Abstract
Mammalian mitochondrial ribosomes synthesize 13 proteins that are essential for oxidative phosphorylation. In addition to their role in protein synthesis, some of the mitochondrial ribosomal proteins have acquired functions in other cellular processes such as apoptosis. Death-associated protein 3 (DAP3), also referred to as mitochondrial ribosomal protein S29 (MRP-S29), is a GTP-binding pro-apoptotic protein located in the small subunit of the ribosome. Previous studies have shown that phosphorylation is one of the most likely regulatory mechanisms for DAP3 function in apoptosis and may be in protein synthesis; however, no phosphorylation sites were identified. In this study, we have investigated the phosphorylation status of ribosomal DAP3 and mapped the phosphorylation sites by tandem mass spectrometry. Mitochondrial ribosomal DAP3 is phosphorylated at Ser215 or Thr216, Ser220, Ser251 or Ser252, and Ser280. In addition, phosphorylation of recombinant DAP3 by Protein kinase A and Protein kinase Cδ at residues that are endogenously phosphorylated in ribosomal DAP3 suggests both of these kinases as potential candidates responsible for the in vivo phosphorylation of DAP3 in mammalian mitochondria. Interestingly, the majority of the phosphorylation sites detected in our study are clustered around the highly conserved GTP-binding motifs, speculating on the significance of these residues on protein conformation and activity. Site-directed mutagenesis studies on selected phosphorylation sites were performed to determine the effect of phosphorylation on cell proliferation and PARP cleavage as indication of caspase activation. Overall, our findings suggest DAP3, a mitochondrial ribosomal small subunit protein, is a novel phosphorylated target.
Keywords: phosphorylation, DAP3, proteomics, mitochondrial ribosome, mass spectrometry, apoptosis
Mitochondria are the powerhouses of eukaryotic cells, generating most of the ATP through oxidative phosphorylation. Although 13 mitochondrially encoded proteins are essential for this process and survival of eukaryotic cells, the mitochondrial translational system responsible for the synthesis of these proteins is one of the least understood systems. Proteomic studies of mitochondrial ribosomes have indicated that some of the ribosomal proteins have acquired unique functions in addition to their essential role in protein synthesis (Koc et al. 2001a,b, c; Sharma et al. 2003; O'Brien et al. 2005). Several mammalian mitochondrial ribosomal proteins (MRPs) have been demonstrated to be involved in apoptotic signaling pathways (Kissil et al. 1999; Koc et al. 2001c; Miyazaki and Reed 2001; Levshenkova et al. 2004; Chintharlapalli et al. 2005). One of these, MRP-S29, more commonly known as DAP3, was first identified as a positive mediator of interferon-γ induced cell death through a functional screen of HeLa cells transfected with an antisense cDNA library (Kissil et al. 1995). Although it is historically linked to cell death, DAP3 is the only GTP-binding protein found in the small subunit of the mammalian and yeast mitochondrial ribosomes (Denslow et al. 1991; Koc et al. 2001a; Saveanu et al. 2001). The presence of this protein accounts for the high specific GTP-binding affinity of small subunits of mitochondrial ribosomes (Denslow et al. 1991). Immuno-EM studies have localized DAP3 to the base of the lower lobe of the small subunit on the solvent side of the ribosome (O'Brien et al. 2005), but its exact function in protein synthesis is still unknown. In yeast, DAP3 is required for mitochondrial DNA synthesis and respiration (Berger et al. 2000). This observation indicates that DAP3 plays a role in protein synthesis, which is required for the maintenance of mitochondrial DNA in yeast. Furthermore, a theme connecting DAP3 to mitochondrial physiology is gaining support as DAP3 deficiency in mice is lethal in utero with abnormal, shrunken mitochondria (Kim et al. 2007).
Mitochondria are highly dynamic, and phosphorylation of Ser, Thr, or Tyr residues is possibly one of the most important regulatory mechanisms employed in this organelle. A large number of phosphorylated mitochondrial proteins have been reported (Liu et al. 2003; Schulenberg et al. 2003; Taylor et al. 2003; Salvi et al. 2005; Hopper et al. 2006). To better understand the mechanism behind the variety of roles that DAP3 plays within the mitochondria, we hypothesized that the function of ribosomal DAP3 can be partially controlled by phosphorylation. Phosphorylation of DAP3 could serve as a molecular switch, impacting protein synthesis as well as apoptosis and mitochondrial physiology. A previous study has shown that DAP3 can be phosphorylated at Thr237 by Akt, resulting in the suppression of anoikis, and mutating this residue resulted in an increased level of cell death as the survival kinase could no longer phosphorylate the site (Miyazaki et al. 2004). Recently, interaction of DAP3 with the Ser/Thr kinase LKB1 and the role of this kinase in DAP3-induced apoptosis have been studied in osteosarcoma cells (Takeda et al. 2007).
To expand on these initial studies, we used mass spectrometry-based proteomics to identify and map the specific phosphorylation sites on ribosomal DAP3 and in vitro phosphorylation sites of recombinant DAP3 using several different kinases localized to the mitochondria. By use of phosphorylated amino acid specific antibodies and tandem mass spectrometry, we provide evidence for phosphorylation of mitochondrial ribosomal DAP3 isolated from bovine mitochondrial 55S ribosomes. Moreover, selected phosphorylated residues were evaluated for their effect on cell proliferation and caspase activation detected by the DNA single-strand repair enzyme poly (ADP-ribose) polymerase (PARP) cleavage. Our findings suggest that phosphorylation is one of the mechanisms modulating the function of ribosomal DAP3 in the progression of apoptosis in mitochondria.
Results
DAP3 is phosphorylated on 55S mitochondrial ribosomes
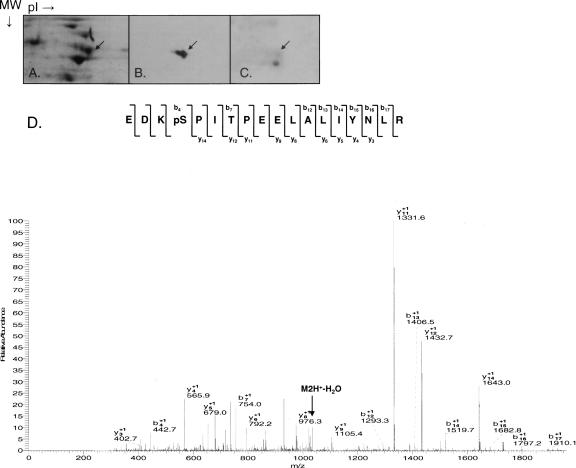
To determine the phosphorylation status of DAP3, crude mitochondrial ribosomes were isolated from bovine liver using previously described methods (Matthews et al. 1982) and fractionated into intact 55S mitochondrial ribosomes. Mammalian mitochondrial ribosomes contain 77 known proteins, of which 29 are in the small subunit and 48 are in the large subunit (O'Brien et al. 1999, 2000; Koc et al. 2001a,b; Suzuki et al. 2001a,b). The proteins of the 55S ribosomes were separated by two-dimensional (2D) gel electrophoresis using NEPHGE (nonequilibrium pH gradient electrophoresis) tube gels rather than the commonly used IPG strips due to the basic characteristic of ribosomal proteins (Cahill et al. 1995). SDS-PAGE separation was used as the second dimension, and the proteins were located by staining with SYPRO Ruby (Fig. 1A). Bovine DAP3 migrates in the 37–50 kDa region, and the exact location of the DAP3 protein was determined by Western blot analysis and mass spectrometric identification of the 2D gel spots as shown by an arrow in Figure 1. The pattern of DAP3 obtained from this analysis showed the characteristic trailing toward a more acidic pI expected for a phosphorylated protein (Fig. 1B). Furthermore, the sample was probed with anti-phospho serine antibody, and DAP3 is serine phosphorylated as noted in Figure 1C.
Figure 1.
DAP3 is phosphorylated on 55S bovine mitochondrial ribosomes. Bovine mitochondrial ribosomal proteins were separated on 2D NEPHGE gels; the DAP3 protein was excised, digested with trypsin, and analyzed by LC/MS/MS for potential phosphorylation sites using an LTQ ion trap mass spectrometer. (A) SYPRO Ruby stained gel. (B, C) Western blot analysis of the 55S bovine mitochondrial ribosomes using anti-DAP3 and phosphoserine antibodies, respectively. The arrow indicates the location of DAP3 on the gel or Western blot. (D) CID MS/MS spectrum for doubly charged phosphorylated peptide EDKpSPITPEELALIYNLR (2084.4, 2+).
In addition, purified 55S ribosome preparations were analyzed by in-solution digests of the whole ribosomal protein mixture, the shotgun approach, and in-gel digests of the 1D-gel separated proteins in order to improve the peptide coverage and, as a result, the likelihood of detecting more phosphopeptides. Despite a high peptide coverage (>70%) obtained using these various approaches, only one phosphorylated peptide was detected for DAP3, among the more common unphosphorylated form of the peptide. We then employed phosphopeptide enrichment of the intact 55S ribosomes by immobilized metal affinity chromatography (IMAC) based on selective binding of phosphopeptides to gallium. This enrichment strategy allowed us to detect additional peptides, which resulted in a higher overall sequence coverage and identification of two new DAP3 phosphopeptides (Table 1).
Table 1.
In vivo and in vitro phosphorylation sites of DAP3 detected and mapped by MS/MS analyses
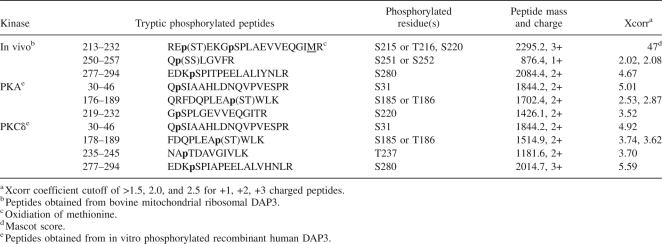
Tandem MS spectra generated by fragmentation of peptides by collision-induced dissociation (CID) were used for both detection and mapping of phosphorylation sites. The spectra matching to phosphopeptides were each carefully validated for overall data quality and signal-to-noise of peaks matching to expected m/z values in addition to applying the Sequest Xcorr coefficient cutoff of >1.5, 2.0, and 2.5 for +1, +2, and +3 charged peptides, respectively. Interestingly, the majority of the phosphorylation sites identified in the bovine DAP3 sequence had been predicted to be phosphorylated using NetPhos prediction tool, including Ser215, Thr216, Ser220, Ser252, and Ser280 (Table 1). Furthermore, most of the phosphorylation sites are located in close proximity to GTP-binding motifs found in the DAP3 sequence except Ser280 (Fig. 2A). Therefore, as a comparison the three-dimensional (3D) structure of the human Ras-21 was modeled in the GTP-bound configuration to illustrate relative conformations of GTP-binding motifs, G1–G5 in Figure 2B (Pai et al. 1990). Interestingly, eight of the phosphorylated residues are found in the vicinity of the GTP-binding motifs. For example, Ser215, Thr216, and Ser220 are evolutionarily conserved and located after G4 motif (NKRE). Similarly, the peptide score for phosphorylation of Ser251 was very close to the score for Ser252; therefore, phosphorylation of Ser251 and Ser252 were not distinguishable (Table 1). Both of these residues are positioned just a short distance after G3 motif, implying their possible role in forming the GTP-binding pocket (Fig. 2A,B). The other phosphorylation site determined was Ser280, and this residue is highly conserved and predicted to be phosphorylated. The tandem mass spectrum of the phosphorylated peptide containing the Ser280 phosphorylation is shown in Figure 1D, indicating a neutral loss of a water (−18 Da) molecule that is known to occur as a result of loss of the phosphate moiety from the Ser280 through β-elimination. One additional potential phosphorylation site, Thr237, was also identified in our analysis, but with a lower MASCOT score and/or Sequest Xcorr coefficient.
Figure 2.
Primary sequence alignment of bovine (Swiss-Prot P82922), human (Swiss-Prot P51398), dog (Gen Bank CK653556 and DR105073), and mouse (Swiss-Prot Q9ER88) DAP3 proteins and the 3D model of the GTP-binding pocket. (A) The four guanine nucleotide binding motifs are indicated by G1, G2, G3, and G4 with thick lines. Asterisks (*) specify some the phosphorylation sites found in this proteomic analysis. The alignment was created with CLUSTALW program in Biology Workbench, and the results are displayed in BOXSHADE. (B) The 3D model of the human Ras-21 in the GTP-bound configuration was generated by PyMOL (DeLano Scientific) using the coordinates obtained from Protein Data Bank (PDB) entry 5P21 (Pai et al. 1990). The five conserved guanine nucleotide-binding motifs and GTP are labeled as G1–G5 and GTP and colored in cyan, brown, green, yellow, purple, and red, respectively.
Ectopically expressed DAP3 is serine and threonine phosphorylated in HEK293T cells
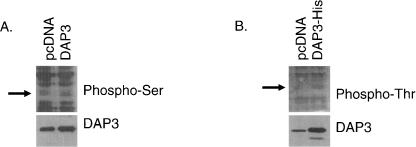
To further support the in vivo phosphorylation analysis of DAP3, the phosphorylation status of ectopically expressed human DAP3 was subjected to Western blot analysis probed with anti-phospho serine and threonine antibodies. In Figure 3, A and B, human DAP3 is clearly overexpressed compared with the cells transfected with the control vector in HEK293T cells. It was evident that ectopically expressed human DAP3 is phosphorylated at both serine and threonine residues. This is also in agreement with the proteomics analysis performed using bovine ribosomal DAP3.
Figure 3.
Ectopically expressed DAP3 is serine and threonine phosphorylated in HEK293T cells. Plasmid DNA for DAP3, DAP3-His, and the control vector pcDNA were transiently transfected into HEK293T cells. (A, B) Each panel is a Western blot probed with phosphoserine, phosphothreonine, and DAP3 antibodies using 50 μg of protein. The arrow indicates the location of DAP3 on the Western blot.
In vitro phosphorylation of recombinant DAP3 by PKA and PKCδ
Web-based tools, including Scan ProSite and NetPhos, were employed to identify possible kinases, which may serve as candidates to phosphorylate DAP3 in vivo and to identify potential serine, threonine, and tyrosine phosphorylation sites in the human DAP3 sequence (Blom et al. 1999). cAMP-dependent protein kinase (PKA) and protein kinase C delta (PKCδ) were chosen as the best candidates to phosphorylate DAP3, as each had predicted sites that corresponded to our in vivo data. For example, PKA was predicted to phosphorylate Ser215 and Ser251. This kinase can be localized to the mitochondria through A-kinase anchoring proteins (AKAPs), and its enzyme activity occurs on the matrix side of the inner membrane (Chen et al. 2004). Furthermore, given the consensus motif for PKCδ, Thr216 is a probable target. PKCδ is found within the mitochondria or is translocated there depending on the cell type (Brodie and Blumberg 2003). Overall, the NetPhos prediction results indicated that eight serine, two threonine, and one tyrosine residues were predicted to be phosphorylated with high probability in the human DAP3 sequence.
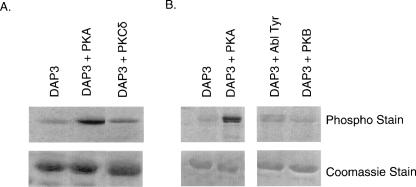
Human recombinant DAP3 was overexpressed in Escherichia coli and purified under native conditions using a Ni-NTA column. Phosphorylation reactions carried out with recombinant DAP3 in the presence of various kinases were analyzed by SDS-PAGE followed by ProQ-Diamond phosphoprotein staining. It was clear that DAP3 was phosphorylated by both PKA and PKCδ compared with the untreated recombinant DAP3 used as a control (Fig. 4A). Since the ProQ-Diamond is not a residue-specific phosphoprotein dye, it was not possible to determine which residue was phosphorylated. In order to determine specific site(s) of phosphorylation by these kinases, LC/MS/MS (Liquid chromatography, tandem mass spectrometry) analyses were performed on the in-gel digests of kinase reaction products. Tandem mass spectrometric analyses revealed phosphorylation of Ser31, Ser185, or Thr186 by both PKA and PKCδ as the CID tandem mass spectra reflected either a neutral loss of phosphoric acid (M2H+-H3PO4) or water (−18 Da) molecule (Table 1). Two additional phosphorylation sites, Thr237 and Ser280, and one additional PKA phosphorylation site, Ser220, were identified in the PKCδ and PKA reactions, respectively (Table 1). The spectra used in mapping of these sites are shown in Figure 5, A and B. These spectra reflected a loss of water (−18 Da) from Ser220 by β-elimination in Figure 5A and an increase of 80 Da in molecular weight of the peptide for Thr237 in Figure 5B. Of interest, neither Ser185 nor Thr237 had been predicted to be phosphorylated, yet each site is evolutionarily conserved. In particular, Thr237 is the same site as previously found to be phosphorylated by Akt suppressing anoikis; however, we were unable to show in vitro phosphorylation by this kinase (Fig. 4B; Miyazaki et al. 2004). This site precedes GTP-binding G3 motif (DAVG), while Ser185 is upstream of G2 motif (Thr193), once again emphasizing this clustering effect of the phosphorylation sites (Fig. 2A,B). In addition to the kinases mentioned, we also tested whether the expressed DAP3 was a substrate for the Abl protein tyrosine kinase since Tyr208 was predicted (96%) to be phosphorylated (Fig. 4B). The ProQ analysis showed that the recombinant DAP3 could be slightly phosphorylated, but we were unable to confidently map any phosphorylated Tyr residues. This was probably because some of the Tyr residues are located in Lys and Arg rich regions and tryptic digestion of DAP3 resulted in peptides that were too small to be detected and phosphorylated residues could potentially be missed. For this reason, proteolytic digestions with Glu C (cleaves at the C-terminal end of glutamic acid) and Lys C (cleaves at the C-terminal end of lysine) were also performed. However, no additional phosphopeptides were detected. Overall, the DAP3 sequence coverage was 84%, 17%, and 32% using trypsin, Glu C, and Lys C, respectively.
Figure 4.
DAP3 is phosphorylated by PKA and PKCδ. The in vitro phosphorylation reactions were run on SDS-PAGE gels using 5 μg of protein, stained with the ProQ Diamond Phospho dye. Phosphorylated DAP3 was excised from a Coomassie-stained gel; digested with trypsin, Lys C, or Glu C; and analyzed with an LTQ ion trap mass spectrometer. (A) Overexpressed DAP3 with and without PKA and PKCδ. (B) Overexpressed DAP3 with and without PKA, Abl tyrosine kinase, and PKB.
Figure 5.
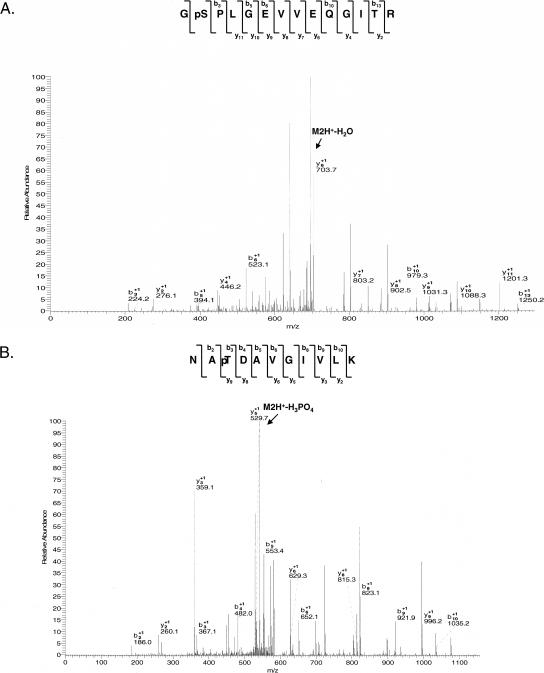
DAP3 is phosphorylated at S31, S185 or T186, and S220 by PKA and at S31, S185 or T186, T237, and S280 by PKCδ. CID MS/MS spectra acquired by LTQ ion trap mass spectrometer were used to map the specific sites of phosphorylation by observing a shift of +80 Da in the masses of b and y ions, which corresponds to addition of a phosphate group, starting at the site of modification. CID MS/MS spectra for tryptic doubly charged phosphoserine-containing peptide GpSPLGEVVEQGITR (1426.1, 2+) (A) and phosphothreonine-containing peptide NApTDAVGIVLK (1181.6, 2+) (B).
Effects of DAP3 mutations on cell proliferation and PARP cleavage
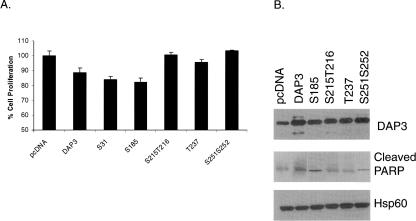
DAP3 was originally identified as a pro-apoptotic protein; therefore, the role of phosphorylated residues on cell proliferation and PARP cleavage as an indication of caspase activation was investigated by site-directed mutagenesis. Residues determined to be phosphorylated and in close proximity to GTP-binding motifs were mutated to alanine residues, and mutants were constructed in the mammalian expression vector pcDNA 3.1(+). Each were transfected into HEK293T cells to compare the expression levels of the wild-type DAP3, DAP3 mutants, and endogenous DAP3 as obtained from the control vector. The WST-1 cell proliferation assay is based on the cleavage of tetrazolium salt by mitochondrial dehydrogenases in viable cells. Therefore, these assays are directly related to mitochondrial function of cells transfected with the wild-type and the mutant DAP3 constructs. As seen in Figure 6A, the ectopically expressed DAP3 reduced the cell proliferation by ∼11%, while the expression of S31A and S185A mutants decreased it by 16%–17%. In particular, the mutations generated for some of the phosphorylation sites found in ribosome-associated DAP3, namely, Ser215, Thr216, Thr237, Ser251, and Ser252, reversed the cell death caused by the wild-type DAP3 (Fig. 6A). Therefore, these Ser and Thr residues might be critical for induction of apoptosis by ectopic expression of DAP3 in these human cell lines.
Figure 6.
Cell proliferation and PARP cleavage is assessed in selected DAP3 phosphorylation mutants. (A) Plasmid DNA for each of the DAP3 constructs and the pcDNA control vector were transiently transfected into HEK293T cells. Cell proliferation was based on mitochondrial dehydrogenase activities using the WST-1 reagent. Results were expressed as mean ±SD and analyzed using the ANOVA's t-test. Values of *P < 0.05 were considered statistically significant. (B) Plasmid DNA for the control vector pcDNA, DAP3, and selected DAP3 phosphorylation mutants were transiently transfected into HEK293T cells. After separating 50 μg of cell lysate by SDS-PAGE, Western blot analyses were performed to detect DAP3 expression, cleaved PARP levels, and Hsp60 levels as a loading control.
In addition to cell proliferation assays, Western blot analysis of PARP cleavage was performed using cell lysates obtained from HEK293T cells transfected with the DAP3 mutants. As observed in Figure 6B, wild-type DAP3 and mutant constructs were overexpressing corresponding protein products compared with the endogenous level of DAP3 expression. Probing the same blot for endogenous Hsp60 levels served as a loading control. In normal cells, PARP is involved in DNA repair, but once cleaved by caspase 3, it is inactivated and can serve as a marker for the induction of apoptosis. As indicated in Figure 6B, more PARP cleavage was detected in cells expressing exogenous DAP3 compared with cells receiving the control vector alone. Moreover, Ser215, Thr216, Thr237, Ser251, and Ser252 DAP3 mutants reduced the PARP cleavage compared with cells overexpressing wild-type DAP3, while the Ser31 mutant (data not shown) and the Ser185 mutant had comparable levels of PARP cleavage. Findings obtained with these mutants near the GTP-binding motifs 2, 3, and 4 of DAP3 in cell proliferation assays suggest the importance of the GTP-binding motifs in DAP3-induced apoptosis. Interestingly, Ser31 and Ser185 phosphorylated in vitro by PKA and PKCδ were not as effective in modulating apoptotic function of DAP3 as detected in cell proliferation and PARP cleavage assays.
Discussion
DAP3 has been described as a pro-apoptotic protein promoting apoptosis induced by interferon γ, tumor necrosis factor α, and Fas as observed by structure-function studies (Kissil et al. 1995, 1999; Mukamel and Kimchi 2004). Further studies, however, demonstrated that the overexpression of DAP3 protects Ataxia telangiectasia and glioblastoma cells from ionizing radiation and streptonigrin-induced and camptothecin-induced apoptosis (Henning 1993; Mariani et al. 2001). These two contradictory effects attributed to the overexpression of DAP3 might be controlled by phosphorylation in apoptosis. In fact, phosphorylation of DAP3 by Akt kinase and LKB1, a Ser/Thr kinase, was reported to be important for suppression of anoikis (Miyazaki et al. 2004) and pro-apoptotic function of DAP3 in induction of apoptosis in osteosarcoma cells (Takeda et al. 2007). However, there are no proteomics data available for identification of specific phosphorylated residues of DAP3 as part of the mammalian mitochondrial small subunit and role of these phosphorylation sites in induction of apoptosis. Here, we report the phosphorylation status of mitochondrial ribosomal DAP3 as a component of 55S ribosomes and construct a list of potential kinases known to be translocated into mitochondria, which may be capable of phosphorylating DAP3 in vivo and the role of some of the phosphorylation sites in induction of apoptosis.
One of the common features of the phosphorylation sites found in this study is their close proximities to the conserved GTP-binding motifs found in DAP3. Mutation of GTP binding, specifically the P-loop motif was previously shown to be important for pro-apoptotic activity of DAP3 (Kissil et al. 1999). GTP-binding proteins are constantly switching between GDP and GTP-bound state, triggering the activation of various effectors. In general, guanine nucleotide binding motif G2 is essential for magnesium coordination; G3 interacts with the magnesium binding site and the γ-phosphate of GTP, while motif G4 is important for recognizing the guanine ring. Especially, DAP3 phosphorylation at residues Ser215, Thr216, Thr237, Ser251, and Ser252 surrounding the G4 and G3 motifs may distort the conformation of the GTP-binding pocket and affect GTP binding/hydrolysis (Fig. 2A,B). For this reason, mutations generated at these residues have possibly caused conformational and/or electrostatic changes in the GTP-binding pocket and affected the apoptotic function of DAP3 as demonstrated by PARP cleavage and cell proliferation assays. On the other hand, mutations at Ser31 and Ser185 did not interfere with pro-apoptotic functions of DAP3, and they have shown decreased cell viability and increased PARP cleavage activity resembling the wild-type DAP3. Therefore, as with many other GTP-binding proteins, DAP3 can also be tightly regulated by phosphorylation, and mutations at these phosphorylated residues may cause changes in its functions in cells.
Many of the phosphorylated serine and threonine residues found in this study are highly conserved, and locations of these residues in various kinase recognition motifs suggest that the DAP3 protein is a substrate for many different kinases. As demonstrated in this study, multiple kinases can phosphorylate the same residue in DAP3, which was evident with the phosphorylation of Ser31, Ser185, Thr186, Thr237, and Ser280 by a kinase endogenously phosphorylating DAP3, PKA, and PKCδ. Phosphorylation of the same residue in the DAP3 sequence by different kinases may also modulate DAP3 function, as some kinases are classified as survival vs. pro-apoptotic. Furthermore, in comparison to other well-known pro-apoptotic proteins, DAP3 is rather unique as a component of the mitochondrial ribosome; therefore, one needs to keep in mind that multiple roles attributed to DAP3 in apoptosis might simply be due to changes in the synthesis of 13 mitochondrially encoded proteins that are essential for oxidative phosphorylation.
Materials and Methods
Purification and in vitro phosphorylation of recombinant DAP3
Human DAP3 was cloned into the pET28a vector using XhoI and NdeI, expressed in the BL21-RIL strain of E. coli for 5 h at 23°C, and purified under native conditions using the Ni-NTA column. In vitro phosphorylation of DAP3 was carried out with kinases PKA the catalytic subunit (New England BioLabs), PKCδ (Sigma-Aldrich, Inc.), protein kinase B (PKB) (New England BioLabs), and Abl protein tyrosine kinase (New England BioLabs). Approximately 5 μg of recombinant DAP3 protein was used for each 20 μL kinase assay, and the reaction conditions were adapted from the protocols provided by the manufacturers using 200 μM cold ATP. Resulting protein samples were run on SDS-PAGE gels, stained with ProQ Diamond Phosphoprotein Gel Stain (Molecular Probes, Inc.), and visualized by a laser scanner at an excitation wavelength of 532 nm. The bands corresponding to DAP3 were excised and in-gel digested with proteases trypsin (cleaves at the C-terminal end of lysine and arginine residues), Lys C (cleaves at the C-terminal end of lysine residues), and Glu C (cleaves at the C-terminal end of glutamic acid) for better coverage. The peptides from in-gel digestion were analyzed by LC/MS/MS for DAP3 identification and mapping of the phosphorylation sites.
Isolation and detection of phosphorylated mitochondrial ribosomal DAP3
Preparation of mitochondrial ribosomes starting from 4 kg of bovine liver using a flow-through tissue homogenizer was adapted from a previously described method (Matthews et al. 1982). In order to preserve phosphorylation status of ribosomal proteins, phosphatase inhibitors (2 mM imizadole, 1 mM sodium orthovanadate, 1.15 mM sodium molybdate, 1 mM sodium fluoride, and 4 mM sodium tartrate dehydrate) were added during the homogenization process. Crude ribosomes were loaded onto a 10%–30% sucrose gradient and fractionated to isolate intact 55S ribosomes. Proteins in the fractions were either pelleted by ultracentrifugation or concentrated using a Microcon Ultracel YM-10 centrifugal unit (Millipore Corp.) and precipitated with acetone. The resulting pellet (∼1.8 A260 units) was resuspended in lysis buffer consisting of 9.8 M urea, 2% (w/v) NP-40, 2% ampholytes pI 3–10 and 8–10, and 100 mM DTT. The samples were loaded on NEPHGE tube gels and equilibrated in buffer containing 60 mM Tris-HCl (pH 6.8), 2% SDS, 100 mM DTT, and 10% glycerol (Cahill et al. 1995). The 14% 2D gel was stained with SYPRO Ruby (Molecular Probes Inc.) or Coomassie blue for total protein content, and the DAP3 gel piece was excised based on Western blot analysis. To complement this, one-dimensional SDS-PAGE and in-solution digestion samples (∼1.8 A260 units) were also prepared from purified 55S fractions. A C4-reverse phase trap column (Michrom Bioresources Inc.) was used to desalt and to concentrate the above protein samples using the procedure provided by the manufacturer. For the in-solution digests, the eluate was dried in a SpeedVac and resuspended in 25 mM NH4HCO3 for trypsin digestion, which was added directly to the sample solution at 1:50 ratio and incubated overnight at 37°C. Peptides obtained from in-gel and in-solution digestions were analyzed by LC/MS/MS, and the peptides generated were used for DAP3 identification and mapping of the phosphorylation sites.
Enrichment of phosphorylated peptides
The PhosphoProfile I phosphopeptide enrichment kit (Sigma-Aldrich, Inc.) was used for enrichment of phosphorylated peptides of DAP3. The protocol provided in the kit was followed to enrich the phosphopeptides obtained from in-solution digestion of intact 55S mitochondrial ribosomes (∼3.6 A260 units). Briefly, the digested protein sample was acidified with glacial acetic acid to a final concentration of 250 mM and loaded on the gallium silica spin column (Sigma-Aldrich, Inc.). The column was washed twice with 50 mM acetic acid in 10% acetonitrile and eluted with 0.4 M NH4OH in 20% acetonitrile. The eluate was dried by SpeedVac and reconstituted in 0.1% formic acid for the LC/MS/MS analysis.
Mass spectrometric mapping of DAP3 phosphorylation sites
Detection and mapping of phosphorylation sites was achieved by database searching of tandem mass spectra of proteolytic peptides searched against protein databases. Tandem MS spectra obtained by fragmenting a peptide by CID were acquired using a capillary LC/MS/MS system that consisted of a Surveyor HPLC pump, a Surveyor Micro AS autosampler, and an LTQ linear ion trap mass spectrometer (ThermoFinnigan). The acquired spectra were analyzed using Xcalibur 2.0 and Bioworks 3.2 software. The raw CID tandem mass spectrometer spectra were also converted to Mascot generic files (.mgf) using the extract msn software (ThermoFinnigan). The .dta and .mgf files were submitted to a site-licensed Sequest and Mascot (version 2.2) search engines to search against in-house-generated sequences of 55S proteins, all known mitochondrial proteins, and proteins in the Swiss-Prot database. Database searches were performed with cysteine carbamidomethylation as a fixed modification. The variable modifications were methionine oxidation (+16 Da) and phosphorylation (+80 Da) of Ser, Thr, and Tyr residues and loss of water (−18 Da) from Ser and Thr due to β-elimination during loss of the phosphate moiety. Up to two missed cleavages were allowed for the protease of choice. Peptide mass tolerance and fragment mass tolerance were set to 3 and 2 Da, respectively. Tandem mass spectra that matched to phosphorylated peptides were manually evaluated at the raw data level with the consideration of overall data quality, signal-to-noise of matched peaks, and the presence of dominant peaks that did not match to any theoretical m/z value.
Site-directed mutagenesis and transient transfection
DAP3 and DAP3-His6 constructs were cloned into the mammalian vector pcDNA 3.1(+) using KpnI and XhoI as the restriction sites, and selected phosphorylation sites (Ser31, Ser185, Ser215, Thr216, Thr237, Ser251, Ser252) were mutated to alanine residues using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, Inc.). DNA from each construct was prepared using either the QIA Prep Spin Miniprep Kit (Qiagen, Inc.) or the QIAfilter Plasmid Midi Kit (Qiagen, Inc.), and further purification was obtained by phenol-chloroform extraction. HEK293T cells (human embryonic kidney cell line) were maintained in DMEM medium supplemented with 10% heat-inactivated FBS, penicillin G, and streptomycin and were cultured at 37°C in a humidified atmosphere containing 5% CO2 and 95% air. HEK293T cells were seeded at 4 × 105 in a six-well plate and transiently transfected with 2 μg of plasmid DNA using Lipofectamine 2000 (Invitrogen Corp.). Upon a 24-h incubation at 37°C, cells were collected, and lysed in buffer consisting of 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% NP-40, 0.5% SDS, and a protease and phosphatase inhibitor cocktail for mammalian cells (Sigma-Aldrich, Inc.). The protein concentration was determined by using the BCA Protein Assay Kit (Pierce Biochemicals, Inc.) for Western blot analysis.
Pulldown assay
HEK293T cells were seeded at 4 × 105 in a six-well plate and transiently transfected with 4 μg of plasmid DNA using Lipofectamine 2000 (Invitrogen Corp.). Upon a 48-h incubation at 37°C, the cells were collected, and lysed in a buffer consisting of 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 0.5% SDS, and protease and phosphatase inhibitor cocktails (Sigma-Aldrich, Inc.). The whole cell lysate was diluted to 0.125% SDS and incubated overnight at 4°C with 50 μL of the Ni-NTA beads (Qiagen, Inc.). The beads were collected by centrifugation and washed twice with a wash buffer containing 40 mM KC1, 20 mM MgCl2, 20 mM Tris-HCl (pH 7.6), 10% glycerol, 7 mM β-mercaptoethanol, 0.125% SDS, and 10 mM imidazole. The beads were eluted three times in elution buffer consisting of 40 mM KC1, 20 mM MgCl2, 20 mM Tris-HCl (pH 7.6), 10% glycerol, 7 mM β-mercaptoethanol, 250 mM imidazole, and protease and phosphatase inhibitor cocktails (Sigma-Aldrich, Inc.), followed by Western blot analysis.
Western blot analysis
Protein samples for either one-dimensional or 2D analysis were loaded on SDS-PAGE gels and transferred to a PVDF membrane. The blot was probed with a monoclonal DAP3 antibody at a 1:5000 dilution (Transduction Laboratories), a monoclonal Hsp60 antibody at a 1:4000 dilution (Transduction Laboratories), or a polyclonal cleaved PARP antibody at a 1:1000 dilution (Cell Signaling Technology, Inc.). The secondary antibody was ImmunoPure Antibody Goat Anti-Mouse IgG (Pierce Biochemicals, Inc.) at a 1:5000 dilution or Goat Anti-Rabbit IgG at a 1:1000 dilution followed by development with the SuperSignal West Pico Chemiluminescent Substrate (Pierce Biochemicals, Inc.) according to the protocol provided by the manufacturer. To determine the phosphorylation state of DAP3, blots were probed with phosphoserine antibody at a 1:20,000 dilution (Sigma-Aldrich, Inc.) and phosphothreonine antibody at a 1:10,000 dilution (Sigma-Aldrich, Inc.). The secondary antibody was streptavidin HRP at a 1:30,000 dilution (Amersham Biosciences, Inc.). The membranes were developed by using the SuperSignal West Femto Max Sensitivity Substrate (Pierce Biochemicals, Inc.) according to the protocol provided by the manufacturer.
Cell proliferation assay
HEK293T cells were seeded at 1 × 104 in a 96-well plate and transiently transfected with 0.2 μg of plasmid DNA using Lipofectamine 2000 (Invitrogen Corp.). Upon a 24-h incubation at 37°C, 10 μL of the Cell Proliferation Reagent WST-1 (Roche Applied Science) was added to each well and incubated for 5 h at 37°C. The absorbance was read at 450 and 650 nm using a microplate reader. Results were expressed as mean ± SD and analyzed using the ANOVA's t-test. Values of P < 0.05 were considered statistically significant.
Acknowledgments
We thank Dr. Linda Spremulli for critical reading and review of the manuscript. We thank Huseyin Cimen, Seung-Kyu Lee, and George Y. Soung for technical assistance in preparation of mitochondrial ribosomes and in-gel digestion of protein bands from 2D and one-dimensional gels. This work was supported by the National Institutes of Health grants R01 GM071034-01A1 and R01 EB005197-01 EB005197-01 to E.C.K.
Footnotes
Reprint requests to: Emine C. Koc, Department of Biochemistry and Molecular Biology, Pennsylvania State University, 103 Althouse, University Park, PA 16802, USA; e-mail: eminekoc@psu.edu; fax: (814) 863-7024.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073185608.
References
- Berger, T., Brigl, M., Herrmann, J.M., Vielhauer, V., Luckow, B., Schlondorff, D., Kretzler, M. The apoptosis mediator mDAP-3 is a novel member of a conserved family of mitochondrial proteins. J. Cell Sci. 2000;113:3603–3612. doi: 10.1242/jcs.113.20.3603. [DOI] [PubMed] [Google Scholar]
- Blom, N., Gammeltoft, S., Brunak, S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Brodie, C., Blumberg, P.M. Regulation of cell apoptosis by protein kinase c δ . Apoptosis. 2003;8:19–27. doi: 10.1023/a:1021640817208. [DOI] [PubMed] [Google Scholar]
- Cahill, A., Baio, D., Cunningham, C. Isolation and characterization of rat liver mitochondrial ribosomes. Anal. Biochem. 1995;232:47–55. doi: 10.1006/abio.1995.9962. [DOI] [PubMed] [Google Scholar]
- Chen, R., Fearnley, I.M., Peak-Chew, S.Y., Walker, J.E. The phosphorylation of subunits of complex I from bovine heart mitochondria. J. Biol. Chem. 2004;279:26036–26045. doi: 10.1074/jbc.M402710200. [DOI] [PubMed] [Google Scholar]
- Chintharlapalli, S.R., Jasti, M., Malladi, S., Parsa, K.V., Ballestero, R.P., Gonzalez Garcia, M. BMRP Is a Bcl-2 binding protein that induces apoptosis. J. Cell. Biochem. 2005;94:611–626. doi: 10.1002/jcb.20292. [DOI] [PubMed] [Google Scholar]
- Denslow, N., Anders, J., O'Brien, T.W. Bovine mitochondrial ribosomes possess a high affinity binding site for guanine nucleotides. J. Biol. Chem. 1991;266:9586–9590. [PubMed] [Google Scholar]
- Henning, K.A. Stanford University; Palo Alto, CA: 1992. “The molecular genetics of human diseases with defective DNA damaging processing.”. Ph.D. thesis. [Google Scholar]
- Hopper, R.K., Carroll, S., Aponte, A.M., Johnson, D.T., French, S., Shen, R.F., Witzmann, F.A., Harris, R.A., Balaban, R.S. Mitochondrial matrix phosphoproteome: Effect of extra mitochondrial calcium. Biochemistry. 2006;45:2524–2536. doi: 10.1021/bi052475e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.R., Chae, H.J., Thomas, M., Miyazaki, T., Monosov, A., Monosov, E., Krajewska, M., Krajewski, S., Reed, J.C. Mammalian dap3 is an essential gene required for mitochondrial homeostasis in vivo and contributing to the extrinsic pathway for apoptosis. FASEB J. 2007;21:188–196. doi: 10.1096/fj.06-6283com. [DOI] [PubMed] [Google Scholar]
- Kissil, J.L., Deiss, L.P., Bayewitch, M., Raveh, T., Khaspekov, G., Kimchi, A. Isolation of DAP3, a novel mediator of interferon-γ-induced cell death. J. Biol. Chem. 1995;270:27932–27936. doi: 10.1074/jbc.270.46.27932. [DOI] [PubMed] [Google Scholar]
- Kissil, J.L., Cohen, O., Raveh, T., Kimchi, A. Structure–function analysis of an evolutionary conserved protein, DAP3, which mediates TNF-α- and Fas-induced cell death. EMBO J. 1999;18:353–362. doi: 10.1093/emboj/18.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc, E.C., Burkhart, W., Blackburn, K., Moseley, A., Spremulli, L.L. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J. Biol. Chem. 2001a;276:19363–19374. doi: 10.1074/jbc.M100727200. [DOI] [PubMed] [Google Scholar]
- Koc, E.C., Burkhart, W., Blackburn, K., Schlatzer, D.M., Moseley, A., Spremulli, L.L. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J. Biol. Chem. 2001b;276:43958–43969. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- Koc, E.C., Ranasinghe, A., Burkhart, W., Blackburn, K., Koc, H., Moseley, A., Spremulli, L.L. A new face on apoptosis: Death-associated protein 3 and PDCD9 are mitochondrial ribosomal proteins. FEBS Lett. 2001c;492:166–170. doi: 10.1016/s0014-5793(01)02250-5. [DOI] [PubMed] [Google Scholar]
- Levshenkova, E.V., Ukraintsev, K.E., Orlova, V.V., Alibaeva, R.A., Kovriga, I.E., Zhugdernamzhilyn, O., Frolova, E.I. The structure and specific features of the cDNA expression of the human gene MRPL37 . Bioorg. Khim. 2004;30:499–506. doi: 10.1023/b:rubi.0000043788.86955.ea. [DOI] [PubMed] [Google Scholar]
- Liu, J., Chen, J., Dai, Q., Lee, R.M. Phospholipid scramblase 3 is the mitochondrial target of protein kinase C δ-induced apoptosis. Cancer Res. 2003;63:1153–1156. [PubMed] [Google Scholar]
- Mariani, L., Beaudry, C., McDonough, W.S., Hoelzinger, D.B., Kaczmarek, E., Ponce, F., Coons, S.W., Giese, A., Seiler, R.W., Berens, M.E. Death-associated protein 3 (Dap-3) is overexpressed in invasive glioblastoma cells in vivo and in glioma cell lines with induced motility phenotype in vitro. Clin. Cancer Res. 2001;7:2480–2489. [PubMed] [Google Scholar]
- Matthews, D.E., Hessler, R.A., Denslow, N.D., Edwards, J.S., O'Brien, T.W. Protein composition of the bovine mitochondrial ribosome. J. Biol. Chem. 1982;257:8788–8794. [PubMed] [Google Scholar]
- Miyazaki, T., Reed, J.C. A GTP-binding adapter protein couples TRAIL receptors to apoptosis-inducing proteins. Nat. Immunol. 2001;2:493–500. doi: 10.1038/88684. [DOI] [PubMed] [Google Scholar]
- Miyazaki, T., Shen, M., Fujikura, D., Tosa, N., Kon, S., Uede, T., Reed, J.C. Functional role of death-associated protein 3 (DAP3) in anoikis. J. Biol. Chem. 2004;279:44667–44672. doi: 10.1074/jbc.M408101200. [DOI] [PubMed] [Google Scholar]
- Mukamel, Z., Kimchi, A. Death-associated protein 3 localizes to the mitochondria and is involved in the process of mitochondrial fragmentation during cell death. J. Biol. Chem. 2004;279:36732–36738. doi: 10.1074/jbc.M400041200. [DOI] [PubMed] [Google Scholar]
- O'Brien, T.W., Fiesler, S., Denslow, N.D., Thiede, B., Wittmann-Liebold, B., Mouge, E., Sylvester, J.E., Graack, H.R. Mammalian mitochondrial ribosomal proteins (2). Amino acid sequencing, characterization, and identification of corresponding gene sequences. J. Biol. Chem. 1999;274:36043–36051. doi: 10.1074/jbc.274.51.36043. [DOI] [PubMed] [Google Scholar]
- O'Brien, T.W., Liu, J., Sylvester, J., Mourgey, E.B., Fischel-Ghodsian, N., Thiede, B., Wittmann-Liebold, B., Graack, H.R. Mammalian mitochondrial ribosomal proteins (4). Amino acid sequencing, characterization, and identification of corresponding gene sequences. J. Biol. Chem. 2000;275:18153–18159. doi: 10.1074/jbc.M909762199. [DOI] [PubMed] [Google Scholar]
- O'Brien, T.W., O'Brien, B.J., Norman, R.A. Nuclear MRP genes and mitochondrial disease. Gene. 2005;354:147–151. doi: 10.1016/j.gene.2005.03.026. [DOI] [PubMed] [Google Scholar]
- Pai, E.F., Krengel, U., Petsko, G.A., Goody, R.S., Kabsch, W., Wittinghofer, A. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: Implications for the mechanism of GTP hydrolysis. EMBO J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi, M., Brunati, A.M., Toninello, A. Tyrosine phosphorylation in mitochondria: A new frontier in mitochondrial signaling. Free Radic. Biol. Med. 2005;38:1267–1277. doi: 10.1016/j.freeradbiomed.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Saveanu, C., Fromont-Racine, M., Harington, A., Ricard, F., Namane, A., Jacquier, A. Identification of 12 new yeast mitochondrial ribosomal proteins including six that have no prokaryotic homologues. J. Biol. Chem. 2001;276:15861–15867. doi: 10.1074/jbc.M010864200. [DOI] [PubMed] [Google Scholar]
- Schulenberg, B., Aggeler, R., Beechem, J.M., Capaldi, R.A., Patton, W.F. Analysis of steady-state protein phosphorylation in mitochondria using a novel fluorescent phosphosensor dye. J. Biol. Chem. 2003;278:27251–27255. doi: 10.1074/jbc.C300189200. [DOI] [PubMed] [Google Scholar]
- Sharma, M.R., Koc, E.C., Datta, P.P., Booth, T.M., Spremulli, L.L., Agrawal, R.K. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Terasaki, M., Takemoto-Hori, C., Hanada, T., Ueda, T., Wada, A., Watanabe, K. Structural compensation for the deficit of rRNA with proteins in the mammalian mitochondrial ribosome. Systematic analysis of protein components of the large ribosomal subunit from mammalian mitochondria. J. Biol. Chem. 2001a;276:21724–21736. doi: 10.1074/jbc.M100432200. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Terasaki, M., Takemoto-Hori, C., Hanada, T., Ueda, T., Wada, A., Watanabe, K. Proteomic analysis of the mammalian mitochondrial ribosome. Identification of protein components in the 28 S small subunit. J. Biol. Chem. 2001b;276:33181–33195. doi: 10.1074/jbc.M103236200. [DOI] [PubMed] [Google Scholar]
- Takeda, S., Iwai, A., Nakashima, M., Fujikura, D., Chiba, S., Li, H.M., Uehara, J., Kawaguchi, S., Kaya, M., Nagoya, S., et al. LKB1 is crucial for TRAIL mediated apoptosis induction in osteosarcoma. Anticancer Res. 2007;27:761–768. [PubMed] [Google Scholar]
- Taylor, S.W., Fahy, E., Zhang, B., Glenn, G.M., Warnock, D.E., Wiley, S., Murphy, A.N., Gaucher, S.P., Capaldi, R.A., Gibson, B.W., et al. Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 2003;21:281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]