments can be efficiently implemented on membrane protein samples is beyond the scope of this work; likely the fast methods developed in solution NMR in the last several years, including GFT, nonlinear sampling, and projection reconstructions (Szyperski and Atreya 2006), will be of benefit to acquire 4D spectra with higher digital resolution. However, the quality of the initial spectrum acquired at 500 MHz 1H frequency leads us to believe that the assignment of larger membrane proteins can benefit from 4D experiments.
Figure 4.
Representative 2D planes from 4D CANCOCX spectrum acquired on a [U-13C, 15N] DsbB C41S sample (∼1 μmol) at 500-MHz 1H frequency, 11.111-kHz MAS frequency, and 223 K; 80-kHz 1H TPPM decoupling (6.1 μs, 17°) was used during indirect evolution periods and acquisition period; 90-kHz CW decoupling was used during 15N-13C and 13C-15N SPECIFIC CP periods. The data were acquired with 0.45-ms 1H-15C CP, 40 points with a dwell time of 90 μs in t1 (13C), 6.0 ms 13Cα-15N CP, 40 points with a dwell time of 180 μs in t2 (15N), 6.0 ms 15N-13CO SPECIFIC CP, 20 points with a dwell time of 270 μs in t3 (13C), 25 ms 13C-13C DARR mixing, 1024 points with a dwell time of 15 μs in t4 (13C). Twelve scans were acquired for each row. The data were processed with 150-Hz, 90-Hz, 100-Hz, and 100-Hz net line broadening (Lorentzian-to-Gausssian apodization) in F1, F2, F3, and F4 dimensions, respectively, and zero filled to 128 (F1) × 80 (F2) × 128 (F3) × 2048 (F4) complex points prior to Fourier transformation.
In summary, we have employed a robust protocol to assign the majority of the signals from the transmembrane helices of DsbB C41S mutant by MAS NMR. Each 3D spectrum can be acquired on a DsbB sample containing ∼1 μmol protein within a few days. With the improved sensitivity and resolution of the data sets presented here, tentatively assigned signals from our previous study were in most cases confirmed; a subset of the previous assignments were determined to be in error, and in all cases the present study should be considered definitive. Our results demonstrate the feasibility of assigning the transmembrane helices with relatively high chemical shift degeneracy using uniformly labeled samples. The approach presented here is generally applicable to membrane proteins that can be efficiently expressed in isotope-enriched growth media.
Materials and Methods
Sample preparation
DsbB was expressed as previously described (Li et al. 2007) from E. coli C43 (DE3) containing a plasmid encoding DsbB C41S with a 6-His tag and mutation of two nonessential cysteines, C8A and C49V (Inaba et al. 2004). DsbB was solubilized from isolated membranes using 1% dodecylmaltoside and purified using metal chelate chromatography on Talon cobalt resin, by elution with 120 mM imidazole (pH 6.0). Fractions containing DsbB were pooled and concentrated, followed by dialysis against 25 mM Tris (pH 7.5) to remove dodecylmaltoside and imidazole. Centrifugation of the dialyzed suspension in a fixed-angle rotor (1 h, 100,000g) produced a hard pellet consisting of contaminating protein and lipid. Further centrifugation in a swinging bucket rotor (20 h, 100,000g) separated the resulting magenta-colored supernatant into a viscous deep red pellet (DsbB C41S) and a colorless solution. The pellet was then packed into a 3.2-mm thin wall rotor with an active volume of ∼36 μL using several low speed centrifugations (5–10 min, 3000g). The 13C and 15N signals detected in 1D CP-MAS experiments correspond to 20 mg (∼1.0 μmol) of packed protein, which was determined by comparing to the total integrated signals of a standard protein of known quantity (GB1) (Franks et al. 2005).
NMR spectroscopy
SSNMR experiments were performed either on a 750 MHz (1H frequency) three-channel Varian Inova spectrometer or a 500 MHz (1H frequency) four-channel Varian InfinityPlus spectrometer. Both spectrometers were equipped with 3.2 mm 1H-13C-15N Balun MAS probes. The typical π/2 pulse widths were 2.5 μs on 1H, 2.9 μs on 13C, and 5.6 μs on 15N for the 750 MHz probe and were 2.4 μs on 1H, 2.4 μs on 13C, and 7.3 μs on 15N for the 500 MHz probe. The flow rate of sample cooling gas was maintained at ∼90 scfh, with the gas temperature measured at the output of the delivery stack. For 3D 15N-13C-13C and 13C-15N-13C correlation experiments, band-selective SPECIFIC CP (Baldus et al. 1998) was used for polarization transfer between 15N and 13C, and DARR (Takegoshi et al. 2001) was employed for 13C-13C mixing. All pulse sequences were implemented with tangent ramped CP (Hediger et al. 1994) and TPPM 1H decoupling (Bennett et al. 1995) at ∼75 kHz during indirect evolution and acquisition periods.
Data were processed with nmrPipe (Delaglio et al. 1995), employing zero filling and Lorentzian-to-Gaussian apodization for each dimension before Fourier transformation. Back linear prediction and polynomial baseline correction (frequency domain) were applied to the direct dimension. Additional acquisition and processing parameters for each spectrum are included in either the figure captions or Supplementary material. Chemical shifts were referenced externally with adamantane, with the downfield 13C resonance referenced to 40.48 ppm (Morcombe and Zilm 2003). Peak picking and assignment were performed with Sparky (http://www.cgl.ucsf.edu/home/sparky/).
Acknowledgments
This work was supported by the National Institutes of Health (NIGMS and Roadmap Initiative R01GM075937). We thank Dr. Kenji Inaba and Professor Koreaki Ito for providing the plasmid encoding DsbB and the University of Illinois School of Chemical Sciences NMR Facility for technical support.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Chad M. Rienstra, Department of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, USA; e-mail: rienstra@scs.uiuc.edu; fax: (217) 244-3186.
Abbreviations: BMRB, biological magnetic resonance data bank; CP, cross-polarization; DARR, dipolar assisted rotational resonance; SPECIFIC CP, spectrally induced filtering in combination with cross-polarization; TPPM, two pulse phase modulation.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073225008.
References
- Andronesi, O.C., Becker, S., Seidel, K., Heise, H., Young, H.S., Baldus, M. Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J. Am. Chem. Soc. 2005;127:12965–12974. doi: 10.1021/ja0530164. [DOI] [PubMed] [Google Scholar]
- Baldus, M., Petkova, A.T., Herzfeld, J., Griffin, R.G. Cross polarization in the tilted frame: Assignment and spectral simplification in heteronuclear spin systems. Mol. Phys. 1998;95:1197–1207. [Google Scholar]
- Bennett, A.E., Rienstra, C.M., Auger, M., Lakshmi, K.V., Griffin, R.G. Heteronuclear decoupling in rotating solids. J. Chem. Phys. 1995;103:6951–6958. [Google Scholar]
- Byrne, B., Iwata, S. Membrane protein complexes. Curr. Opin. Struct. Biol. 2002;12:239–243. doi: 10.1016/s0959-440x(02)00316-0. [DOI] [PubMed] [Google Scholar]
- Cornilescu, G., Delaglio, F., Bax, A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Egorova-Zachernyuk, T.A., Hollander, J., Fraser, N., Gast, P., Hoff, A.J., Cogdell, R., de Groot, H.J.M., Baldus, M. Heteronuclear 2D-correlations in a uniformly [C-13, N-15] labeled membrane-protein complex at ultra-high magnetic fields. J. Biomol. NMR. 2001;19:243–253. doi: 10.1023/a:1011235417465. [DOI] [PubMed] [Google Scholar]
- Etzkorn, M., Martell, S., Andronesi, O.C., Seidel, K., Engelhard, M., Baldus, M. Secondary structure, dynamics, and topology of a seven-helix receptor in native membranes, studied by solid-state NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2007;46:459–462. doi: 10.1002/anie.200602139. [DOI] [PubMed] [Google Scholar]
- Fernandez, C., Wider, G. TROSY in NMR studies of the structure and function of large biological macromolecules. Curr. Opin. Struct. Biol. 2003;13:570–580. doi: 10.1016/j.sbi.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Franks, W.T., Zhou, D.H., Wylie, B.J., Money, B.G., Graesser, D.T., Frericks, H.L., Sahota, G., Rienstra, C.M. Magic-angle spinning solid-state NMR spectroscopy of the beta1 immunoglobulin binding domain of protein G (GB1): 15N and 13C chemical shift assignments and conformational analysis. J. Am. Chem. Soc. 2005;127:12291–12305. doi: 10.1021/ja044497e. [DOI] [PubMed] [Google Scholar]
- Franks, W.T., Kloepper, K.D., Wylie, B.J., Rienstra, C.M. Four-dimensional heteronuclear correlation experiments for chemical shift assignment of solid proteins. J. Biomol. NMR. 2007;39:107–131. doi: 10.1007/s10858-007-9179-1. [DOI] [PubMed] [Google Scholar]
- Frericks, H.L., Zhou, D.H., Yap, L.L., Gennis, R.B., Rienstra, C.M. Magic-angle spinning solid-state NMR of a 144 kDa membrane protein complex: E. coli cytochrome bo(3) oxidase. J. Biomol. NMR. 2006;36:55–71. doi: 10.1007/s10858-006-9070-5. [DOI] [PubMed] [Google Scholar]
- Gammeren, A.J., Hulsbergen, F.B., Hollander, J.G., de Groot, H.J. Residual backbone and side-chain 13C and 15N resonance assignments of the intrinsic transmembrane light-harvesting 2 protein complex by solid-state magic angle spinning NMR spectroscopy. J. Biomol. NMR. 2005;31:279–293. doi: 10.1007/s10858-005-1604-8. [DOI] [PubMed] [Google Scholar]
- Hediger, S., Meier, B.H., Kurur, N.D., Bodenhausen, G., Ernst, R.R. NMR cross-polarization by adiabatic passage through the Hartmann-Hahn condition (APHH) Chem. Phys. Lett. 1994;223:283–288. [Google Scholar]
- Hologne, M., Chevelkov, V., Reif, B. Deuterated peptides and proteins in MAS solid-state NMR. Prog. NMR Spec. 2006;48:211–232. [Google Scholar]
- Inaba, K., Takahashi, Y.H., Fujieda, N., Kano, K., Miyoshi, H., Ito, K. DsbB elicits a red-shift of bound ubiquinone during the catalysis of DsbA oxidation. J. Biol. Chem. 2004;279:6761–6768. doi: 10.1074/jbc.M310765200. [DOI] [PubMed] [Google Scholar]
- Inaba, K., Takahashi, Y.H., Ito, K. Reactivities of quinone-free DsbB from Escherichia coli . J. Biol. Chem. 2005;280:33035–33044. doi: 10.1074/jbc.M506189200. [DOI] [PubMed] [Google Scholar]
- Inaba, K., Murakami, S., Suzuki, M., Nakagawa, A., Yamashita, E., Okada, K., Ito, K. Crystal structure of DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell. 2006;127:789–801. doi: 10.1016/j.cell.2006.10.034. [DOI] [PubMed] [Google Scholar]
- Kadokura, H., Beckwith, J. Four cysteines of the membrane protein DsbB act in concert to oxidize its substrate DsbA. EMBO J. 2002;21:2354–2363. doi: 10.1093/emboj/21.10.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadokura, H., Katzen, F., Beckwith, J. Protein disulfide bond formation in prokaryotes. Annu. Rev. Biochem. 2003;72:111–135. doi: 10.1146/annurev.biochem.72.121801.161459. [DOI] [PubMed] [Google Scholar]
- Li, Y., Berthold, D.A., Frericks, H.L., Gennis, R.B., Rienstra, C.M. Partial 13C and 15N chemical-shift assignments of the disulfide-bond-forming enzyme DsbB by 3D magic-angle spinning NMR spectroscopy. ChemBioChem. 2007;8:434–442. doi: 10.1002/cbic.200600484. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Engelman, D.M., Gerstein, M. Genomic analysis of membrane protein families: Abundance and conserved motifs. Genome Biol. 2002;3:0054.1–0054.12. doi: 10.1186/gb-2002-3-10-research0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstrom, K. Structural genomics of GPCRs. Trends Biotechnol. 2005;23:103–108. doi: 10.1016/j.tibtech.2004.12.006. [DOI] [PubMed] [Google Scholar]
- McDermott, A.E. Structural and dynamic studies of proteins by solid-state NMR spectroscopy: Rapid movement forward. Curr. Opin. Struct. Biol. 2004;14:554–561. doi: 10.1016/j.sbi.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Morcombe, C.R., Zilm, K.W. Chemical shift referencing in MAS solid state NMR. J. Magn. Reson. 2003;162:479–486. doi: 10.1016/s1090-7807(03)00082-x. [DOI] [PubMed] [Google Scholar]
- Opella, S.J., Marassi, F.M. Structure determination of membrane proteins by NMR spectroscopy. Chem. Rev. 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson, E.K., Morcombe, C.R., Gaponenko, V., Dancheck, B., Byrd, R.A., Zilm, K.W. Sensitive high resolution inverse detection NMR spectroscopy of proteins in the solid state. J. Am. Chem. Soc. 2003;125:15831–15836. doi: 10.1021/ja037315+. [DOI] [PubMed] [Google Scholar]
- Pervushin, K., Riek, R., Wider, G., Wuthrich, K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintacuda, G., Giraud, N., Pierattelli, R., Bockmann, A., Bertini, I., Emsley, L. Solid-state NMR spectroscopy of a paramagnetic protein: Assignment and study of human dimeric oxidized CuII-ZnII superoxide dismutase (SOD) Angew. Chem. Int. Ed. Engl. 2007;46:1079–1082. doi: 10.1002/anie.200603093. [DOI] [PubMed] [Google Scholar]
- Seavey, B.R., Farr, E.A., Westler, W.M., Markley, J.L. A relational database for sequence-specific protein NMR data. J. Biomol. NMR. 1991;1:217–236. doi: 10.1007/BF01875516. [DOI] [PubMed] [Google Scholar]
- Szyperski, T., Atreya, H.S. Principles and applications of GFT projection NMR spectroscopy. Magn. Reson. Chem. 2006;44:S51–S60. doi: 10.1002/mrc.1817. [DOI] [PubMed] [Google Scholar]
- Takegoshi, K., Nakamura, S., Terao, T. C-13-H-1 dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 2001;344:631–637. doi: 10.1063/1.2364503. [DOI] [PubMed] [Google Scholar]
- Tamm, L.K., Liang, B.Y. NMR of membrane proteins in solution. Prog. NMR Spec. 2006;48:201–210. [Google Scholar]
- Torres, J., Stevens, T.J., Samso, M. Membrane proteins: The “wild west” of structural biology. Trends Biochem. Sci. 2003;28:137–144. doi: 10.1016/S0968-0004(03)00026-4. [DOI] [PubMed] [Google Scholar]
- von Heijne, G. Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- Zhou, D.H., Shah, G., Cormos, M., Mullen, C., Sandoz, D., Rienstra, C.M. Proton-detected solid-state NMR spectroscopy of fully protonated proteins at 40 kHz magic-angle spinning. J. Am. Chem. Soc. 2007a;129:11791–11801. doi: 10.1021/ja073462m. [DOI] [PubMed] [Google Scholar]
- Zhou, D.H., Shea, J.J., Nieuwkoop, A.J., Franks, W.T., Wylie, B.J., Mullen, C., Sandoz, D., Rienstra, C.M. Solid-state protein structure determination with proton-detected triple resonance 3D magic-angle spinning spectroscopy. Angew. Chem. Int. Ed. Engl. 2007b;46:8380–8383. doi: 10.1002/anie.200702905. [DOI] [PMC free article] [PubMed] [Google Scholar]