Abstract
Buried water molecules (having no contact with bulk solvent) in 30 helical transmembrane (TM) protein structures were identified. The average amount of buried water in helical TM proteins is about the same as for all water-soluble (WS) proteins, but it is greater than the average for helical WS proteins. Buried waters in TM proteins make more polar contacts, and are more frequently found contacting helices than in WS proteins. The distribution of the buried water binding sites across the membrane profile shows that the sites to some extent reflect protein function. There is also evidence for asymmetry of the sites, with more in the extracellular half of the membrane. Many of the buried water contact sites are conserved across families of proteins, including family members having different functions. This suggests that at least some buried waters play a role in structural stabilization. Disease-causing mutations, which are known to result in misfolded TM proteins, occur at buried water contact sites at a higher than random frequency, which also supports a stabilizing role for buried water molecules.
Keywords: membrane protein structure, bacteriorhodopsin, aquaporin, cytochrome c oxidase, rhodopsin
In protein crystal structures, water molecules which have no contact with bulk solvent are called “buried.” A detailed survey of buried water in proteins was done by Williams et al. (1994). This study examined 75 water-soluble proteins (WS). However, no comparable survey is available for integral membrane proteins. Membrane-embedded proteins which transport polar solutes across the membrane are expected to have water-filled channels. However, some high-resolution structures of helical transmembrane proteins show helix-bridging water molecules (Luecke et al. 1999), suggesting a possible structural role of buried water. In our recent studies of acid-induced unfolding of the membrane protein bacterio-opsin (Valluru et al. 2006) we found evidence that helix-bridging water molecules contribute to the stabilization of the folded structure. In order to determine whether buried water is common in helical transmembrane (TM) proteins, and to compare these structures with water-soluble proteins, I surveyed 30 high-resolution crystal structures from the Protein Data Bank (Table 1).
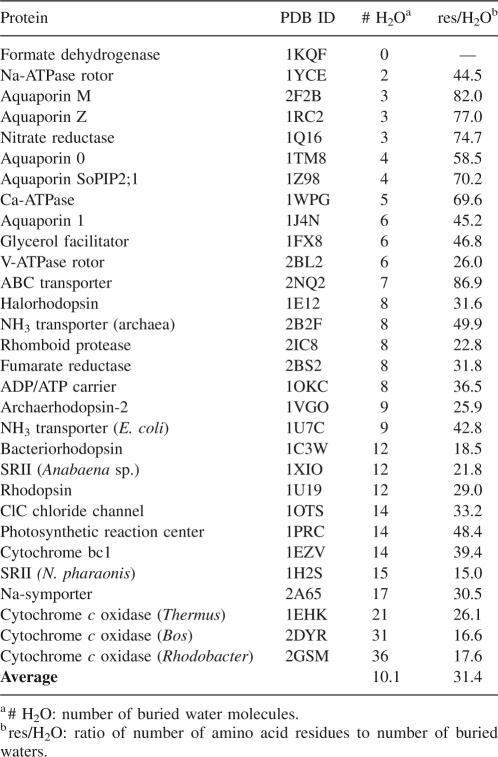
Table 1.
Proteins included in this study
Results and Discussion
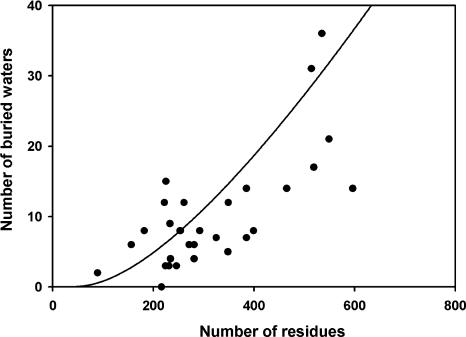
The number of buried water molecules in helical TM proteins increases with protein size (Fig. 1), as found in previous surveys of buried water in WS proteins (Williams et al. 1994; Park and Saven 2005). The buried water content in helical TM proteins is one buried water per 31 amino acids (Table 1), about the same as the average of one per 27 amino acids for all WS proteins examined by Williams et al. (1994) but a much higher frequency than their average of one per 91 amino acids for α-helical WS proteins. The difference could result from: (1) the small size of the WS helical proteins in their study (all were 20 kDa or smaller); and (2) the ion transport function of many of the helical TM proteins in Figure 1: for example, buried water in the microbial rhodopsin family could have a dual role in both stabilizing the structure and participating in the transport function, as discussed below; or (3) inherent differences in the stability of WS and TM proteins.
Figure 1.
Number of buried waters in helical TM proteins. Number of buried water molecules generally increases with size of transmembrane domain (line represents fit to data for water-soluble proteins) (Park and Saven 2005). Proteins are listed in Table 1.
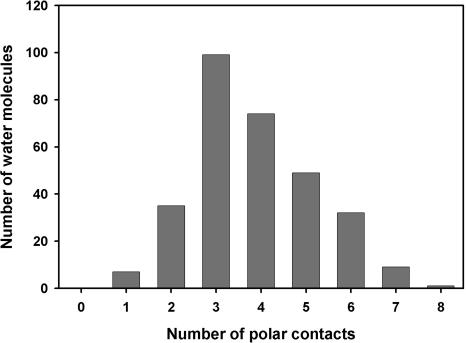
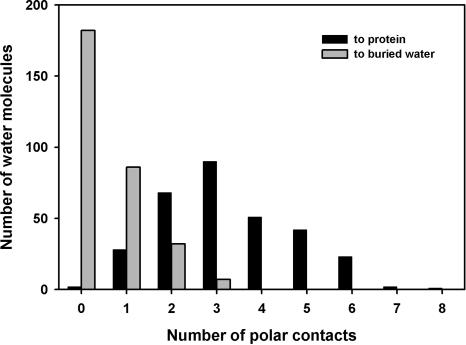
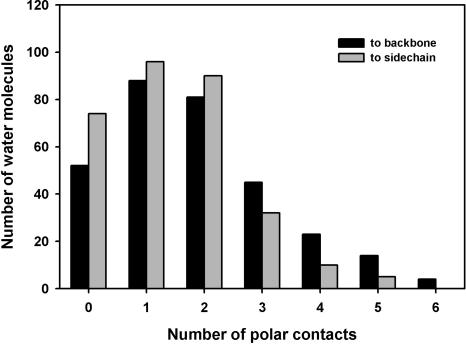
The distribution of polar contacts of buried water in helical TM proteins has a maximum at three (Fig. 2), the same as for buried waters in WS proteins (cf. Fig. 5A in Williams et al. 1994). However, the distribution is broader in TM proteins, with relatively more buried waters having five or six polar contacts. The distribution of buried water polar contacts to protein or to other water molecules (Fig. 3) shows that there are relatively more waters with more than three polar contacts to protein in TM than in WS proteins (cf. Fig. 5B in Williams et al. 1994). The distribution of polar contacts between backbone and side chain (Fig. 4) is similar to WS proteins for zero and one polar contact. However, water molecules that make more than one polar contact with protein have relatively more contacts to side chains in TM than in WS proteins (cf. Fig. 5C in Williams et al. 1994). These differences may reflect the transport function of many of the TM proteins in the data set, in which buried water molecules interact with protein side chains through hydrogen-bonded networks.
Figure 2.
Number of polar contacts made by water molecules buried in TM proteins. Polar contacts defined as nitrogen or oxygen atoms within 3.6 Å of buried water oxygen atom. Most common number is 3, the same as for WS proteins (cf. Williams et al. 1994).
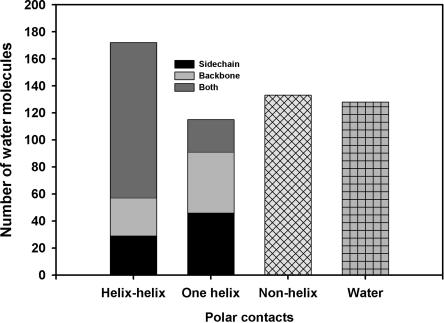
Figure 5.
Polar contacts classified by helix interaction. Number of waters bridging between helices, contacting a single helix, contacting nonhelical regions, or contacting other waters. Substantial numbers of polar contacts to helix backbone are observed.
Figure 3.
Number of polar contacts made by buried waters to protein and to other buried water molecules. Three contacts to protein are most common, but some waters make three polar contacts by interacting with other waters. Waters with more than three polar contacts to protein are relatively more abundant in TM proteins compared to WS proteins (cf. Williams et al. 1994).
Figure 4.
Distribution of polar contacts between buried water and protein side chains or backbone. There are relatively more polar contacts to side chains in TM proteins than in WS proteins (cf. Williams et al. 1994).
The number of buried water polar contacts to helices (Fig. 5) is much larger than might be expected from the number of buried water hydrogen bonds previously observed in WS proteins by Park and Saven (2005). At least one polar contact is made to a helix by 94% of the buried water molecules in TM helical proteins; 44% to a nonhelical region; and 42% to another water molecule. By contrast, only 24% of buried waters were found by Park and Saven (2005) to hydrogen bond to helices in WS proteins. The TM results in Figure 5 are polar contacts, and the WS results of Park and Saven (2005) are calculated H-bonds, which are not directly comparable. However, the number of polar contacts sets an upper limit for the number of H-bonds to water in TM proteins. Another difference is the higher helix frequency in the TM data set of this paper compared with Dunbrack's WS protein data set used by Park and Saven (2005). Nevertheless, as pointed out above, the TM helical proteins have more buried water per amino acid than the WS helical proteins examined by Williams et al. (1994). Therefore, it seems likely that at least part of the higher amount of buried water contacting helices is a real difference between WS and TM proteins. A substantial number of the buried water contacts to TM helices are made to backbone O or NH. This is surprising, because these interactions should weaken the helix H-bonds. However, 69% of the water molecules making polar contacts to backbone atoms in TM helices link two helices together. This additional stabilization may be an acceptable trade-off for weakening the intrahelix H-bonding.
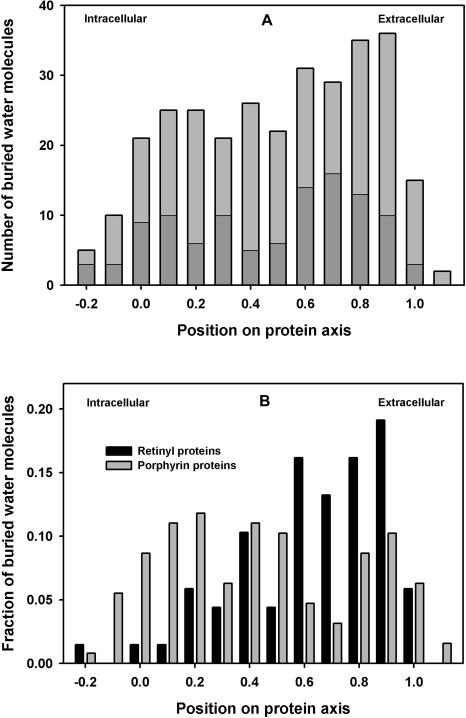
If water randomly occupies polar cavities in TM proteins, then the profile of water binding sites projected on the membrane normal would simply parallel the water activity in the bilayer (assuming that cavities occur randomly). This would result in a symmetric bilobed profile, with the highest water content near the two membrane surfaces and the lowest in the center (Zaccai et al. 1975; Gawrisch et al. 2007). The transmembrane positions of the water molecules in 30 TM proteins are shown in Figure 6A. The position corresponding to the bilayer center has a slight decline in buried water positions compared with the membrane surfaces, but not as pronounced as the water profile in protein-free lipid bilayers, which have very low water activity near the center of the membrane. In addition, there is a surprising asymmetry. More water molecules are found in the extracellular half of the proteins. To some extent, the asymmetry probably reflects protein function. When separated by function (Fig. 6B), the results show six retinal-containing proteins have high water content on the extracellular half, and eight porphyrin-containing proteins have high water content on the cytosolic half. When these 14 proteins are separated in Figure 6A, the profile of the remaining proteins (dark shaded bars) still shows more buried water on the extracellular half.
Figure 6.
Locations of buried water positions in the transmembrane profile. A transmembrane protein axis was created by calculating protein poles at the intracellular and extracellular membrane surfaces (see text). Water positions were projected onto the protein axis. (A) Histogram of all waters in proteins from Table 1. Shaded bars exclude proteins in Figure 6B. (B) Separate histograms of two functional groups, retinyl proteins and heme- or chlorophyll-binding proteins.
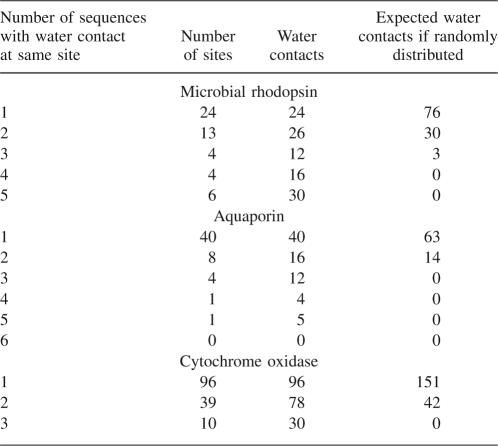
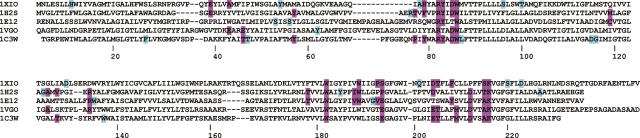
If buried water molecules have a role in stabilizing the folded structure of TM proteins, then water-binding sites would be subject to evolutionary selection. One way to test for selection would be to compare bound water positions in protein families with similar folds. Within the data set in Table 1, there are three protein families with three or more members: microbial rhodopsin (five), aquaporin (six), and cytochrome oxidase (three). Aligned sequences of the three families show the water contact positions are generally conserved. For microbial rhodopsins, 51 sequence positions have water in close contact with either the side chain or backbone. Of these, 27 are found at identical positions in at least two sequences (Table 2; Fig. 7). These positions account for 84 of 108 water contacts (78%). If water binds randomly to each of the five sequences, only 30% of the water contacts would occur at the same position in two or more sequences. Similar results were obtained for six aquaporins (48% of water contacts identical in two or more sequences, compared with 18% in a random binding model) and three cytochrome oxidases (53% identical contacts, compared with 22% in a random model). The high degree of conservation of these sites indicates selection, but it is not clear what is being selected. The proton pumps included in these sequences have internally bound water molecules which are involved in the H+ transport function. Therefore, some of the conserved water-binding sites reflect conserved function. However, Anabaena SR-II is not an ion transporter (Sineshchekov and Spudich 2004), but many of the same water-binding sites of bacteriorhodopsin are conserved in this photo-sensor. Thus, buried water may simultaneously serve roles for both structural stabilization by interhelix H-bonding, and also for participation in ion transport.
Table 2.
Comparison of buried water contact sites across protein families
Figure 7.
Conservation of water contact sites across the microbial rhodopsin family. Water contact positions (either side chain or backbone) are shaded with colored blocks. Cyan: sites only found in one sequence. Magenta: sites found in two or more sequences. Clustal alignment with sequence numbering for bacteriorhodopsin (1C3W). See Table 1 for other PDB codes.
The idea of structural stabilization by buried water can be stringently tested using site-directed mutagenesis. Side chains which participate in interhelix H-bonds could be replaced by non-H-bonding side chains. The resulting mutants should be less stable to unfolding conditions, as has been shown for water-soluble proteins (Griffin et al. 2002; Takano et al. 2003). Unfortunately, a methodology for unequivocal reversible unfolding of helical TM proteins is not available (Renthal 2006). Nevertheless, mutations which cause misfolding are known for some membrane proteins. Two misfolding mutations of aquaporin-0 are known to cause cataracts (Francis et al. 2000). A carboxyl oxygen in one site, E134 is about 4 Å from a conserved buried water binding site. The E134G mutation would alter the steric and electrostatic environment of the binding site for water 413, which consists of four polar backbone contacts and a side chain H-bond (see Supplemental Table A). It may be possible to separate the contributions of water 413 and E134 to the stabilization of aquaporin-0 by generating an S188A mutant, which would remove the hydrogen bond between S188 and water 413.
Many rhodopsin mutations are known to result in autosomal dominant retinitis pigmentosa (ADRP), a retinal degenerative disease. Fifty-seven rhodopsin sequence positions are known to have ADRP-causing missense mutations (www.retina-international.com/sci-news/rhomut.htm). Of these, 20 have been characterized as mutations which destabilize rhodopsin folding, either by in vitro measurements of stability or by studies of cellular localization (Sung et al. 1993; Kaushal and Khorana 1994). Five of the 12 buried waters in rhodopsin contact protein residues at these sites of destabilizing mutations (Fig. 8). There are 20 known sequence positions for which misfolding mutations are known, out of 348 sequence positions in rhodopsin. This implies that, if randomly distributed, only one of the 12 water contact sites should coincide with a misfolding mutation site. Therefore, retinitis pigmentosa mutations of rhodopsin support the concept of buried water molecules providing stabilization to helical TM proteins.
Figure 8.
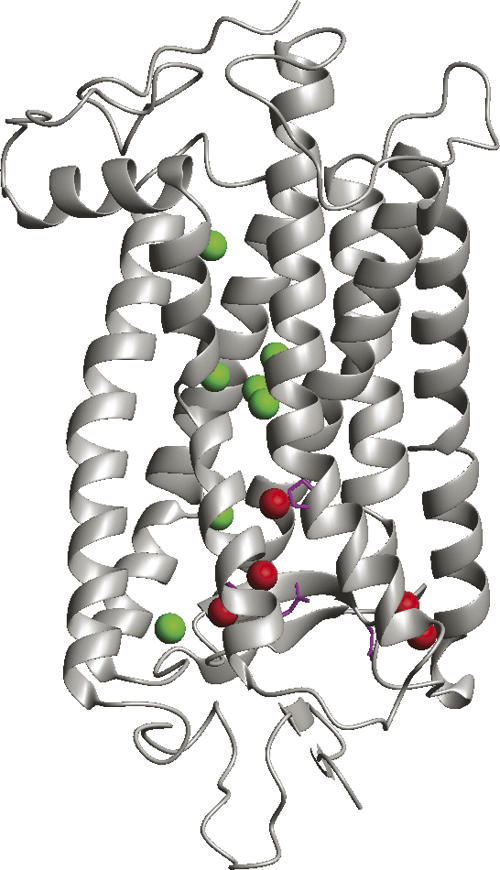
Structure of bovine rhodopsin showing positions of mutations associated with autosomal dominant retinitis pigmentosa (ADRP). Red spheres mark positions of five buried waters which contact sites of ADRP mutations thought to cause rhodopsin misfolding. Green spheres show seven other buried waters. Side chains shown in magenta. Figure generated from PDB file 1U19 using MOLMOL (Koradi et al. 1996).
Materials and Methods
PDB files of atomic coordinates of helical transmembrane proteins were selected from Stephen White's Web site “Membrane proteins of known 3D structure” (http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html), according to the following criteria. Only proteins with resolutions of 2.5 Å or better were considered. In cases where multiple structures are known for the same protein, generally the highest resolution structure was chosen. I excluded single TM helix proteins and proteins with large water channels. In proteins with narrow channels or half-channels, (e.g., aquaporins or microbial rhodopsins), waters which primarily contacted other waters in the channel were not included in the analysis. All protein domains extending into the aqueous regions outside the membrane were omitted. For structures with more than one protein molecule in the asymmetric unit, usually the chain containing more buried waters was selected for analysis. For the ATPase rotors (1YCE and 2BL2) only one chain of the multimer was analyzed. Six of the proteins in Table 1 are hetero-oligomers; in four (1H2S, 1EZV, 2GSM, and 1EHK), all buried waters were found in one subunit. Water that was shared between chains was not included, except for three waters shared by the L and M chains in 1PRC. In 2DYR, only subunit I was analyzed.
Identification of buried water molecules
MSMS (Sanner et al. 1996) was used to calculate points located on the solvent-excluded surface of the protein (vertices). Cavity surfaces, if present, were not included. Surface normals for each vertex were also calculated using MSMS. The closest vertex to each water molecule was identified, and the angle between the surface normal and the vector from the vertex to the water oxygen was calculated. Waters with angles >90° were assumed to be interior. Each water identified in this way was examined with molecular display software to verify that it was buried (i.e., not visible from outside the molecular surface; a few waters located in occluded crevices on the surface were included). A complete list of the buried water molecules is available as Supplementary electronic material (Table A).
Polar contacts
For each buried water, the distances to the neighboring atoms were measured, and all atoms within 3.6 Å were tabulated (Supplementary material, Table A). Polar contacts were defined as N or O atoms within 3.6 Å of a water molecule. (Sulfur was not included as a polar atom, since its electronegativity is about the same as carbon.) Contacts with helices were determined based on the helix boundaries listed in the PDB files.
For comparisons of protein families (Table 2), water contacts were calculated by randomly allocating the average number of buried water molecule contact sites per chain (22 for microbial rhodopsin, 13 for aquaporin, 68 for cytochrome oxidase) and averaging the number of positional identities found in 10 groups of five chains (microbial rhodopsin), six chains (aquaporin), or three chains (cytochrome oxidase).
Protein axis normal to the membrane
The protein axis normal to the membrane was constructed as follows. The axis was defined by creating intracellular and extracellular poles. The poles were defined by averaging the coordinates of the Cα atoms at the beginning and end of each transmembrane helix (or each transmembrane cross, in the case of proteins with half-membrane helices, such as aquaporins). For some proteins the “extracellular” side was the periplasmic or luminal side of the protein. For mitochondrial proteins, the matrix side was considered to be the intracellular side. The protein axis is the transmembrane line connecting the intracellular and extracellular poles. The position of each water molecule was projected on the protein axis by calculating its projected distance d from the intracellular pole:
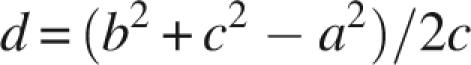 |
where a is the distance from the water molecule to the extracellular pole, b is the distance from the water molecule to the intracellular pole, and c is the length of the protein axis. Coordinates of the protein poles are listed in the Supplementary material (Table B).
Electronic supplemental material
Table A contains a list of buried water molecules and the atoms within 3.6 Å. Table B consists of coordinates of intracellular and extracellular poles used for transmembrane profile.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (S06 GM 08194). I thank Isaac Peña for assistance.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Robert Renthal, Department of Biology, University of Texas at San Antonio, 1 UTSA Circle, San Antonio, TX 78249, USA; e-mail: Robert.Renthal@UTSA.edu; fax: (210) 458-4467.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073237508.
References
- Francis, P., Chung, J.J., Yasui, M., Berry, V., Moore, A., Wyatt, M.K., Wistow, G., Bhattacharya, S.S., Agre, P. Functional impairment of lens aquaporin in two families with dominantly inherited cataracts. Hum. Mol. Genet. 2000;9:2329–2334. doi: 10.1093/oxfordjournals.hmg.a018925. [DOI] [PubMed] [Google Scholar]
- Gawrisch, K., Gaede, H.C., Mihailescu, M., White, S.H. Hydration of POPC bilayers studied by 1H-PFG-MAS-NOESY and neutron diffraction. Eur. Biophys. J. 2007;36:281–291. doi: 10.1007/s00249-007-0142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin, S., Vitello, A., Wittung-Stafshede, P. Buried water molecules contribute to cytochrome f stability. Arch. Biochem. Biophys. 2002;404:335–337. doi: 10.1016/s0003-9861(02)00333-8. [DOI] [PubMed] [Google Scholar]
- Kaushal, S., Khorana, H.G. Structure and function in rhodopsin. 7. Point mutations associated with autosomal dominant retinitis pigmentosa. Biochemistry. 1994;33:6121–6128. doi: 10.1021/bi00186a011. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., Wüthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Luecke, H., Schobert, B., Richter, H.-T., Cartailler, J.P., Lanyi, J.K. Structure of bacteriorhodopsin at 1.55 Å resolution. J. Mol. Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- Park, S., Saven, J.G. Statistical and molecular dynamics studies of buried waters in globular proteins. Proteins. 2005;60:450–463. doi: 10.1002/prot.20511. [DOI] [PubMed] [Google Scholar]
- Renthal, R. An unfolding story of helical transmembrane proteins. Biochemistry. 2006;45:14559–14566. doi: 10.1021/bi0620454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanner, M.F., Spehner, J.-C., Olson, A.J. Reduced surface: An efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Sineshchekov, O.A., Spudich, J.L. Light-induced intramolecular charge movements in microbial rhodopsins in intact E. coli cells. Photochem. Photobiol. Sci. 2004;3:548–554. doi: 10.1039/b316207a. [DOI] [PubMed] [Google Scholar]
- Sung, C.-H., Davenport, C.M., Nathans, J. Rhodopsin mutations responsable for autosomal dominant retinitis pigmentosa. Clustering of functional classes along the polypeptide chain. J. Biol. Chem. 1993;268:26646–26649. [PubMed] [Google Scholar]
- Takano, K., Yamagata, Y., Yutani, K. Buried water molecules contribute to the conformational stability of a protein. Protein Eng. 2003;16:5–9. doi: 10.1093/proeng/gzg001. [DOI] [PubMed] [Google Scholar]
- Valluru, N., Silva, F., Dhage, M., Rodriguez, G., Alloor, S.R., Renthal, R. Transmembrane helix-helix association: Relative stabilities at low pH. Biochemistry. 2006;45:4371–4377. doi: 10.1021/bi0525268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, M.A., Goodfellow, J.M., Thornton, J.M. Buried waters and internal cavities in monomeric proteins. Protein Sci. 1994;3:1224–1235. doi: 10.1002/pro.5560030808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccai, G., Blasie, J.K., Schoenborn, B.P. Neutron diffraction studies on the location of water in lecithin bilayer model membranes. Proc. Natl. Acad. Sci. 1975;72:376–380. doi: 10.1073/pnas.72.1.376. [DOI] [PMC free article] [PubMed] [Google Scholar]