Abstract
Plant lipid-transfer proteins (LTPs) are abundant, small, lipid binding proteins that are capable of exchanging lipids between membranes in vitro. Despite their name, a role in intracellular lipid transport is considered unlikely, based on their extracellular localization. A number of other biological roles, including antimicrobial defense, signaling, and cell wall loosening, have been proposed, but conclusive evidence is generally lacking, and these functions are not well correlated with in vitro activity or structure. A survey of sequenced plant genomes suggests that the two biochemically characterized families of LTPs are phylogenetically restricted to seed plants and are present as substantial gene families. This review aims to summarize the current understanding of LTP biochemistry, as well as the evidence supporting the proposed in vivo roles of these proteins within the emerging post-genomic framework.
Keywords: lipid-transfer proteins, lipid binding, plant defense, extracellular signaling, cuticle
Lipids play an assortment of essential roles in biology: They define the interfaces between organelles, cells, and organisms; provide a rich energy resource of highly reduced carbon; and can act as important signaling molecules and hormones. Much has been learned about the fundamental lipid biosynthetic pathways, and it has been shown that various types are synthesized and accumulate in different subcellular compartments (Kent 1995; Dowhan 1997; Vance and Vance 2004; Jouhet et al. 2007). It is also known that there is considerable trafficking of lipids between different cellular membranes, and in eukaryotes, this occurs via both vesicular and nonvesicular pathways (Maxfield and Mondal 2006). The isolation of soluble animal proteins that could transfer phospholipids between membranes in vitro (Kamp et al. 1973; Bloj and Zilversmit 1977) leads to efforts to identify similar proteins in plants. This resulted in the characterization of a small, basic protein from spinach with similar in vitro activity (Kader et al. 1984), which in turn marked the beginning of the study of a new class of proteins in plants. These proteins, now known as lipid-transfer proteins (LTPs), were later found to be common to flowering plants and evolutionarily distinct from the LTPs previously identified in animals. Given their clear in vitro activity, plant LTPs were originally hypothesized to be involved in intracellular lipid trafficking, but this idea was subsequently abandoned when they were shown to be synthesized as preproteins containing signal peptides (Bernhard et al. 1991) that direct their secretion into the extracellular matrix (Meijer et al. 1993; Thoma et al. 1993). Over the past 15 years, despite much study, no clear evidence has emerged to suggest a common alternative mechanistic hypothesis, and LTPs largely remain proteins in search of a clear biological function.
Nonetheless, LTPs have attracted considerable interest due to their importance in industrial applications and human health: They represent a stable and abundant protein in barley malt, contributing to beer quality and foam formation (Jegou et al. 2000), and have also been identified as major human allergens, particularly in fruits of the Rosaceae family (Salcedo et al. 2007), such as apple and peach. The practical value of LTPs has even developed to the extent that hypoallergenic tomatoes have been developed through suppressing the expression of LTP genes in transgenic fruit (Le et al. 2006; Lorenz et al. 2006), yet there is still no generalized theory as to their in vivo function. In recent years, plant genome sequencing efforts have revealed LTP and “LTP-like” sequences to comprise large, ubiquitous gene families in higher plants, and the stage is therefore set to elucidate the breadth of LTP functional diversity. The aim of this review is to summarize the current understanding of plant LTP biochemistry and their potential functions in the emerging post-genome framework.
LTP biochemistry: Structure and lipid-binding properties
Biochemically characterized LTPs belong to two distinct families, which share the ability to transfer phospholipids between membranes in vitro. While both families are characterized by a high pI (∼9) and a characteristic eight-cysteine motif (C…C…CC…CXC…C…C), LTPs from families 1 and 2 are typically ∼9 kDa or ∼7 kDa, respectively, and they exhibit low overall amino acid sequence similarity (∼30% identity). The two families also share a common structural architecture of a hydrophobic cavity enclosed by four α-helices, held in a compact fold by four disulfide bonds.
Family 1 LTPs
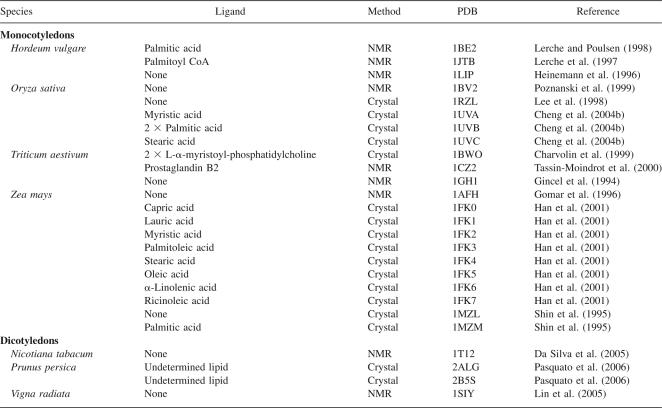
Several three-dimensional structures of family 1 LTPs have been solved by NMR and X-ray crystallography, both unassociated with a ligand and in a complex with a variety of lipid ligands (Table 1). In general, the structural features of LTPs in this family are very similar (Fig. 1). The first three α-helices are amphiphilic and parallel to the central lipid binding pocket, and in all solved structures, the first helix has a pronounced kink near the second cysteine that, in most structures, corresponds with a conserved proline. The remaining sides of the lipid binding cavity are defined by part of the fourth α-helix and the C-terminal loop (Fig. 1B).
Table 1.
Summary of family 1 LTP structures from the PDB
Figure 1.
Family 1 LTP sequence and structure. (A) Cutaway view showing the lipid binding pocket of rice LTP1 occupied by myristic acid (redrawn from the coordinates of PDB 1UVA) (Cheng et al. 2004b). (B) Stereoview of the same model highlighting the four helix structure of family 1 LTPs with disulfide bridges in red. Both structures were rendered using UCSF Chimera (Pettersen et al. 2004) with surfaces calculated by the MSMS package (Sanner et al. 1996); (C) Sequence logo showing the conserved features of family 1 LTPs from Arabidopsis thaliana, rice, and poplar. Disulfide pairs are indicated above the logo. Residue numbers and helices according to the rice LTP1 structure are shown below the sequence. Mature peptides were predicted using SignalP 3.0 (Bendtsen et al. 2004) and aligned using ClustalW (Chenna et al. 2003). Logo was calculated using WebLogo (Crooks et al. 2004) after manually removing nonconserved gaps in the alignment. The sequences considered were translated from the following gene models: A. thaliana At2g15050, At2g15325, At2g18370, At2g38530, At2g38540, At3g08770, At3g51590, At3g51600, At4g33355, At5g01870, At5g59310, At5g59320; rice Os01g0219500, Os01g0822900, Os03g0808500, Os08g0131200, Os11g0114900, Os11g0115100, Os11g0115400, Os11g0427800, Os12g0114500, Os12g0115000, Os12g0115100, Os12g0115300, Os12g0115500; poplar (JGI assembly v 1.1) estExt_fgenesh4_pg.C_65530001, estExt_Genewise1_v1.C_LG_IV3602, estExt_Genewise1_v1.C_LG_VI1197, estExt_Genewise1_v1.C_LG_XVI0292, fgenesh4_pg.C_LG_XI000197, fgenesh4_pg.C_LG_XI000199, fgenesh4_pg.C_LG_XII001150, fgenesh4_pm.C_LG_IX000642, fgenesh4_pm.C_scaffold_148000018, grail3.0035029301, grail3.0038010001, grail3.0076005601, grail3.0076005701, grail3.0148004401, gw1.1437.5.1, gw1.40.321.1, gw1.I.5890.1, gw1.XVI.307.1.
Aside from the perfectly conserved eight-cysteine motif that contributes to the four disulfide bonds of all LTPs, those of family 1 have several other specific conserved residues (Fig. 1C). Small hydrophobic amino acids (Ile, Val, Leu, Ala) appear throughout the LTP sequence and help to define the hydrophobic binding tunnel. Tyr16 is well conserved, but the side chain's orientation away from the binding cavity and site-directed mutagenesis experiments (Lullien-Pellerin et al. 1999) indicates that it is not involved in lipid binding. Tyr79 is semi-conserved and is responsible for experimentally observed intrinsic fluorescence that is enhanced upon lipid binding (Lullien-Pellerin et al. 1999). Some structures show this residue hydrogen bonding to the polar head group of lipid ligands (Charvolin et al. 1999; Tassin-Moindrot et al. 2000), but this is not always the case (Han et al. 2001; Cheng et al. 2004b). Another well-conserved feature of the family 1 LTP sequence is the presence of two oppositely charged residues at the beginning of the third α-helix (Asp43 Arg44 in rice LTP1) (Fig. 1B,C). The acid typically points away from the binding cavity, while the positively charged residue points inward at the opening of the LTP binding cavity, and despite its location near the entrance, it is usually not seen forming a hydrogen bond with the lipid head group (Han et al. 2001).
In addition to the ligand binding properties suggested by structural studies, the affinity of LTPs for a variety of lipids has been confirmed by equilibrium titration experiments. Binding of substrates such as glycerolipids (Lerche et al. 1997; Guerbette et al. 1999; Douliez et al. 2000), fatty acids (Tsuboi et al. 1992; Lerche et al. 1997; Douliez et al. 2000), and acyl-CoA (Ostergaard et al. 1993; Lerche et al. 1997) has been demonstrated. In cases where dissociation constants (Kd) have been determined, LTPs have been shown to have affinity for their ligands in the micromolar range. When binding curves were analyzed, assuming noncooperative independent binding sites, the number of binding sites per monomer has been found to be between one and two. Intermediate values greater than one likely indicate that the interaction with two ligands occurs with some cooperativity, but the techniques employed thus far (primarily enhancement of intrinsic tyrosine fluorescence) do not provide the free ligand concentration, which is a requirement for determining the cooperativity (Douliez et al. 2000). The hypothesis of two cooperative binding sites is also suggested by the crystal structures of family 1 LTPs from rice (Oryza sativa) and wheat (Triticum aestivum) bound to palmitic acid and L-α-myristoyl-phosphatidylcholine, respectively (Protein Data Bank [PDB] 1UVB and 1BWO). In these structures, the two ligands share a common binding tunnel, but with a “tail-to-tail” arrangement, presumably having entered the tunnel from opposite ends, leaving their polar ends exposed to the solvent (Charvolin et al. 1999; Cheng et al. 2004b).
Family 2 LTPs
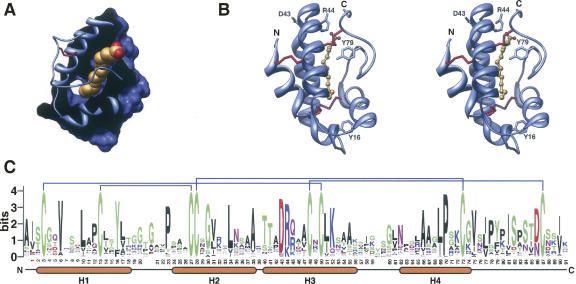
Compared with family 1 LTPs, there have been considerably fewer structural studies of those from family 2. Structures have only been solved for an LTP2 from rice (Samuel et al. 2002) and the LTP2 from wheat in complex with L-α-palmitoyl-phosphatidyl glycerol (Pons et al. 2003; Hoh et al. 2005). Family 2 LTPs also have four helices bound by four disulfide bonds enclosing a hydrophobic lipid binding cavity (Fig. 2). However, the pairing partners of cysteines 5 and 6 (CXC) are switched: In family 1 LTPs, C5 forms a disulfide with C8 and C6 with C1, while in family 2 LTPs, C5 forms a disulfide with C1 and C6 bonds with C8 (Cheng et al. 2004a). Although smaller than the hydrophobic cavity of LTP1s, the cavity of family 2 LTPs is more flexible, allowing for the accommodation of a wider variety of lipids, including sterols (Samuel et al. 2002; Pons et al. 2003; Cheng et al. 2004a; Hoh et al. 2005).
Figure 2.
Family 1 LTP sequence and structure. (A) Cutaway image showing wheat LTP2 bound to L-α-palmitoyl-phosphatidyl glycerol (redrawn from the coordinates of PDB 1N89) (Pons et al. 2003). (B) Stereoview of the same model highlighting the four helix structure of family 2 LTPs with disulfide bridges in red. Images were constructed as described for Figure 1. (C) Sequence logo showing the conserved features of family 2 LTPs from Arabidopsis thaliana, rice, and poplar. Disulfide pairs are indicated above the logo. Residue numbers and helices according to the wheat LTP2 structure are shown below. Logo was prepared as for Figure 1. The sequences considered were translated from the following gene models: A. thaliana At1g66850, At5g38160, At1g48750, At5g38195, At3g18280, At5g38170, At1g43667, At2g14846, At3g57310, At1g73780, At5g38180, At1g43666, At4g12825, At1g07747, At3g29152; rice Os03g0111300, Os10g0505700, Os10g0504900, Os05g0550300, Os10g0505500, Os10g0505000, Os05g0550600, Os06g0705400, Os11g0620300, Os01g0691100, Os01g0691300; poplar (JGI assembly v 1.1) grail3.0021013901, estExt_Genewise1_v1.C_LG_IV3319, estExt_Genewise1_v1.C_LG_XV0799, gw1.XII.565.1.
Two well-conserved proline residues define tight turns between helix 1 and 2 (Pro20, wheat LTP2 numbering) (Fig. 2B) and between the third and fourth helix (Pro42). Recent mutational studies have revealed residues that are important to both the LTP2 structure and ligand binding. Site-directed mutagenesis of Leu8, Phe36, and Val49 of rice LTP2 (numbers corresponding to wheat Leu7, Phe35, and Ile48, respectively) lead to significant alteration of LTP2 structure and lipid binding and transfer activity. Conversely, the mutation of Tyr45 to Ala (corresponding to wheat Tyr44) resulted in reduced lipid binding and transfer, without major effect on the protein structure. Interestingly, the Y48A mutant (corresponding to wheat Tyr47) showed reduced affinity for planar sterol molecules but not for linear lysophospholipids (Cheng et al. 2007), which given the conservation of aromaticity at this position perhaps provides some indication of the nature of the in vivo substrate (Fig. 2C).
The biological roles of LTPs
Although initially identified as potential mediators of intracellular membrane lipid transport, the demonstration of LTP extracellular localization suggested that this was not the case. In place of this simple hypothesis, based on their in vitro activity, numerous studies over the last two decades have resulted in an association with a myriad of biological processes. As described below, these include direct antimicrobial defense, defensive signaling, cuticle deposition, and cell wall loosening.
LTPs and defense against pathogens
LTPs may be important components of direct defense against bacterial and fungal pathogens, and early evidence of this role came from in vitro screens of protein extracts from radish (Raphanus sativus) (Terras et al. 1992), barley (Hordeum vulgare) (Molina et al. 1993), Arabidopsis thaliana (Segura et al. 1993), spinach (Spinacia oleracea) (Segura et al. 1993), and onion (Allium cepa) seeds (Cammue et al. 1995). Each of these isoforms inhibited growth of bacterial and fungal pathogens to varying degrees, depending on the isoform and the pathogen. The presence of calcium ions leads to a decrease in the antimicrobial effectiveness in vitro (Terras et al. 1992; Cammue et al. 1995). Curiously, an especially effective antimicrobial LTP isolated from onion seeds, Ace-AMP1, had no detectable inter-membrane lipid transfer activity, (Cammue et al. 1995), although it possesses a similar structure to the “true” LTPs (Tassin et al. 1998). While the mechanism of LTP toxicity is not entirely clear, it appears that it depends at least in part on the ability of these LTP isoforms to promote membrane permeabilization in pathogen, but not host, cells (Cammue et al. 1995; Regente et al. 2005). To our knowledge, the antimicrobial properties of family 2 LTPs have not been reported.
Support for a defensive role of LTPs also comes from analysis of their expression patterns, since many isoforms are induced upon pathogen challenge, thus leading to the definition of LTPs as a family of pathogenesis-related (PR) proteins, PR-14 (Van Loon and Van Strien 1999). Additional evidence has been provided through use of transgenic plants overexpressing LTPs. A barley family 1 LTP overexpressed in Arabidopsis or tobacco (Nicotiana tabacum) led to enhanced resistance to Pseudomonas syringae (Molina and Garcia-Olmedo 1997), while, as is the case for other PR proteins, the Ace-AMP1 LTP overexpressed in combination with another PR protein (a chitinase) showed synergistic enhancement of resistance to fungal pathogens (Jayaraj and Punja 2007).
The first function for an LTP protein clearly defined by a genetic lesion was a role in defensive signaling. The DIR1 gene of Arabidopsis was identified in a screen for mutants that were deficient in systemic acquired resistance (SAR), but not local defense responses. The predicted sequence of this protein has some similarity to family 2 LTPs, sharing such features as the eight-cysteine motif, a predicted signal peptide, and small size. However, the mature peptide has a pI of 4.5, which is much lower than is typical for family 2 LTPs (Maldonado et al. 2002). Since overexpression of DIR1 in transgenic plants did not lead to constitutive SAR, the investigators of this study proposed that DIR1 interacts with a second, unidentified signaling component in order to function in the SAR signaling pathway. The crystallization of DIR1 has been reported, and its structure is currently being solved (Lascombe et al. 2006).
Evidence that family 1 LTPs are also involved in defensive signaling came from studies showing that wheat LTP1 can compete for a high affinity receptor binding site in tobacco cells, with elicitin, a defense response inducing elicitor protein from oomycetes (Buhot et al. 2001). Later it was shown that a family 1 tobacco LTP could similarly act as a binding competitor and that the strength of this interaction was increased by LTP in complex with the phytohormone jasmonic acid. Furthermore, treatment of plants with this complex lead to increased resistance against the oomycete Phytophthora parasitica (Buhot et al. 2004). Thus, it appears that, in this case, a family 1 LTP and elicitin act on the same, currently unidentified, receptor in order to modulate plant defense responses.
Further evidence corroborating a role in signaling comes from reports that several family 1 LTPs can bind calmodulin, albeit in a calcium-independent manner (Liu et al. 2001; Wang et al. 2005), and by the recent characterization of a covalently modified form of LTP1 (LTP1b) that has been observed in barley seeds. Bakan et al. (2006) demonstrated that the LTP1b consisted of an allene oxide that is specifically adducted to barley LTP1 at Asp7 by the action of 9-lipoxygenase and allene oxide synthase. The similarity of the adduct structure to precursors of jasmonic acid suggests a role in signaling, although a role in trapping of toxic oxylipins may also be possible. It should be noted that oxylipin adduction is not likely a universal feature of plant LTPs as Asp7 is not well conserved in the LTP1 family (see Fig. 1C).
LTP function in cuticle synthesis
The aerial organs of all land plants are covered with a structure, termed the cuticle, consisting of cutin, a polyester of hydroxy-fatty acids (Heredia 2003), and a variety of high melting point organic soluble compounds that are generically referred to as waxes (Jetter et al. 2006). Since the lipid precursors of the cuticle are synthesized within epidermal cells and must pass through the hydrophilic cell wall to the developing cuticle, LTPs have been proposed as carriers of these compounds. While no direct evidence has been produced for this role, the expression of family 1 LTPs in the epidermis (Sterk et al. 1991; Pyee and Kolattukudy 1995; Suh et al. 2005) and the correlation of wax accumulation with increased LTP expression (Hollenbach et al. 1997; Cameron et al. 2006) support this hypothesis. In vitro binding of acyl-CoA has been shown for family 1 LTPs (see above), but binding of the predicted hydroxyacyl-CoA precursors of cutin, or the very-long chain aliphatic compounds of waxes, has not been demonstrated. An alternative model for LTPs as structural components of the cuticle has also been proposed, in which an LTP binds two acyl moieties from neighboring cutin domains in order to form noncovalent bridges (Renan et al. 2003).
LTPs as modulators of plant growth and development
LTPs are also purported to be involved in reproductive and vegetative growth and development. A family 1 LTP from lily styles (SCA) was purified and shown to be necessary, along with pectin polysaccharides, for pollen tube adhesion on an artificial stylar matrix (Park et al. 2000; Park and Lord 2003). Moreover, when the expression of orthologous AtLTP3 and AtLTP4 genes was reduced in transgenic Arabidopsis, defects in both vegetative and reproductive growth were seen, since the resulting plants were dwarfed, with sterile anthers and semi-sterile pistils when pollinated with wild-type pollen (K. Chae and E.M. Lord, pers. comm.).
An interesting and unexpected finding was that family 1 LTPs can promote plant cell wall loosening and thus may play a role in cell expansion and plant growth. A family 1 LTP isolated from tobacco stigma exudate was shown to promote extension in cell walls under tension. This appears to be a general property of family 1 LTPs, as a family 1 LTP from wheat showed a similar effect. The mechanism of action is not clear, although it appears to depend on the availability of the hydrophobic cavity of LTPs (Nieuwland et al. 2005).
In general, although there has been significant progress in both the study of LTP biochemistry and biology, there remains a significant gap between the understanding of LTP structure and proposed functions in vivo. Their affinity for lipids is presumably fundamental to their biological function, but many critical questions will remain until in vivo substrates are clearly identified.
Gene evolution and LTP-like sequences
LTPs were first identified through assaying a biochemical activity, but it is clear from the analysis of completed plant genomes that the two biochemically characterized families are large and that there are additional sequences that share some characteristics with these “true” LTPs. The eight-cysteine motif that is typical of LTPs is also found in plant 2S-albumins, some protease inhibitors, and hybrid proline-rich proteins (Jose-Estanyol et al. 2004). In a 2003 survey of A. thaliana genes that may be involved in acyl lipid metabolism, Beisson et al. (2003) identified 71 LTP-like sequences on the basis of the conserved eight-cysteine motif. However, a more recent analysis of the Arabidopsis genome has shown that in the past, small cysteine-rich peptides, such as LTPs, have been underpredicted in genome annotation efforts (Silverstein et al. 2007).
With the availability of complete genome sequences from several plant species, it is now possible to investigate how LTPs are distributed throughout the plant kingdom. In the three angiosperm species for which full genome sequence is available (rice, Arabidopsis, and poplar) approximately a dozen family 1 LTP genes are present in each genome, while sequences with clear homology with family 1 LTPs are also present in the gymnosperms, as seen from EST sequences in pine (Pinus taeda) and other species. However, BLAST searches of current Physcomitrella patens (a moss) and Selaginella moellendorffii (a lycophyte) genome drafts shows no family 1 LTP sequences. A similar phylogenetic distribution is seen for the family 2 LTPs, and so, both family 1 and family 2 LTPs appear to be restricted to seed plants and absent in lower plants.
Conclusions
Plant LTPs represent a case where an understanding of the biochemical activity of a protein class and its association with structure is relatively sophisticated, while equivalent knowledge of in vivo function remains elusive. With evidence of varying quality for a plethora of different functions, it seems likely that there is not a single function that can be assigned to LTPs in general. Rather, individual isoforms likely play specific or multiple biological roles. The phylogenetic distribution of the two biochemically characterized LTP families suggests that they are unique to higher plants, but this in itself is not particularly informative, as the understanding of plant biochemistry and molecular biology is also biased toward higher plants. What is lacking is biochemical characterization of the less abundant isoforms of LTPs, and LTP-like proteins, and in vivo confirmation of function, especially through genetic phenotypes that can be ascribed to LTP genes.
Acknowledgments
Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081). Research support in the area was provided to J.K.C.R. by grants from the National Science Foundation's Plant Genome Program (award DBI 0606595) and New York State Office of Science, Technology and Academic Research (NYSTAR). T.H.Y. is supported in part by a NIH chemistry/biology interface training grant (T32 GM008500). We thank E. Lord (UC Riverside) for sharing prepublication data.
Footnotes
Reprint requests to: Jocelyn K.C. Rose, Department of Plant Biology, 228 Plant Science Building, Cornell University, Ithaca, NY 14853, USA; e-mail: jr286@cornell.edu; fax: (607) 255-5470.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073300108.
References
- Bakan, B., Hamberg, M., Perrocheau, L., Maume, D., Rogniaux, H., Tranquet, O., Rondeau, C., Blein, J.P., Ponchet, M., Marion, D. Specific adduction of plant lipid transfer protein by an allene oxide generated by 9-lipoxygenase and allene oxide synthase. J. Biol. Chem. 2006;281:38981–38988. doi: 10.1074/jbc.M608580200. [DOI] [PubMed] [Google Scholar]
- Beisson, F., Koo, A.J., Ruuska, S., Schwender, J., Pollard, M., Thelen, J.J., Paddock, T., Salas, J.J., Savage, L., Milcamps, A., et al. Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol. 2003;132:681–697. doi: 10.1104/pp.103.022988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen, J.D., Nielsen, H., von Heijne, G., Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bernhard, W.R., Thoma, S., Botella, J., Somerville, C.R. Isolation of a cDNA clone for spinach lipid transfer protein and evidence that the protein is synthesized by the secretory pathway. Plant Physiol. 1991;95:164–170. doi: 10.1104/pp.95.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloj, B., Zilversmit, D.B. Rat liver proteins capable of transferring phosphatidylethanolamine. Purification and transfer activity for other phospholipids and cholesterol. J. Biol. Chem. 1977;252:1613–1619. [PubMed] [Google Scholar]
- Buhot, N., Douliez, J.P., Jacquemard, A., Marion, D., Tran, V., Maume, B.F., Milat, M.L., Ponchet, M., Mikes, V., Kader, J.C., et al. A lipid transfer protein binds to a receptor involved in the control of plant defense responses. FEBS Lett. 2001;509:27–30. doi: 10.1016/s0014-5793(01)03116-7. [DOI] [PubMed] [Google Scholar]
- Buhot, N., Gomes, E., Milat, M.L., Ponchet, M., Marion, D., Lequeu, J., Delrot, S., Coutos-Thevenot, P., Blein, J.P. Modulation of the biological activity of a tobacco LTP1 by lipid complexation. Mol. Biol. Cell. 2004;15:5047–5052. doi: 10.1091/mbc.E04-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, K.D., Teece, M.A., Smart, L.B. Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol. 2006;140:176–183. doi: 10.1104/pp.105.069724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammue, B.P., Thevissen, K., Hendriks, M., Eggermont, K., Goderis, I.J., Proost, P., Van Damme, J., Osborn, R.W., Guerbette, F., Kader, J.C., et al. A potent antimicrobial protein from onion seeds showing sequence homology to plant lipid transfer proteins. Plant Physiol. 1995;109:445–455. doi: 10.1104/pp.109.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvolin, D., Douliez, J.P., Marion, D., Cohen-Addad, C., Pebay-Peyroula, E. The crystal structure of a wheat nonspecific lipid transfer protein (ns-LTP1) complexed with two molecules of phospholipid at 2.1 Å resolution. Eur. J. Biochem. 1999;264:562–568. doi: 10.1046/j.1432-1327.1999.00667.x. [DOI] [PubMed] [Google Scholar]
- Cheng, C.S., Samuel, D., Liu, Y.J., Shyu, J.C., Lai, S.M., Lin, K.F., Lyu, P.C. Binding mechanism of nonspecific lipid transfer proteins and their role in plant defense. Biochemistry. 2004a;43:13628–13636. doi: 10.1021/bi048873j. [DOI] [PubMed] [Google Scholar]
- Cheng, H.C., Cheng, P.T., Peng, P., Lyu, P.C., Sun, Y.J. Lipid binding in rice nonspecific lipid transfer protein-1 complexes from Oryza sativa . Protein Sci. 2004b;13:2304–2315. doi: 10.1110/ps.04799704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, C.S., Chen, M.N., Lai, Y.T., Chen, T., Lin, K.F., Liu, Y.J., Lyu, P.C. Mutagenesis study of rice nonspecific lipid transfer protein 2 reveals residues that contribute to structure and ligand binding. Proteins. 2007 doi: 10.1002/prot.21520. [DOI] [PubMed] [Google Scholar]
- Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T.J., Higgins, D.G., Thompson, J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks, G.E., Hon, G., Chandonia, J.M., Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva, P., Landon, C., Industri, B., Marais, A., Marion, D., Ponchet, M., Vovelle, F. Solution structure of a tobacco lipid transfer protein exhibiting new biophysical and biological features. Proteins. 2005;59:356–367. doi: 10.1002/prot.20405. [DOI] [PubMed] [Google Scholar]
- Douliez, J., Michon, T., Marion, D. Steady-state tyrosine fluorescence to study the lipid-binding properties of a wheat nonspecific lipid-transfer protein (nsLTP1) Biochim. Biophys. Acta. 2000;1467:65–72. [PubMed] [Google Scholar]
- Dowhan, W. Molecular basis for membrane phospholipid diversity: Why are there so many lipids? Annu. Rev. Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- Gincel, E., Simorre, J.P., Caille, A., Marion, D., Ptak, M., Vovelle, F. Three-dimensional structure in solution of a wheat lipid-transfer protein from multidimensional 1H-NMR data. A new folding for lipid carriers. Eur. J. Biochem. 1994;226:413–422. doi: 10.1111/j.1432-1033.1994.tb20066.x. [DOI] [PubMed] [Google Scholar]
- Gomar, J., Petit, M.C., Sodano, P., Sy, D., Marion, D., Kader, J.C., Vovelle, F., Ptak, M. Solution structure and lipid binding of a nonspecific lipid transfer protein extracted from maize seeds. Protein Sci. 1996;5:565–577. doi: 10.1002/pro.5560050402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerbette, F., Grosbois, M., Jolliot-Croquin, A., Kader, J.C., Zachowski, A. Lipid-transfer proteins from plants: Structure and binding properties. Mol. Cell. Biochem. 1999;192:157–161. [PubMed] [Google Scholar]
- Han, G.W., Lee, J.Y., Song, H.K., Chang, C., Min, K., Moon, J., Shin, D.H., Kopka, M.L., Sawaya, M.R., Yuan, H.S., et al. Structural basis of nonspecific lipid binding in maize lipid-transfer protein complexes revealed by high-resolution X-ray crystallography. J. Mol. Biol. 2001;308:263–278. doi: 10.1006/jmbi.2001.4559. [DOI] [PubMed] [Google Scholar]
- Heinemann, B., Andersen, K.V., Nielsen, P.R., Bech, L.M., Poulsen, F.M. Structure in solution of a four-helix lipid binding protein. Protein Sci. 1996;5:13–23. doi: 10.1002/pro.5560050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heredia, A. Biophysical and biochemical characteristics of cutin, a plant barrier biopolymer. Biochim. Biophys. Acta. 2003;1620:1–7. doi: 10.1016/s0304-4165(02)00510-x. [DOI] [PubMed] [Google Scholar]
- Hoh, F., Pons, J.L., Gautier, M.F., de Lamotte, F., Dumas, C. Structure of a liganded type 2 nonspecific lipid-transfer protein from wheat and the molecular basis of lipid binding. Acta Crystallogr. D Biol. Crystallogr. 2005;61:397–406. doi: 10.1107/S0907444905000417. [DOI] [PubMed] [Google Scholar]
- Hollenbach, B., Schreiber, L., Hartung, W., Dietz, K.J. Cadmium leads to stimulated expression of the lipid transfer protein genes in barley: Implications for the involvement of lipid transfer proteins in wax assembly. Planta. 1997;203:9–19. doi: 10.1007/s00050159. [DOI] [PubMed] [Google Scholar]
- Jayaraj, J., Punja, Z.K. Combined expression of chitinase and lipid transfer protein genes in transgenic carrot plants enhances resistance to foliar fungal pathogens. Plant Cell Rep. 2007;26:1539–1546. doi: 10.1007/s00299-007-0368-x. [DOI] [PubMed] [Google Scholar]
- Jegou, S., Douliez, J.P., Molle, D., Boivin, P., Marion, D. Purification and structural characterization of LTP1 polypeptides from beer. J. Agric. Food Chem. 2000;48:5023–5029. doi: 10.1021/jf000075m. [DOI] [PubMed] [Google Scholar]
- Jetter, R., Kunst, L., Samuels, A.L. Composition of plant cuticular waxes. In: Reider M., Müller C., editors. Biology of the plant cuticle. Blackwell; Oxford, UK: 2006. pp. 145–181. [Google Scholar]
- Jose-Estanyol, M., Gomis-Ruth, F.X., Puigdomenech, P. The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiol. Biochem. 2004;42:355–365. doi: 10.1016/j.plaphy.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Jouhet, J., Marechal, E., Block, M.A. Glycerolipid transfer for the building of membranes in plant cells. Prog. Lipid Res. 2007;46:37–55. doi: 10.1016/j.plipres.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Kader, J.C., Julienne, M., Vergnolle, C. Purification and characterization of a spinach-leaf protein capable of transferring phospholipids from liposomes to mitochondria or chloroplasts. Eur. J. Biochem. 1984;139:411–416. doi: 10.1111/j.1432-1033.1984.tb08020.x. [DOI] [PubMed] [Google Scholar]
- Kamp, H.H., Wirtz, K.W.A., Van Deenen, L.L. Some properties of phosphatidylcholine exchange protein purified from beef liver. Biochim. Biophys. Acta. 1973;318:313–325. [Google Scholar]
- Kent, C. Eukaryotic phospholipid biosynthesis. Annu. Rev. Biochem. 1995;64:315–343. doi: 10.1146/annurev.bi.64.070195.001531. [DOI] [PubMed] [Google Scholar]
- Lascombe, M.B., Buhot, N., Bakan, B., Marion, D., Blein, J.P., Lamb, C.J., Prange, T. Crystallization of DIR1, a LTP2-like resistance signalling protein from Arabidopsis thaliana . Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006;62:702–704. doi: 10.1107/S1744309106023748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, L.Q., Lorenz, Y., Scheurer, S., Fötisch, K., Enrique, E., Bartra, J., Biemelt, S., Vieths, S., Sonnewald, U. Design of tomato fruits with reduced allergenicity by dsRNAi-mediated inhibition of ns-LTP (Lyc e 3) expression. Plant Biotechnol. J. 2006;4:231–242. doi: 10.1111/j.1467-7652.2005.00175.x. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y., Min, K., Cha, H., Shin, D.H., Hwang, K.Y., Suh, S.W. Rice nonspecific lipid transfer protein: The 1.6 Å crystal structure in the unliganded state reveals a small hydrophobic cavity. J. Mol. Biol. 1998;276:437–448. doi: 10.1006/jmbi.1997.1550. [DOI] [PubMed] [Google Scholar]
- Lerche, M.H., Poulsen, F.M. Solution structure of barley lipid transfer protein complexed with palmitate. Two different binding modes of palmitate in the homologous maize and barley nonspecific lipid transfer proteins. Protein Sci. 1998;7:2490–2498. doi: 10.1002/pro.5560071202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerche, M.H., Kragelund, B.B., Bech, L.M., Poulsen, F.M. Barley lipid-transfer protein complexed with palmitoyl CoA: The structure reveals a hydrophobic binding site that can expand to fit both large and small lipid-like ligands. Structure. 1997;5:291–306. doi: 10.1016/s0969-2126(97)00186-x. [DOI] [PubMed] [Google Scholar]
- Lin, K.F., Liu, Y.N., Hsu, S.T., Samuel, D., Cheng, C.S., Bonvin, A.M., Lyu, P.C. Characterization and structural analyses of nonspecific lipid transfer protein 1 from mung bean. Biochemistry. 2005;44:5703–5712. doi: 10.1021/bi047608v. [DOI] [PubMed] [Google Scholar]
- Liu, H., Xue, L., Li, C., Zhang, R., Ling, Q. Calmodulin-binding protein BP-10, a probable new member of plant nonspecific lipid transfer protein superfamily. Biochem. Biophys. Res. Commun. 2001;285:633–638. doi: 10.1006/bbrc.2001.5219. [DOI] [PubMed] [Google Scholar]
- Lorenz, Y., Enrique, E., Lequynh, L., Fotisch, K., Retzek, M., Biemelt, S., Sonnewald, U., Vieths, S., Scheurer, S. Skin prick tests reveal stable and heritable reduction of allergenic potency of gene-silenced tomato fruits. J. Allergy Clin. Immunol. 2006;118:711–718. doi: 10.1016/j.jaci.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Lullien-Pellerin, V., Devaux, C., Ihorai, T., Marion, D., Pahin, V., Joudrier, P., Gautier, M.F. Production in Escherichia coli and site-directed mutagenesis of a 9-kDa nonspecific lipid transfer protein from wheat. Eur. J. Biochem. 1999;260:861–868. doi: 10.1046/j.1432-1327.1999.00229.x. [DOI] [PubMed] [Google Scholar]
- Maldonado, A.M., Doerner, P., Dixon, R.A., Lamb, C.J., Cameron, R.K. A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis . Nature. 2002;419:399–403. doi: 10.1038/nature00962. [DOI] [PubMed] [Google Scholar]
- Maxfield, F.R., Mondal, M. Sterol and lipid trafficking in mammalian cells. Biochem. Soc. Trans. 2006;34:335–339. doi: 10.1042/BST0340335. [DOI] [PubMed] [Google Scholar]
- Meijer, E.A., de Vries, S.C., Sterk, P., Gadella, D.W.J., Wirtz, K.W., Hendriks, T. Characterization of the nonspecific lipid transfer protein EP2 from carrot (Daucus carota L.) Mol. Cell. Biochem. 1993;123:159–166. doi: 10.1007/BF01076488. [DOI] [PubMed] [Google Scholar]
- Molina, A., Garcia-Olmedo, F. Enhanced tolerance to bacterial pathogens caused by the transgenic expression of barley lipid transfer protein LTP2. Plant J. 1997;12:669–675. doi: 10.1046/j.1365-313x.1997.00669.x. [DOI] [PubMed] [Google Scholar]
- Molina, A., Segura, A., Garcia-Olmedo, F. Lipid transfer proteins (nsLTPs) from barley and maize leaves are potent inhibitors of bacterial and fungal plant pathogens. FEBS Lett. 1993;316:119–122. doi: 10.1016/0014-5793(93)81198-9. [DOI] [PubMed] [Google Scholar]
- Nieuwland, J., Feron, R., Huisman, B.A., Fasolino, A., Hilbers, C.W., Derksen, J., Mariani, C. Lipid transfer proteins enhance cell wall extension in tobacco. Plant Cell. 2005;17:2009–2019. doi: 10.1105/tpc.105.032094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard, J., Vergnolle, C., Schoentgen, F., Kader, J.C. Acyl-binding/lipid-transfer proteins from rape seedlings, a novel category of proteins interacting with lipids. Biochim. Biophys. Acta. 1993;1170:109–117. doi: 10.1016/0005-2760(93)90059-i. [DOI] [PubMed] [Google Scholar]
- Park, S.Y., Lord, E.M. Expression studies of SCA in lily and confirmation of its role in pollen tube adhesion. Plant Mol. Biol. 2003;51:183–189. doi: 10.1023/a:1021139502947. [DOI] [PubMed] [Google Scholar]
- Park, S.Y., Jauh, G.Y., Mollet, J.C., Eckard, K.J., Nothnagel, E.A., Walling, L.L., Lord, E.M. A lipid transfer-like protein is necessary for lily pollen tube adhesion to an in vitro stylar matrix. Plant Cell. 2000;12:151–164. doi: 10.1105/tpc.12.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquato, N., Berni, R., Folli, C., Folloni, S., Cianci, M., Pantano, S., Helliwell, J.R., Zanotti, G. Crystal structure of peach Pru p 3, the prototypic member of the family of plant nonspecific lipid transfer protein pan-allergens. J. Mol. Biol. 2006;356:684–694. doi: 10.1016/j.jmb.2005.11.063. [DOI] [PubMed] [Google Scholar]
- Pettersen, E.F., Goddard, T.D., Huang, C.C., Couch, G.S., Greenblatt, D.M., Meng, E.C., Ferrin, T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pons, J.L., de Lamotte, F., Gautier, M.F., Delsuc, M.A. Refined solution structure of a liganded type 2 wheat nonspecific lipid transfer protein. J. Biol. Chem. 2003;278:14249–14256. doi: 10.1074/jbc.M211683200. [DOI] [PubMed] [Google Scholar]
- Poznanski, J., Sodano, P., Suh, S.W., Lee, J.Y., Ptak, M., Vovelle, F. Solution structure of a lipid transfer protein extracted from rice seeds. Comparison with homologous proteins. Eur. J. Biochem. 1999;259:692–708. doi: 10.1046/j.1432-1327.1999.00093.x. [DOI] [PubMed] [Google Scholar]
- Pyee, J., Kolattukudy, P.E. The gene for the major cuticular wax-associated protein and three homologous genes from broccoli (Brassica oleracea) and their expression patterns. Plant J. 1995;7:49–59. doi: 10.1046/j.1365-313x.1995.07010049.x. [DOI] [PubMed] [Google Scholar]
- Regente, M.C., Giudici, A.M., Villalain, J., de la Canal, L. The cytotoxic properties of a plant lipid transfer protein involve membrane permeabilization of target cells. Lett. Appl. Microbiol. 2005;40:183–189. doi: 10.1111/j.1472-765X.2004.01647.x. [DOI] [PubMed] [Google Scholar]
- Renan, M., Francois, J., Didier, M., Axelos, M., Douliez, J.P. Study of the interaction between end-capped telechelic polymers and the wheat lipid transfer protein LTP1, in solution and in the air/water interface. Coloids Surf., B. 2003;32:213–221. [Google Scholar]
- Salcedo, G., Sanchez-Monge, R., Barber, D., Diaz-Perales, A. Plant nonspecific lipid transfer proteins: An interface between plant defence and human allergy. Biochim. Biophys. Acta. 2007;1771:781–791. doi: 10.1016/j.bbalip.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Samuel, D., Liu, Y.J., Cheng, C.S., Lyu, P.C. Solution structure of plant nonspecific lipid transfer protein-2 from rice (Oryza sativa) J. Biol. Chem. 2002;277:35267–35273. doi: 10.1074/jbc.M203113200. [DOI] [PubMed] [Google Scholar]
- Sanner, M.F., Olson, A.J., Spehner, J.C. Reduced surface: An efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Segura, A., Moreno, M., Garcia-Olmedo, F. Purification and antipathogenic activity of lipid transfer proteins (LTPs) from the leaves of Arabidopsis and spinach. FEBS Lett. 1993;332:243–246. doi: 10.1016/0014-5793(93)80641-7. [DOI] [PubMed] [Google Scholar]
- Shin, D.H., Lee, J.Y., Hwang, K.Y., Kim, K.K., Suh, S.W. High-resolution crystal structure of the nonspecific lipid-transfer protein from maize seedlings. Structure. 1995;3:189–199. doi: 10.1016/s0969-2126(01)00149-6. [DOI] [PubMed] [Google Scholar]
- Silverstein, K.A., Moskal, W.A.J., Wu, H.A., Underwood, B.A., Graham, M.A., Town, C.D., Vanden Bosch, K.A. Small cystein-rich peptides resembling antimicrobial peptides have been underpredicted in plants. Plant J. 2007;51:262–280. doi: 10.1111/j.1365-313X.2007.03136.x. [DOI] [PubMed] [Google Scholar]
- Sterk, P., Booij, H., Schellekens, G.A., Van Kammen, A., De Vries, S.C. Cell-specific expression of the carrot EP2 lipid transfer protein gene. Plant Cell. 1991;3:907–921. doi: 10.1105/tpc.3.9.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh, M.C., Samuels, A.L., Jetter, R., Kunst, L., Pollard, M., Ohlrogge, J., Beisson, F. Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 2005;139:1649–1665. doi: 10.1104/pp.105.070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin, S., Broekaert, W.F., Marion, D., Acland, D.P., Ptak, M., Vovelle, F., Sodano, P. Solution structure of Ace-AMP1, a potent antimicrobial protein extracted from onion seeds. Structural analogies with plant nonspecific lipid transfer proteins. Biochemistry. 1998;37:3623–3637. doi: 10.1021/bi9723515. [DOI] [PubMed] [Google Scholar]
- Tassin-Moindrot, S., Caille, A., Douliez, J.P., Marion, D., Vovelle, F. The wide binding properties of a wheat nonspecific lipid transfer protein. Solution structure of a complex with prostaglandin B2. Eur. J. Biochem. 2000;267:1117–1124. doi: 10.1046/j.1432-1327.2000.01109.x. [DOI] [PubMed] [Google Scholar]
- Terras, F.R., Goderis, I.J., Van Leuven, F., Vanderleyden, J., Cammue, B.P., Broekaert, W.F. In vitro antifungal activity of a radish (Raphanus sativus L.) seed protein homologous to nonspecific lipid transfer proteins. Plant Physiol. 1992;100:1055–1058. doi: 10.1104/pp.100.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma, S., Kaneko, Y., Somerville, C. A nonspecific lipid transfer protein from Arabidopsis is a cell wall protein. Plant J. 1993;3:427–436. doi: 10.1046/j.1365-313x.1993.t01-25-00999.x. [DOI] [PubMed] [Google Scholar]
- Tsuboi, S., Osafune, T., Tsugeki, R., Nishimura, M., Yamada, M. Nonspecific lipid transfer protein in castor bean cotyledon cells: Subcellular localization and a possible role in lipid metabolism. J. Biochem. 1992;111:500–508. doi: 10.1093/oxfordjournals.jbchem.a123787. [DOI] [PubMed] [Google Scholar]
- Vance, J.E., Vance, D.E. Phospholipid biosynthesis in mammalian cells. Biochem. Cell Biol. 2004;82:113–128. doi: 10.1139/o03-073. [DOI] [PubMed] [Google Scholar]
- Van Loon, L.C., Van Strien, E.A. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999;55:85–97. [Google Scholar]
- Wang, Z., Xie, W., Chi, F., Li, C. Identification of nonspecific lipid transfer protein-1 as a calmodulin-binding protein in Arabidopsis . FEBS Lett. 2005;579:1683–1687. doi: 10.1016/j.febslet.2005.02.024. [DOI] [PubMed] [Google Scholar]