Abstract
We present a detailed investigation of unfolded and partially folded states of a mutant apomyoglobin (apoMb) where the distal histidine has been replaced by phenylalanine (H64F). Previous studies have shown that substitution of His64, located in the E helix of the native protein, stabilizes the equilibrium molten globule and native states and leads to an increase in folding rate and a change in the folding pathway. Analysis of changes in chemical shift and in backbone flexibility, detected via [1H]-15N heteronuclear nuclear Overhauser effect measurements, indicates that the phenylalanine substitution has only minor effects on the conformational ensemble in the acid- and urea-unfolded states, but has a substantial effect on the structure, dynamics, and stability of the equilibrium molten globule intermediate formed near pH 4. In H64F apomyoglobin, additional regions of the polypeptide chain are recruited into the compact core of the molten globule. Since the phenylalanine substitution has negligible effect on the unfolded ensemble, its influence on folding rate and stability comes entirely from interactions within the compact folded or partly folded states. Replacement of His64 with Phe leads to favorable hydrophobic packing between the helix E region and the molten globule core and leads to stabilization of helix E secondary structure and overall thermodynamic stabilization of the molten globule. The secondary structure of the equilibrium molten globule parallels that of the burst phase kinetic intermediate; both intermediates contain significant helical structure in regions of the polypeptide that comprise the A, B, E, G, and H helices of the fully folded protein.
Keywords: protein folding, protein stability, hydrophobic interaction, chain dynamics, NMR spectroscopy, myoglobin, molten globule
It is commonly accepted that the primary sequence encodes the three-dimensional structure of proteins. Knowledge concerning preferred amino acid side chain and backbone contacts, residue packing, and rotamer distributions has expanded to a point where the ab initio design of novel proteins now appears possible (Kuhlman et al. 2003). However, the order of assembly of individual chain elements into a stable, native fold still presents an obstacle in protein design as well as in the understanding of many biological processes, including protein misfolding diseases. In particular, the fine balance of forces contributing to the formation of transient and intermediate structures in the various stages of the protein folding process is not well understood. A more detailed understanding of the underlying principles that govern protein folding requires extensive investigations of the kinetics and thermodynamics of folding, as well as structural studies at high resolution of unfolded and partially folded states. One approach, used in the present work, is to compare the effects of carefully selected mutations on both equilibrium and kinetic folding behavior.
The folding of apomyoglobin, the heme-free form of the oxygen binding protein myoglobin, is one of the best understood. The structure of myoglobin consists of eight α-helices (designated helices A through H), of which helices A, B, C, D, E, G, and H are folded in a native-like compact state in the apoprotein (Eliezer and Wright 1996; Lecomte et al. 1996). Helix F, which forms the lid on the heme-binding pocket in the holoprotein, undergoes conformational exchange in the apoprotein between closed and open forms (Eliezer and Wright 1996).
The folding pathway of apomyoglobin involves an obligatory on-pathway kinetic intermediate (Jennings and Wright 1993; Tsui et al. 1999; Nishimura et al. 2002, 2003, 2005a,b, 2006). The kinetic intermediate is formed rapidly, during the burst phase of the stopped-flow folding experiment. A very similar molten globule intermediate is formed under equilibrium conditions at pH 4.1 and is amenable to study by a variety of biophysical techniques, including high-resolution nuclear magnetic resonance (NMR) spectroscopy (Eliezer et al. 1998, 2000). In the equilibrium molten globule, helices A, B, G, and H are populated at almost native-like levels (Eliezer et al. 1998, 2000).
Acidification of apomyoglobin to pH 2.3 results in an acid-denatured state that has also been extensively studied by NMR. At this pH, most of the helical structure present in the molten globule state is lost, except for some residual helix propensity in the regions corresponding to the A, D/E, and H helices, which corresponds quite well with the high helix-forming propensity of these sequences studied as short peptides (Reymond et al. 1997; Yao et al. 2001). Transient contacts between the N- and C-terminal helices of the pH 2.3 acid-unfolded state, inferred from T2 measurements (Yao et al. 2001), were substantiated by a site-directed spin label study (Lietzow et al. 2002). Specific long-range contacts thus appear to occur transiently even in acid-denatured apomyoglobin.
Addition of 8 M urea to the pH 2.3 state eliminates the residual helicity in the A, D/E, and H helix regions and abolishes the transient contacts between the termini of the polypeptide chain (Lietzow et al. 2002; Schwarzinger et al. 2002). However, small preferences for residual extended structure could be detected in parts of the protein with neighboring bulky and branched side chains. Relaxation measurements also indicated variations in the backbone mobility of the sequence: Increased backbone motions could be linked to clusters of small residues, while slowed motions could be linked to local hydrophobic cluster structures that were persistent even in acidic 8 M urea (Schwarzinger et al. 2002).
Here, we investigate the structural differences in unfolded and partially folded states of the apomyoglobin mutant H64F by high-resolution NMR spectroscopy. The distal histidine residue in position 64 (His E7) plays a key role in the binding of oxygen and is highly conserved among globins for functional reasons. Replacement of the rather polar histidine residue by a hydrophobic phenylalanine results in significant changes in the thermodynamic stability as well as the folding kinetics of apomyoglobin (Garcia et al. 2000). The replacement of His64 with Phe is expected to have little effect on the helix propensity of the apomyoglobin E helix, according to the prediction program AGADIR (Muñoz and Serrano 1994). Nevertheless, a significantly increased helix content is observed in the native and molten globule states by CD, fluorescence, and NMR spectroscopy, and the mutation also results in an increased thermodynamic stability (Garcia et al. 2000). Stopped-flow CD experiments reveal a higher helix content in the burst phase intermediate (Garcia et al. 2000), and quench-flow hydrogen exchange experiments show that helix E, which is partially populated in the conformational ensemble of the wild-type kinetic intermediate (Nishimura et al. 2002), is now protected on the same timescale as helices A, B, G, and H, which form the molten globule in wild-type apomyoglobin (Garcia et al. 2000). To elucidate the causes of the dramatic effect of this single-point mutation on folding kinetics and stability of apomyoglobin, we have used NMR measurements to investigate residual structure and dynamics of the unfolded and partially folded states of the H64F mutant protein.
Results
Urea-denatured state at pH 2.3
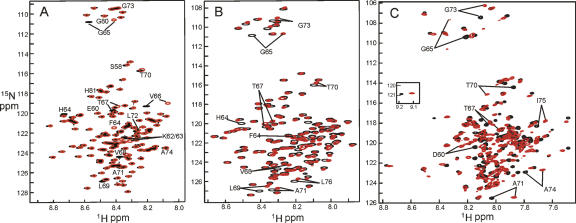
1H-15N correlation (HSQC) spectra of urea-denatured apoMb H64F are essentially identical to the spectra of the wild-type protein (Schwarzinger et al. 2002), except for residues in the immediate neighborhood of the mutation site (Fig. 1A). The changes are most pronounced for Gly65, Val66, and Lys63. Minor changes in the 1HN chemical shift can be observed for residues Ser58, Thr67, Val68, Leu69, and Thr70. Other changes are less than 0.01 ppm, indicating that the effect of substituting His64 by a phenylalanine residue is strictly local, with only a very small effect on the neighboring hydrophobic cluster, consistent with the behavior of the wild-type protein in the urea-denatured state (Schwarzinger et al. 2002).
Figure 1.
Overlay of the 750-MHz 1H-15N correlation spectra of wild-type apomyoglobin (black) and H64F-apomyoglobin (red) in 8 M urea at pH 2.3, 20°C (A), pH 2.3 in the absence of urea at 25°C (B), and pH 4.0 (WT) and pH 3.6 (H64F) at 50°C (C).
Acid-unfolded state at pH 2.3
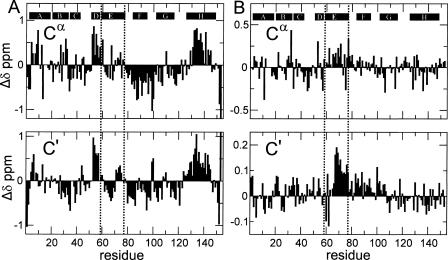
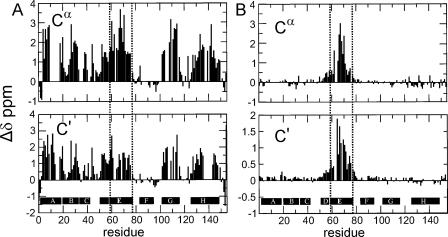
In the absence of urea, the NMR spectrum of H64F apomyoglobin at pH 2.3 displays more differences from the corresponding spectrum of the wild-type protein (Fig. 1B). A small amount of line broadening is observed in some parts of the spectra. Resonance assignments for the acid-unfolded state of H64F were obtained by analyzing HNCA, HN(CO)CA, HNCO, and HN(CA)CO spectra of deuterated samples and by using samples of the protein selectively labeled with single amino acid types. Sequence-corrected secondary chemical shifts display a trace very similar to wild-type apoMb (Fig. 2). There is some residual helicity in helices A, D, and H, as observed for the wild-type protein (Yao et al. 2001). The remainder of the protein remains unstructured, although a very slight increase in helicity in the center of the E helix region is suggested when the chemical shifts of H64F and wild type are compared directly (Fig. 2B).
Figure 2.
(A) Secondary chemical shifts of apoMb H64 (Cα, C′). (B) Chemical shift difference between H64F and wild-type apomyoglobin. The random coil chemical shifts used to obtain the data in A were corrected for sequence-dependent effects; for 13Cα, corrections were also made for shift contributions from attached deuterium atoms. The location of Helix E (residues 59–78) harboring the site of the mutation is indicated by black dotted lines. Manual adjustment for deuterium isotope effects has been made.
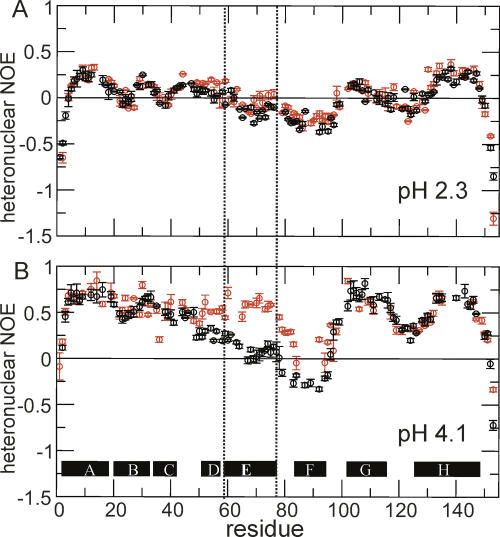
Backbone flexibility was investigated by measuring the [1H]-15N heteronuclear nuclear Overhauser effect (NOE), which monitors motions of the amide bond vector on a ps–ns timescale. These values are plotted for the wild-type and H64F proteins at pH 2.3 in Figure 3A. A very small increase is observed in the heteronuclear NOE in the C-terminal half of the E helix, but the magnitude of the measured heteronuclear NOE remains close to zero, indicating that the helix E region of the polypeptide still moves with the characteristics of a highly flexible polypeptide chain. The remaining parts of the protein showed comparable heteronuclear NOE values to wild-type apoMb.
Figure 3.
[1H]-15N heteronuclear NOE data for the acid-unfolded state of wild-type (black) and H64F mutant (red) apomyoglobin, both at pH 2.3 and 25°C (A), and the intermediate state of wild-type (pH 4.0, black) and H64F (pH 3.6, red) apomyoglobin at 50°C (B). The location of Helix E (residues 59–78) harboring the site of the mutation is indicated by dotted lines.
Molten globule state at pH 3.6
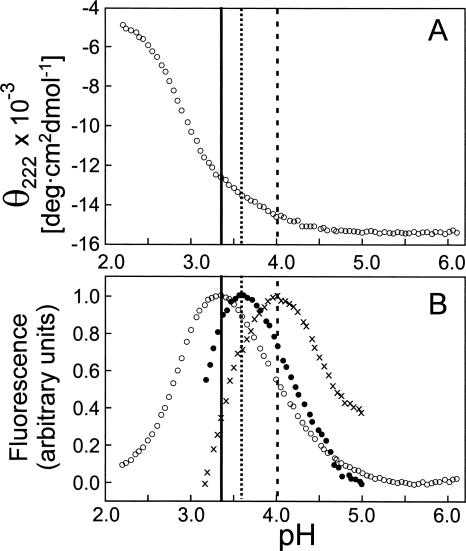
To determine the pH at which the molten globule of H64F is maximally populated, an automated pH titration was performed at 25°C, with detection of CD and fluorescence changes. The use of the automatic titrator allowed collection of a large number of data points, with a separation of only 0.05 pH units, which in turn allowed clear detection of the molten globule state, characterized by a change of the slope at pH 3.4 in the CD titration (Fig. 4A). For precise localization of the pH with the maximum population of the molten globule state, fluorescence (Fig. 4B) is superior to CD. In apomyoglobin, the tryptophan fluorescence is solvent-quenched at low pH and in the native folded state at pH 6 by specific side chain interactions and reaches a maximum in the molten globule state. At 25°C, the maximum of the fluorescence signal agrees with the CD data in indicating a pH of 3.4 for the maximal population of the molten globule. At 50°C, the temperature at which NMR experiments are performed, the H64F molten globule state is slightly destabilized and the fluorescence maximum is shifted to pH 3.6 (Fig. 4B). This value is still 0.4 pH units below the maximum for the wild-type molten globule. NMR data were therefore acquired at pH 3.6 and 50°C (Fig. 1C).
Figure 4.
(A) pH titration of H64F apomyoglobin at 25°C, monitored by ellipticity at 222 nm in the CD experiment. (B) Fluorescence-detected pH titration of H64F apomyoglobin at 25°C (open circles), titration of H64F at 50°C (the conditions of the NMR experiments) (solid circles), and pH titration of wild-type apoMb at 50°C (crosses). The solid vertical line indicates the maximum population of the H64F intermediate at 25°C (pH 3.35), the dotted line shows the maximum population at 50°C (pH 3.6), and the broken line the maximum population for the wild-type protein at 50°C (pH 4.0) for comparison.
Molten globule intermediates usually display very little chemical shift dispersion and broad lines, due to flexibility of the polypeptide chain and motions in the μs–ms timescale. Sharper resonance lines can be obtained in the NMR spectra by using elevated temperatures, and 10% ethanol is generally added to pH 4 samples of apoMb to ensure the long-term stability required for NMR experiments (Eliezer et al. 2000). Even with the increase in temperature, the molten globule state of H64F showed increased line broadening compared with the wild-type protein, which hampered analysis of the spectra. This is illustrated in Figure 1C, which shows that a number of the cross peaks for the H64F mutant protein are at considerably lower intensities than for the wild-type protein. The marked decrease in intensity for glycines 65 and 73 (both part of helix E) indicates the presence of complex motions on an intermediate timescale. Approximately 2–3 weeks after the sample was prepared, signal loss was observed throughout the spectrum, probably due to the formation of soluble aggregates, since no precipitation was visible.
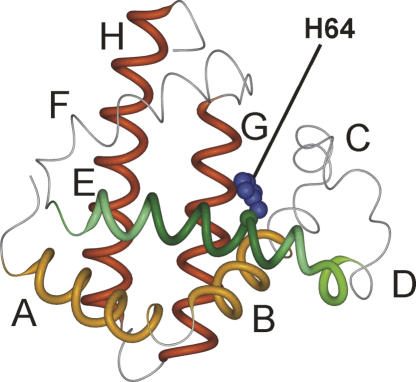
Comparison of the chemical shifts of the pH 4 equilibrium intermediate of the H64F mutant protein to those of wild-type apomyoglobin shows a pronounced increase in the helix content of helix E and a slight increase in helix D (Fig. 5A). The changes in chemical shift caused by the mutation are approximately an order of magnitude larger in the molten globule state (Fig. 5B) than in the pH 2.3 acid-unfolded state (Fig. 2B). The deviations of 13Cα and 13C′ chemical shifts from sequence corrected random coil values (Schwarzinger et al. 2001) show that the population of helical structure in the helix E region (residues 59–77) increases to ∼67% from ∼23% population in the pH 4 molten globule of the wild-type protein (Cavagnero et al. 2001). The increase in helicity for the E helix makes this region of the protein comparable in helical content to the regions that form the core of the wild-type molten globule, namely A (78%), G (60%), and H (53%) (Cavagnero et al. 2001). Interestingly, the helix content of the A, G, and H helices is not significantly affected in H64F, but a clear increase of ∼10% in helix content can be detected for residues 50–58, which include helix D. In summary, regions of helices A, D, E, G, and H are now populated to a similar high extent, while helices B and C remain unchanged and helix F remains disordered. The location of the mutation site at His64 is shown in Figure 6.
Figure 5.
(A) Secondary chemical shifts of H64F apomyoglobin. (B) Chemical shift differences between its intermediate state (pH 3.6) and wild-type (pH 4.1) apomyoglobin at 50°C. The data were manually corrected for 2H isotope effects and minor differences resulting from the difference in pH. The location of Helix E (residues 59–78) harboring the site of the mutation is indicated by dotted lines.
Figure 6.
Backbone structure of myoglobin (Kuriyan et al. 1986) showing the location of helical structure in the apomyoglobin molten globule state of the wild-type protein (A, B helices in orange, G, H helices in red) and the location of the additional helical structure observed in the equilibrium molten globule of H64F (D helix in light green, E helix in dark green, with darker colors denoting a greater helical propensity, according to Fig. 5). The side chain of His64 of the wild-type protein is shown in blue. The figure was prepared in MOLMOL (Koradi et al. 1996).
This picture is further supported by a comparison of the [1H]-15N heteronuclear NOE for the wild-type and the H64F molten globule (Fig. 3B). As seen for the chemical shift data, the most pronounced changes are detected for residues in helix E, whose motions on the ps–ns timescale are now restricted to the same extent as residues in the core helices A, G, H, and part of B. This provides evidence that in the mutant protein the helix E region is recruited into the compact core of the molten globule. Interestingly, the effects of the mutation extend beyond the boundaries of helix E into neighboring segments of the polypeptide chain.
Discussion
Effect of the His64Phe substitution on the unfolded states
For apomyoglobin, the urea-unfolded (pH 2.3, 8 M urea) and the acid-unfolded (pH 2.3, absence of urea) states represent models for the earliest stages of the folding process, in the upper reaches of the folding landscape. The H64F substitution causes only very small changes in the 1H-15N correlation spectrum of the urea-denatured state at pH 2.3, most likely due to strictly local effects arising from the introduction of the aromatic side chain. Likewise, the chemical shifts of the acid-unfolded state indicate only a very small change in the conformational ensemble due to the phenylalanine substitution. The small changes in chemical shift are suggestive of a very slight increase in intrinsic helicity (<5%). The acid-unfolded state of H64F still represents a largely disordered, extremely flexible polypeptide chain sampling a large conformational space. Since the effects of the mutation on the unfolded states of apomyoglobin are negligibly small, we turn to the compact molten globule to seek the source of the pronounced effects on the folding pathway and stability.
Effect of the His64Phe mutation on the pH 4 intermediate
The H64F substitution causes substantial changes in the molten globule state, as judged by both chemical shift differences and backbone heteronuclear NOE data. The region corresponding to helix E is significantly more helical, and the effect even extends weakly to residue 50, thus including helix D. The helical structure in the E region is now populated to the same extent in the pH 4 intermediate as helices A, G, and H (Fig. 5A). We infer that substitution of His64, which would probably be protonated in the intermediate state (Geierstanger et al. 1998) by the hydrophobic phenylalanine side chain, enhances packing between residues in the E helix region and the hydrophobic ABGH core and leads to stabilization of helical structure in the D and E helices.
The picture that emerges for the H64F pH 3.6 equilibrium molten globule is consistent with the characteristics of the kinetic burst phase intermediate derived from kinetic refolding experiments at pH 6 (Garcia et al. 2000). The secondary structure of the equilibrium intermediate precisely parallels that of the kinetic intermediate, with stable helical structure in the A, B, E, G, and H helix regions. H64F shows interesting differences in folding behavior compared with the wild-type protein: The burst phase amplitude in a stopped-flow CD experiment is greater and the observable folding phase is 2.5-fold faster (Garcia et al. 2000). These results indicate that additional helical structure is formed in the burst phase intermediate, which can evolve more rapidly to the fully folded protein. The intermediate formed in the burst phase of the quench-flow experiment for H64F shows enhanced protection in the E helix, which is only slightly protected in the burst phase of the wild-type protein under the same conditions (pH 6) (Jennings and Wright 1993; Nishimura et al. 2002). However, interestingly, several areas of the A and H helices that are highly protected in the wild-type burst phase show decreased protection in H64F. Thus, stabilization of helical structure in the E helix region is accompanied by slight destabilization of the A and H helices in the kinetic intermediate.
The effects of the single residue mutation described here yield important insights into the basic mechanisms of protein folding and protein stabilization and add a new dimension to recent studies of the apomyoglobin folding mechanism (Nishimura et al. 2002, 2003, 2005a,b, 2006). Quench-flow NMR studies of apomyoglobin folding have identified three kinetic phases: fast folding (complete protection within the dead time, seen for amides in the A, G, and H helices), slow folding (complete protection within a timescale of seconds), and biphasic, which show fast and slow components with rates that correspond to those of the “fast” and “slow” amides (Jennings and Wright 1993; Garcia et al. 2000; Nishimura et al. 2002). These results imply that the ensemble of partly folded states that make up the kinetic molten globule is heterogeneous: At sites showing biphasic kinetics, amides in some molecules in the folding ensemble are protected at the fastest folding rates, while in the remaining molecules in the ensemble they are protected only as the rest of the protein folds. Most interestingly, part of the (wild type) E helix exhibits biphasic kinetics, implying that this helix is docked onto the intermediate in some but not all molecules in the folding ensemble (Nishimura et al. 2002).
Mutagenesis of hydrophobic sites in the B helix (Nishimura et al. 2003) and the A, E, G, and H helices (Nishimura et al. 2006) further demonstrated that the folding pathway is not only heterogeneous but that it is overall extraordinarily robust. Of the total of 13 mutant proteins, every one showed a similar folding pathway, incorporating a burst phase intermediate and a slower folding phase. Detectable changes in the protection factors in local regions of each burst phase intermediate gave important information on the presence of native and nonnative long-range contacts. Evidence was seen, for example, for native-like contacts between the B and G helices, by comparison of the wild-type protein and the L32A mutant (Nishimura et al. 2003), while nonnative interactions, including a translocation of the H helix by one helical turn, were inferred from the behavior of the other core mutant proteins (Nishimura et al. 2006). These studies implicated a key role for the E helix in the kinetic intermediate: During the burst phase, an intermediate is formed where the A, B, E, G, and H helices are in an overall correct topology, but where docking of the E helix is disordered, with the corollary that the E helix must be correctly docked prior to the transition state of the folding process (Nishimura et al. 2006).
In all of the aforementioned kinetic studies, the E helix is seen as a key structural element that must be correctly docked before the final stages of folding can occur. In the equilibrium (pH 4) molten globule intermediate formed by wild-type apomyoglobin, the E helix is largely unstructured (Eliezer et al. 1998, 2000). It appears that the lower pH has an adverse effect on the stability of the E helix and the N terminus of the G helix (Nishimura et al. 2006). This effect is mitigated or abolished in the H64F mutant. Published data (Garcia et al. 2000) show that the equilibrium molten globule intermediate of H64F is significantly stabilized compared with the wild-type protein. The present work shows that the mutation has negligible effect on the conformational ensemble of the unfolded state (both pH 2.3, and urea-unfolded), but the replacement of the charged His side chain by Phe leads to enhanced hydrophobic interactions in the collapsed molten globule intermediate, thereby stabilizing helical structure in the E helix region. Consistent with this stabilizing effect, the E helix participates measurably in the kinetic burst phase intermediate (Garcia et al. 2000).
We have previously shown a correlation between the average surface area buried upon folding (termed AABUF) and the earliest events observed in apomyoglobin folding (Lietzow et al. 2002; Nishimura et al. 2005b). Regions of the polypeptide with greater than average values of AABUF, a parameter that can be determined from the amino acid sequence (Rose et al. 1985), show a propensity toward local hydrophobic cluster formation in unfolded states (Yao et al. 2001) and drive transient long-range interactions that lead to collapse and folding (Lietzow et al. 2002; Nishimura et al. 2005b). The AABUF profile of apomyoglobin provides an explanation for the observed folding pathway and for the structure of the burst phase folding intermediate. It was also shown that the AABUF profile could be changed by mutagenesis to alter the kinetic folding pathway by design (Nishimura et al. 2005b; Dyson et al. 2006). However, for the H64F mutant, the observed change in the structure of the folding intermediate does not seem to arise from changes in the AABUF as a result of the mutation of His to Phe but rather from stabilizing hydrophobic interactions in the compact molten globule states. The changes in the AABUF profile are very small, yet the changes in the structure of the kinetic folding intermediate (Garcia et al. 2000) and the equilibrium molten globule are profound. Substitution of His64 by Phe has little effect on the conformational ensemble in the unfolded states; all effects are local, as would be expected since the mutation does not significantly alter the AABUF profile. However, in compact states, replacement of the polar histidine by phenylalanine enhances hydrophobic packing and leads to substantial changes in structure and stability. In the equilibrium intermediate, the mutation leads to recruitment of the E helix region into the compact core of the molten globule, resulting in significant stabilization of its secondary structure. Thus, the effect of the H64F mutation is primarily on the structure of the molten globule intermediate. In the wild-type protein, the kinetic molten globule is heterogeneous but is capable of correct docking of the E helix once the translocation of the H helix has been reversed. The E helix must be correctly folded and packed prior to formation of the protein folding transition state; mutations that interfere with docking of the E helix onto the preformed A, B, G, H core decrease the rate of folding (Nishimura et al. 2006). Thus, the role of the H64F mutation in enhancing the rate of apomyoglobin folding probably arises from early recruitment of the E helix region into compact folding intermediates with concomitant stabilization of its secondary structure. This stabilization of structure is directly evident in the increased helicity of the burst phase intermediate (Garcia et al. 2000).
Materials and Methods
Protein preparation
Unlabeled samples for automated CD- and fluorescence titrations as well as uniformly 15N- and 13C/15N-labeled samples of H64F for NMR studies of the denatured state in acidic urea were prepared as previously described (Jennings et al. 1995; Garcia et al. 2000). Triply labeled samples (2H, 13C, 15N) for spectroscopy of the pH 2.3 state and the pH 3.6 molten globule state were grown as described, except that sterile-filtered D2O (98%) was used to prepare buffers and M9-media. Freshly transformed cells were picked from a plate and stepwise adapted to deuterated M9 (25, 50, 75% D2O) for initial growth tests. However, as no significant difference in expression was observed upon direct inoculation into 100% deuterated M9-medium, this step was omitted. The main culture (1 L) was inoculated to OD ∼ 0.1 from a 50 mL culture in deuterated M9-medium. Cell growth was observed to be ∼50% compared with normal M9. Cells were grown at 37°C, induced at OD 1.0 to a final concentration of 2 mM IPTG, and harvested 20 h after induction. Cells had a slightly reddish color indicative of an increased content of soluble myoglobin. Therefore, in addition to the inclusion body fraction, the 60%–90% ammonium sulfate precipitate of the inclusion body supernatant was also applied to purification on a C4-reversed phase HPLC column, thus increasing the total yield by up to 20% (60 mg/L total). The degree of deuterium incorporation was found to be between 75% and 80% by mass spectrometry.
Amino acid selectively labeled samples were prepared to reduce signal overlap and to facilitate assignment of residues with resonances that were too broad to be detected in triple-resonance experiments. Typically, two different amino acids, which appear in different parts of the 15N-HSQC-spectrum of the urea-denatured state, were combined into one sample. Samples selectively 15N-labeled in histidine and lysine were prepared by supplementing unlabeled minimal M9-medium by addition of a filter-sterilized solution containing 70 mg of each 15N-enriched amino acid upon induction with 2 mM IPTG. Samples selectively 15N-labeled with [Ala/Gly], [Phe/Tyr], [Met/Tyr], and [Arg] were grown in media containing all twenty amino acids (per liter: Ala: 0.5 g; Arg: 0.4 g; Asn: 0.4 g; Asp: 0.4 g; Cys: 0.06 g; Gln: 0.65 g; Glu: 0.65 g; Gly: 0.55 g; His: 0.1 g; Ile: 0.23 g; Leu: 0.12 g; Lys: 0.21 g; Met: 0.25 g; Phe: 0.13 g; Pro: 0.1 g; Ser: 2.1 g; Thr: 0.23 g; Trp: 0.05 g; Tyr: 0.17 g; Val: 0.23 g; glucose: 2 g; (NH4)2SO4: 0.5 g; KH2PO4: 3 g; Na2HPO4: 6.8 g; NaCl: 0.5 g). Only 50% of the amount of the 15N-labeled amino acids was supplied from start. The remaining portions were supplied upon induction with 2 mM IPTG upon a cell density of OD600 = 1. Cells were grown, harvested, and purified as described above. Typical yields were 50–60 mg of pure protein. In general, no scrambling of the 15N label was observed, except for glycine, which resulted in minor enrichment of 15N in serine, and to some extent for phenylalanine and tyrosine.
Automated pH titrations detected by CD and fluorescence spectroscopy
Automated CD and fluorescence titrations were performed to determine the optimal pH for studying the H64F molten globule state by NMR, which requires experiments to be performed at 50°C. Measurements were performed using an Aviv model 202 CD-spectrometer with an additional channel for total fluorescence detection and an automated titration module. Samples for automated pH titration were prepared as 5 μM solutions in pH 10 mM acetate buffer brought to pH 2.3 with the minimum amount of HCl. The concentration of stock solutions was determined as described by Edelhoch (1967) (ε 280nm = 15,200/M · cm, ε 288nm = 11,800/M · cm). Titrations were performed using a square quartz cuvette (Hellma QS 101) with a 1-cm path length in combination with an Orion Ross-Sure-Flow pH electrode in an in-house designed cuvette-adapter. The pH electrode was calibrated between pH 2 and pH 7 using a three-point calibration procedure. The temperature in the spectrometer was fed back to the pH meter for the purpose of temperature correction of the pH using a customized version of the Aviv spectrometer software (version 2.75). A volume of 2.4 mL of pH 2.3 H64F-apomyoglobin was placed in the cuvette. The titration was carried out in the increasing volume mode using 0.5 M NaOH to increase the pH in 0.05 pH units. Reagent mixing occurred immediately using mixer speeds of 80%. Mixing was turned off 5 sec before measurements to allow the mixing vortex to disappear. Circular dichroism spectra were recorded at 222 nm with a 1 nm bandwidth for 5 sec. The instrument was calibrated with D (+) camphor-sulfonic acid prior to titrations. Fluorescence was measured in the same titration step after collection of CD data and was calibrated to the acid denatured sample using the autoset function at an excitation wavelength of 280 nm. Total fluorescence was counted for 5 sec using a 305-nm high-pass filter and a quantum-counter to correct for variations in the intensity of the light source. During the titration, the slits were closed using the auto-slit closure feature to avoid sample degradation by prolonged exposure to intense UV light. A reference titration from pH 2.3 to pH 6.1 was performed at 25°C to optimize the experimental setup in terms of time required per titration set. At 50°C, a reduced range of pH 3.2–5.0 was used to minimize evaporation in the sample compartment causing changes in protein concentration over prolonged periods of time. For the same reason, ethanol, which is added at a concentration of 10% for long-term stability of NMR samples (Eliezer et al. 1997), was omitted from samples used for the automatic CD and fluorescence titrations at 50°C, and the cuvettes were tightly sealed with Parafilm. Total fluorescence data of the titrations at different temperatures were normalized for better comparison (0 = smallest signal, 1 = largest signal in every titration).
NMR spectroscopy
NMR samples were prepared as described previously (Eliezer et al. 2000; Yao et al. 2001; Schwarzinger et al. 2002). Protein concentrations were between 200 μM and 300 μM. Adjustment of the pH with an accuracy of less than 0.05 pH units was critical for molten globule samples. Samples for the molten globule displayed a pronounced tendency to aggregate and could not be used for more than two weeks after preparation. Experiments were recorded on a 750-MHz Bruker DRX spectrometer equipped with a triple-resonance gradient probe. The temperature was calibrated using either methanol or ethylene glycol (Van Geet 1968). Spectral widths were adjusted to the minimum possible for each state (Eliezer et al. 2000; Yao et al. 2001; Schwarzinger et al. 2002). For the pH 2.3 unfolded state and the molten globule state triply labeled samples were used. Assignment was achieved using deuterium-decoupled versions of HNCA, HN(CO)CA, HNCO, and HN(CA)CO (Kay et al. 1990; Bax and Ikura 1991). A deuterium-decoupled version of the HNCACB with delays optimized for evolution of Cβ resonances was recorded for the pH 3.6 molten globule state (Wittekind and Mueller 1993). All experiments for assignment were completed using the same sample. Because of difficulties in assignment of the molten globule state caused by extensive line broadening, a titration was performed to transfer assignments from the pH 2.3 spectrum. To accomplish this, ethanol was added to a sample of H64F apomyoglobin at pH 2.3 in steps of 2% to a final concentration of 10%. Subsequently, the temperature was increased in steps of 5°C from 25°C to 50°C, followed by titration to pH 2.6, 2.8, 3.0, 3.2, 3.4, 3.5, 3.6, and 3.7. To reduce ambiguities caused by spectral overlap, titrations were also carried out with the selective amino acid labeled samples (10% ethanol; 25°C, 33°C, 42°C, 50°C; pH 2.3, 2.6, 2.8, 3.0, 3.2, 3.4, 3.6). Steady state [1H]-15N-heteronuclear NOE spectra were recorded for the acid unfolded state and the molten globule state as described (Eliezer et al. 2000; Yao et al. 2001). For each state, three sets of saturated and unsaturated spectra were recorded in an interleaved manner. NMR spectra were processed with NMRPipe (Delaglio et al. 1995) and analyzed with NMRView (Johnson and Blevins 1994).
Acknowledgments
We thank Linda Tennant for expert technical assistance and Drs. Chiaki Nishimura, David Eliezer, Silvia Cavagnero, and Mike Lietzow for invaluable discussions. This research was supported by grant DK34909 from the National Institutes of Health. S.S. acknowledges an Erwin Schrödinger Fellowship of the Austrian Science Funds (J-1736 CHE) and thanks Professor Paul Rösch for his support. R.M.B. is a Pew Latin American Fellow.
Footnotes
Reprint requests to: Peter E. Wright, Department of Molecular Biology, MB 2, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA; e-mail: wright@scripps.edu; fax: (858) 784-9822.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073187208.
References
- Bax, A., Ikura, M. An efficient 3D NMR technique for correlating the proton and 15N backbone amide resonances with the α-carbon of the preceding residue in uniformly 15N/13C enriched proteins. J. Biomol. NMR. 1991;1:99–104. doi: 10.1007/BF01874573. [DOI] [PubMed] [Google Scholar]
- Cavagnero, S., Nishimura, C., Schwarzinger, S., Dyson, H.J., Wright, P.E. Conformational and dynamic characterization of the molten globule state of an apomyoglobin mutant with an altered folding pathway. Biochemistry. 2001;40:14459–14467. doi: 10.1021/bi011500n. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Guang, Z., Pfeifer, J., Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dyson, H.J., Wright, P.E., Scheraga, H.A. The role of hydrophobic interactions in initiation and propagation of protein folding. Proc. Natl. Acad. Sci. 2006;103:13057–13061. doi: 10.1073/pnas.0605504103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelhoch, H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- Eliezer, D., Wright, P.E. Is apomyoglobin a molten globule? Structural characterization by NMR. J. Mol. Biol. 1996;263:531–538. doi: 10.1006/jmbi.1996.0596. [DOI] [PubMed] [Google Scholar]
- Eliezer, D., Jennings, P.A., Dyson, H.J., Wright, P.E. Populating the equilibrium molten globule state of apomyoglobin under conditions suitable for characterization by NMR. FEBS Lett. 1997;417:92–96. doi: 10.1016/s0014-5793(97)01256-8. [DOI] [PubMed] [Google Scholar]
- Eliezer, D., Yao, J., Dyson, H.J., Wright, P.E. Structural and dynamic characterization of partially folded states of myoglobin and implications for protein folding. Nat. Struct. Biol. 1998;5:148–155. doi: 10.1038/nsb0298-148. [DOI] [PubMed] [Google Scholar]
- Eliezer, D., Chung, J., Dyson, H.J., Wright, P.E. Native and nonnative structure and dynamics in the pH 4 intermediate of apomyoglobin. Biochemistry. 2000;39:2894–2901. doi: 10.1021/bi992545f. [DOI] [PubMed] [Google Scholar]
- Garcia, C., Nishimura, C., Cavagnero, S., Dyson, H.J., Wright, P.E. Changes in the apomyoglobin folding pathway caused by mutation of the distal histidine residue. Biochemistry. 2000;39:11227–11237. doi: 10.1021/bi0010266. [DOI] [PubMed] [Google Scholar]
- Geierstanger, B., Jamin, M., Volkman, B.F., Baldwin, R.L. Protonation behavior of histidine 24 and histidine 119 in forming the pH 4 folding intermediate of apomyoglobin. Biochemistry. 1998;37:4254–4265. doi: 10.1021/bi972516+. [DOI] [PubMed] [Google Scholar]
- Jennings, P.A., Wright, P.E. Formation of a molten globule intermediate early in the kinetic folding pathway of apomyoglobin. Science. 1993;262:892–896. doi: 10.1126/science.8235610. [DOI] [PubMed] [Google Scholar]
- Jennings, P.A., Stone, M.J., Wright, P.E. Overexpression of myoglobin and assignment of the amide, Cα and Cβ resonances. J. Biomol. NMR. 1995;6:271–276. doi: 10.1007/BF00197808. [DOI] [PubMed] [Google Scholar]
- Johnson, B.A., Blevins, R.A. NMRView: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:604–613. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kay, L.E., Ikura, M., Tschudin, R., Bax, A. Three-dimensional triple-resonance NMR spectroscopy of isotopically enriched proteins. J. Magn. Reson. 1990;89:496–514. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., Wüthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Kuhlman, B., Dantas, G., Ireton, G.C., Varani, G., Stoddard, B.L., Baker, D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- Kuriyan, J., Wilz, S., Karplus, M., Petsko, G.A. X-ray structure and refinement of carbon-monoxy (Fe II)-myoglobin at 1.5 Å resolution. J. Mol. Biol. 1986;192:133–154. doi: 10.1016/0022-2836(86)90470-5. [DOI] [PubMed] [Google Scholar]
- Lecomte, J.T.J., Kao, Y.H., Cocco, M.J. The native state of apomyoglobin described by proton NMR spectroscopy: The A-B-G-H interface of wild-type sperm whale apomyoglobin. Proteins. 1996;25:267–285. doi: 10.1002/(SICI)1097-0134(199607)25:3<267::AID-PROT1>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Lietzow, M.A., Jamin, M., Dyson, H.J., Wright, P.E. Mapping long-range contacts in a highly unfolded protein. J. Mol. Biol. 2002;322:655–662. doi: 10.1016/s0022-2836(02)00847-1. [DOI] [PubMed] [Google Scholar]
- Muñoz, V., Serrano, L. Elucidating the folding problem of helical peptides using empirical parameters. Nat. Struct. Bio. 1994;1:399–409. doi: 10.1038/nsb0694-399. [DOI] [PubMed] [Google Scholar]
- Nishimura, C., Dyson, H.J., Wright, P.E. The apomyoglobin folding pathway revisited: Structural heterogeneity in the kinetic burst phase intermediate. J. Mol. Biol. 2002;322:483–489. doi: 10.1016/s0022-2836(02)00810-0. [DOI] [PubMed] [Google Scholar]
- Nishimura, C., Wright, P.E., Dyson, H.J. Role of the B helix in early folding events in apomyoglobin: Evidence from site-directed mutagenesis for native-like long range interactions. J. Mol. Biol. 2003;334:293–307. doi: 10.1016/j.jmb.2003.09.042. [DOI] [PubMed] [Google Scholar]
- Nishimura, C., Dyson, H.J., Wright, P.E. Enhanced picture of protein-folding intermediates using organic solvents in H/D exchange and quench-flow experiments. Proc. Natl. Acad. Sci. 2005a;102:4765–4770. doi: 10.1073/pnas.0409538102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, C., Lietzow, M.A., Dyson, H.J., Wright, P.E. Sequence determinants of a protein folding pathway. J. Mol. Biol. 2005b;351:383–392. doi: 10.1016/j.jmb.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Nishimura, C., Dyson, H.J., Wright, P.E. Identification of native and nonnative structure in kinetic folding intermediates of apomyoglobin. J. Mol. Biol. 2006;355:139–156. doi: 10.1016/j.jmb.2005.10.047. [DOI] [PubMed] [Google Scholar]
- Reymond, M.T., Merutka, G., Dyson, H.J., Wright, P.E. Folding propensities of peptide fragments of myoglobin. Protein Sci. 1997;6:706–716. doi: 10.1002/pro.5560060320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, G.D., Geselowitz, A.R., Lesser, G.J., Lee, R.H., Zehfus, M.H. Hydrophobicity of amino acid residues in globular proteins. Science. 1985;229:834–838. doi: 10.1126/science.4023714. [DOI] [PubMed] [Google Scholar]
- Schwarzinger, S., Kroon, G.J.A., Foss, T.R., Chung, J., Wright, P.E., Dyson, H.J. Sequence dependent correction of random coil NMR chemical shifts. J. Am. Chem. Soc. 2001;123:2970–2978. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- Schwarzinger, S., Wright, P.E., Dyson, H.J. Molecular hinges in protein folding: The urea-denatured state of apomyoglobin. Biochemistry. 2002;41:12681–12686. doi: 10.1021/bi020381o. [DOI] [PubMed] [Google Scholar]
- Tsui, V., Garcia, C., Cavagnero, S., Siuzdak, G., Dyson, H.J., Wright, P.E. Quench-flow experiments combined with mass spectrometry show apomyoglobin folds through an obligatory intermediate. Protein Sci. 1999;8:45–49. doi: 10.1110/ps.8.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Geet, A.L. Calibration of the methanol and glycol nuclear magnetic resonance thermometers with a static thermistor probe. Anal. Chem. 1968;40:2227–2229. [Google Scholar]
- Wittekind, M., Mueller, L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the α- and β-carbon resonances in proteins. J. Magn. Reson. 1993;101:201–205. [Google Scholar]
- Yao, J., Chung, J., Eliezer, D., Wright, P.E., Dyson, H.J. NMR structural and dynamic characterization of the acid-unfolded state of apomyoglobin provides insights into the early events in protein folding. Biochemistry. 2001;40:3561–3571. doi: 10.1021/bi002776i. [DOI] [PubMed] [Google Scholar]