Abstract
Ole e 9 is an olive pollen allergen belonging to group 2 of pathogenesis-related proteins. The protein is composed of two immunological independent domains: an N-terminal domain (NtD) with 1,3-β-glucanase activity, and a C-terminal domain (CtD) that binds 1,3-β-glucans. We have determined the three-dimensional structure of CtD-Ole e 9 (101 amino acids), which consists of two parallel α-helices forming an angle of ∼55°, a small antiparallel β-sheet with two short strands, and a 3–10 helix turn, all connected by long coil segments, resembling a novel type of folding among allergens. Two regions surrounded by aromatic residues (F49, Y60, F96, Y91 and Y31, H68, Y65, F78) have been localized on the protein surface, and a role for sugar binding is suggested. The epitope mapping of CtD-Ole e 9 shows that B-cell epitopes are mainly located on loops, although some of them are contained in secondary structural elements. Interestingly, the IgG and IgE epitopes are contiguous or overlapped, rather than coincident. The three-dimensional structure of CtD-Ole e 9 might help to understand the underlying mechanism of its biochemical function and to determine possible structure–allergenicity relationships.
Keywords: Ole e 9, allergy, olive pollen, glucanase
Olive tree (Olea europaea) pollen is a main cause of seasonal respiratory allergy in Mediterranean countries and other areas (Liccardi et al. 1996). To date, 10 allergens from olive pollen, Ole e 1–Ole e 10, have been characterized (Rodríguez et al. 2002; Barral et al. 2004), but only the three-dimensional (3D) structure of Ole e 6 has been determined (Treviño et al. 2004). Among them, Ole e 9 is important because it has been described as a potential cause of adverse reactions in specific immunotherapy (SIT) (Duffort et al. 2006). Ole e 9 is composed of two structurally and immunologically independent domains: N- and C-terminal domains (NtD and CtD, 36 kDa and 10 kDa, respectively), which can be separately produced by recombinant DNA technology (Palomares et al. 2003, 2005). The use of these recombinant domains has been proven to be effective for the in vitro identification of Ole e 9-allergic patients (Palomares et al. 2006).
Herein, we have solved the high-resolution 3D structure in solution of CtD-Ole e 9 by NMR methods. The comparison of the structure obtained with those in the RCSB Protein Data Bank (Berman et al. 2000) showed a novel type of folding between allergenic proteins. We have also mapped the putative IgG and IgE epitopes using peptides and localized them in the 3D structure, showing that they are located mainly in coil-structured and solvent-accessible regions. In some cases, these epitopes include secondary structure regions, which could indicate that the peptides are able to mimic the conformational disposition in the folded domain. These results might enable the design of hypoallergenic peptides or derivates to be used in SIT protocols at a future time. Also, they might help to elucidate the basis of the allergenicity and why some highly homologous proteins are allergenic while others are not.
Results and Discussion
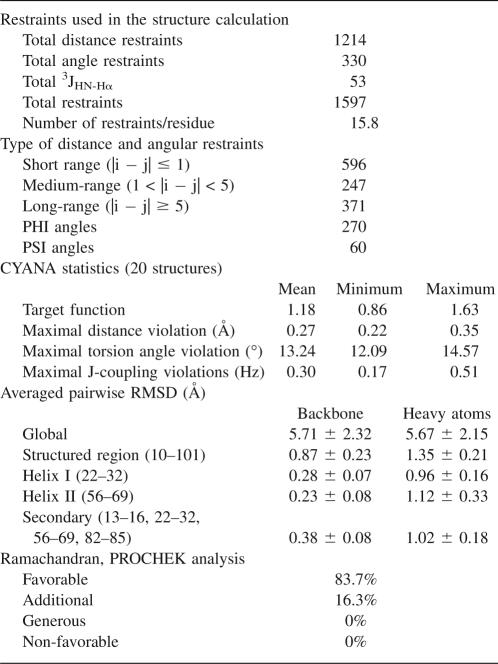
NMR methods were used to determine the solution structure of CtD-Ole e 9. The structural statistics for the 20 lowest energy structures are summarized in Table 1. The coordinates of the 20 conformers (Fig. 1A) have been deposited in the RSCB Protein Data Bank under the accession number 2JON.
Table 1.
NMR structural calculations summary
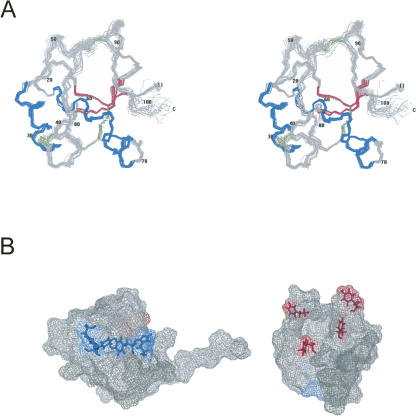
Figure 1.
Details of the solution structure of CtD-Ole e 9. (A) Stereoscopic view of the superimposition of the backbone atoms corresponding to the structured region (11–101) of the 20 lowest energy structures. (Blue) α-Helices, (magenta) β-strands, (green) disulfide bridges. (B) Two views of the protein surface showing two regions surrounded by aromatic amino acid residues, F96, Y91, F49, and Y60 (magenta) and Y31, Y65, H68, and F78 (blue).
Disulfide pairings were found by searching for Hβ to Hβ and Hα to Hβ NOEs between different cysteines. At least one intercysteine NOE could be found for each one of the three pairs (C14–C76, C33–C39, and C48–C94). The long-range NOEs between residues of the vicinity of the cysteines confirm the cysteine pairing defined here, and no NMR evidence for other pairings was found. The pattern of disulfide bonds is different from the one defined in the literature for CtD-Ole e 9 (Palomares et al. 2003). This difference can be a consequence of the aging of the sample, as it has been described in vitro and in vivo for other proteins, like albumin (Wallevik 1976). We have tested the sample used for structure determination by ELISA using the same sera that had been previously employed to characterize CtD-Ole e 9 (Palomares et al. 2003). Both proteins showed equivalent IgE recognition, indicating that the two CtD-Ole e 9 forms are biologically and medically relevant.
The global fold of CtD-Ole e 9 consists of two parallel α-helices spanning residues 22–32 and 56–69 forming an angle of ∼55°, and a small antiparallel β-sheet composed of two short strands, 13–16 and 82–85. Furthermore, a 3–10 helix turn (73–75) can be found in the 3D structure. Each element is connected to the others by long coil regions. The N-terminal region (1–10), corresponding with the peptide linker between both domains of the complete protein, is highly disordered. We have compared CtD-Ole e 9 with other published protein structures using the DALI server (Holm and Sander 1995). The comparison reveals a low degree of homology (Z-score <2.0) with two protein fragments holding α-helices with similar relative orientation as in CtD-Ole e 9. One of them, a DNA binding domain of Arabidopsis thaliana ethylene-insensitive3-like3 (Yamasaki et al. 2005), shows a reasonable superposition with CtD-Ole e 9.
The majority of pollen allergens belongs to a relatively small number of protein families (Radauer and Breiteneder 2006), and to date the Protein Data Bank contains only about 40 3D structures of allergens (Chapman et al. 2007). CtD-Ole e 9 shows no structural similarities to any of them and defines a novel type of folding among allergenic proteins. At the present, it is a generalized idea that belonging to the same homology group or family provides the molecular basis for the existence of allergic cross-response (Chapman et al. 2007). Despite that, not all the members of a family are allergenic, nor is the response to them identical between allergic individuals. In addition, it seems that belonging to the same family is not enough to explain the phenomenon, and instead the determining factor is the similarity or difference in the surface and accessible areas of each allergen of the homology group (Breiteneder and Ebner 2001). In this context, the determination of the first structure of one group acquires an additional importance because it can be used as a base for the modeling of the rest of the members of the family and helps define which areas are differentially oriented or accessible and can be responsible for the putative cross-responses.
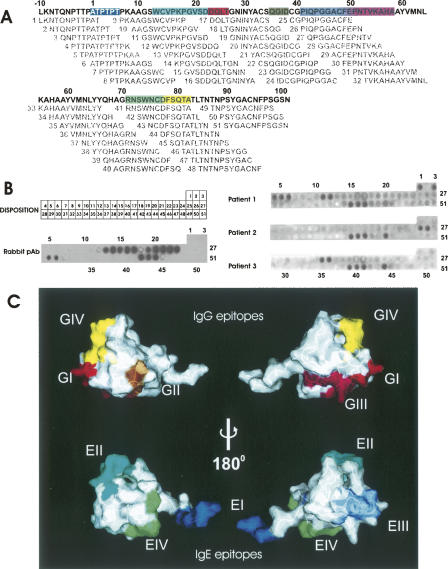
In order to define the epitopes recognized by both IgG and IgE, a set of overlapping synthetic peptides was used in a dot blot and immunostained with sera from eight allergic patients, as well as with a specific antiserum raised against CtD-Ole e 9 in rabbits. The set of synthetic dodecapeptides covers all the putative epitopes of the CtD-Ole e 9, including the possible ones formed by the peptide linker between both domains of Ole e 9. Four minimal IgG epitopes were detected using this method (Fig. 2), comprising residues 23–26 (epitope GI), 35–38 (epitope GII), 49–58 (epitope GIII), and 77–84 (epitope GIV). After locating these epitopes in the 3D structure, it is worth emphasizing that two of the epitopic regions are in large loops of the protein, which is expected for a nearly random structure in a peptide, whereas the other two are part of a secondary structure element: epitope GI is at the beginning of the first α-helix, and epitope GIV comprises part of the second β-strand and its preceding loop. The same method was carried out to determine IgE epitopes: residues 1–6 (epitope EI), 13–22 (epitope EII), 41–50 (epitope EIII), and 71–77 (epitope EIV). Similar to the IgG epitopes, two of the IgE epitopes are located in coil regions (epitopes EI and EIII), and the others comprise structured and unstructured regions: epitope EI spreads out over the first β-strand and a loop, and epitope EIV occupies the 3–10 turn and a loop. A comparison between IgG and IgE epitopes shows that they are located in the same regions of the molecule, but are shifted. Interestingly, the average surface of the IgG epitopes is smaller than the surface of IgE epitopes, and, in fact, this average surface of IgG epitopes is smaller than the 600 Å2 considered as the minimum surface necessary for an antigenic recognition (Davies et al. 1990). This might indicate a suboptimal recognition by the IgGs that would point to the idea that two or more of these mapped epitopes are actually defining one conformational epitope. In fact, regions GI and GIII occupy spatially adjacent positions, and GIII and GIV are relatively close, too.
Figure 2.
IgE and IgG epitopes of CtD-Ole e 9. (A) Location of the synthetic peptides used for the epitope mapping, in the CtD-Ole e 9 sequence. The different epitopes are shown in different colors: (blue) EI, (cyan) EII, (magenta) EIII, (green) EIV, (red) GI, (brown) GII, (pink) GIII, (yellow) GIV. (B) Disposition of the synthetic peptides in the cellulose membrane and Western blot using different sera. Results for rabbit serum and for sera from three representative patients are presented. (C) Location of the epitopes in the CtD-Ole e 9 protein surface; same color code used in A.
Ole e 9 is a protein with 1,3-β-glucanase activity, and as the catalytic site is located at the N-terminal domain (Huecas et al. 2001), a regulatory role was suggested for CtD-Ole e 9 (Palomares et al. 2005). However, some data point to a certain relationship with sugar recognition. In fact, it has been demonstrated that this domain displays the capability of interacting with the 1,3-β-polysaccharide laminarin (Rodríguez et al. 2007). Regarding this subject, two cavities surrounded by aromatic residues are located in the 3D structure of CtD-Ole e 9 (Fig. 1B), and they could be involved in the contact with the carbohydrate as reported for sugar-binding proteins (Quiocho 1986). One is a groove limited by two handles close to the aromatic rings of F96, Y91, F49, and Y60. Another is the surface upholstered by Y31, Y65, H68, and F78. Interestingly, six out of the eight aromatic residues forming these two clusters in CtD-Ole e 9 are conserved in the amino acid sequence of its homologous allergen Ole e 10, another olive pollen allergen, that presents glycan-binding activity. Moreover, other vegetal proteins containing a homologous domain (Barral et al. 2004) also conserve these positions.
In conclusion, we have solved the 3D structure of CtD-Ole e 9, an immunological independent domain of Ole e 9, and we have located the epitopes defined by peptide mapping in the structure. We can deduce that the peptides are, at least partially, capable of mimicking regions of the protein containing some secondary structure elements, even though a complete mapping of the conformational epitopes will require molecular biology techniques to mutate some residues on the basis of the accessibilities and 3D disposition. These results may lead to the design of more effective and safer tools for allergy diagnosis and therapy.
Materials and Methods
CtD-Ole e 9 (comprising Ala334–Asn434 of Ole e 9) was produced in Pichia pastoris as previously described (Palomares et al. 2003). In the case of 15N- or 15N-13C tagged proteins, the same slight modifications described previously (Treviño et al. 2004) were introduced. All samples were analyzed by amino acid analysis, N-terminal end sequencing, and mass spectroscopy. Samples were prepared for NMR experiments at 0.7 mM in 90% H2O/10% D2O or in D2O containing sodium-4,4-dimethyl-4-silapentane-1-sulfonate (DSS) at pH 6.0. NMR spectra were carried out at 10°C or 25°C on a Bruker AV 600 and AV 800 spectrometer. Assignments were performed using standard methods in two steps. First, the automatic backbone assignment was performed with a home-written version of MARS (Jung and Zweckstetter 2004) using 3D HNCA, HN(CO)CA, CBCANH, CBCA(CO)NH, and HNHA experiments. Then, side-chain assignments were extended with the help of 2D NOESY, 2D TOCSY, 3D HCCH-TOCSY, and 13C-edited NOESY data. The final assignments of the 1H, 15N, and 13C resonances have been deposited in the BioMagResBank (Seavey et al. 1991) under accession number 7187 and have been published (Castrillo et al. 2006).
After chemical-shift assignment, NOE cross-peaks assignment and structure calculation were performed by CYANA/CANDID (Herrmann et al. 2002). Assignments provided 1214 upper-limit distance constraints. Additionally, standard upper and lower limits for each of the three disulfide bonds were introduced during the rounds of calculations. The distance constraints were supplemented with angle restraints derived from the cross-correlated dipole–dipole relaxation between 1HN-15N and 1HN-1Hα (Crowley et al. 2000), 3JNHHA coupling constants, and an analysis of 13C shifts using the program PREDITOR (Berjanskii et al. 2006). No stereospecific assignments were introduced initially. In the final steps, 48 pairs of stereospecific limits were introduced by CYANA for the calculations. PROCHECK-NMR version 3.4.4 (Laskowski et al. 1996) was used to analyze the quality of the refined structures, and MOLMOL (Koradi et al. 1996) and PyMOL (DeLano Scientific) were used to visualize them, calculate accessibilities, and prepare the figures of the molecules.
Polyclonal antiserum against CtD-Ole e 9 was obtained by immunizing New Zealand white rabbits as described (Huecas et al. 1999). Sera were obtained from eight patients that fulfilled the following criteria: (1) seasonal rhinitis and/or bronchial asthma from late April to June; (2) positive skin prick test to O. europaea pollen extract (ALK), and specific IgE values >0.7 kU/L (determined by CAP System of Pharmacia Diagnostics); and (3) no previous O. europaea immunotherapy. Furthermore, all sera were reactive to Ole e 9.
Overlapping dodecapeptides corresponding to the complete sequence of CtD-Ole e 9 plus the 10 residues previous to the domain were synthesized on an amino-cellulose membrane and tested by immunoblotting against the sera and polyclonal antibodies. The equilibrated membrane was blocked with 2% milk during 3 h at room temperature. It was then incubated for 2 h at ambient temperature in the same buffer but containing individual sera from allergic patients (dilution 1:5), polyclonal antibodies from rabbit (dilution 1:50,000), or monoclonal antibodies from mouse (dilution 1:5000). After extensive washing, the membrane was incubated 1 h with peroxidase-conjugated antibodies: goat anti-rabbit IgG for polyclonal antibodies or goat anti-mouse IgG for monoclonal antibodies. For detection of human IgE, membranes were incubated with a mouse anti-human IgE monoclonal antibody (dilution 1:5000) and then with the goat anti-mouse IgG. After extensive washing, the membrane was developed with ECL (Amersham).
Acknowledgments
This paper was supported by projects GEN2003-20642-C09-03, SAF02-02711, SAF05-01847, and BFU2005-01855/BMC from the Spanish Ministerio de Educación y Ciencia. We thank F. Roncal (Centro Nacional de Biotecnología, Madrid) for performing peptide synthesis.
Footnotes
Reprint requests to: Marta Bruix, Departamento de Espectroscopía y Estructura Molecular, Instituto de Química Física “Rocasolano,” Serrano 119, 28006 Madrid, Spain; e-mail: mbruix@iqfr.csic.es; fax: 34-91-564-24-31.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073230008.
References
- Barral, P., Batanero, E., Palomares, O., Quiralte, J., Villalba, M., Rodríguez, R. A major allergen from pollen defines a novel family of plant proteins and shows intra- and interspecies cross-reactivity. J. Immunol. 2004;172:3644–3651. doi: 10.4049/jimmunol.172.6.3644. [DOI] [PubMed] [Google Scholar]
- Berjanskii, M.V., Neal, S., Wishart, D.S. PREDITOR: A Web server for predicting protein torsion angle restraints. Nucleic Acids Res. 2006;34:W63–W69. doi: 10.1093/nar/gkl341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H.M., Bhat, T.N., Bourne, P.E., Feng, Z., Gilliland, G., Weissig, H., Westbrook, J. The Protein Data Bank and the challenge of structural genomics. Nat. Struct. Biol. 2000;7 Suppl:957–959. doi: 10.1038/80734. [DOI] [PubMed] [Google Scholar]
- Breiteneder, H., Ebner, C. Atopic allergens of plant foods. Curr. Opin. Allergy Clin. Immunol. 2001;1:261–267. doi: 10.1097/01.all.0000011024.76416.01. [DOI] [PubMed] [Google Scholar]
- Castrillo, I., Treviño, M.A., Palomares, O., Rico, M., Santoro, J., Bruix, M. NMR assignment of the C-terminal domain of Ole e 9, a major allergen from the olive tree pollen. J. Biomol. NMR. 2006;36 Suppl 5:67. doi: 10.1007/s10858-006-9053-6. [DOI] [PubMed] [Google Scholar]
- Chapman, M.D., Pomés, A., Breiteneder, H., Ferreira, F. Nomenclature and structural biology of allergens. J. Allergy Clin. Immunol. 2007;119:414–420. doi: 10.1016/j.jaci.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Crowley, P., Ubbink, M., Otting, G. φ angle restraints in protein backbones from dipole–dipole cross-correlation between 1HN-15N and 1HN-1Hα vectors. J. Am. Chem. Soc. 2000;122:2968–2969. [Google Scholar]
- Davies, D.R., Padlan, E.A., Sheriff, S. Antibody–antigen complexes. Annu. Rev. Biochem. 1990;59:439–473. doi: 10.1146/annurev.bi.59.070190.002255. [DOI] [PubMed] [Google Scholar]
- Duffort, O., Palomares, O., Lombardero, M., Villalba, M., Barber, D., Rodríguez, R., Polo, F. Variability of Ole e 9 allergen in olive pollen extracts: Relevance of minor allergens in immunotherapy treatments. Int. Arch. Allergy Immunol. 2006;140:131–138. doi: 10.1159/000092532. [DOI] [PubMed] [Google Scholar]
- Herrmann, T., Güntert, P., Wüthrich, K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- Holm, L., Sander, C. DALI: A network tool for protein structure comparison. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- Huecas, S., Villalba, M., González, E., Martínez-Ruiz, A., Rodríguez, R. Production and detailed characterization of biologically active olive pollen allergen Ole e 1 secreted by the yeast Pichia pastoris . Eur. J. Biochem. 1999;261:539–546. doi: 10.1046/j.1432-1327.1999.00307.x. [DOI] [PubMed] [Google Scholar]
- Huecas, S., Villalba, M., Rodríguez, R. Ole e 9, a major olive pollen allergen is a 1,3-β-glucanase. Isolation, characterization, amino acid sequence, and tissue specificity. J. Biol. Chem. 2001;276:27959–27966. doi: 10.1074/jbc.M103041200. [DOI] [PubMed] [Google Scholar]
- Jung, Y.S., Zweckstetter, M. Mars: Robust automatic backbone assignment of proteins. J. Biomol. NMR. 2004;30:11–23. doi: 10.1023/B:JNMR.0000042954.99056.ad. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., Wüthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:29–32. 51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., Rullmannn, J.A., MacArthur, M.W., Kaptein, R., Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Liccardi, G., D'Amato, M., D'Amato, G. Oleaceae pollinosis: A review. Int. Arch. Allergy Immunol. 1996;111:210–217. doi: 10.1159/000237370. [DOI] [PubMed] [Google Scholar]
- Palomares, O., Villalba, M., Rodríguez, R. The C-terminal segment of the 1,3-β-glucanase Ole e 9 from olive (Olea europaea) pollen is an independent domain with allergenic activity: Expression in Pichia pastoris and characterization. Biochem. J. 2003;369:593–601. doi: 10.1042/BJ20020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomares, O., Villalba, M., Quiralte, J., Polo, F., Rodríguez, R. 1,3-β-glucanases as candidates in latex–pollen–vegetable food cross-reactivity. Clin. Exp. Allergy. 2005;35:345–351. doi: 10.1111/j.1365-2222.2004.02186.x. [DOI] [PubMed] [Google Scholar]
- Palomares, O., Batanero, E., Cañamero, M., Villalba, M., Rodríguez, R. Prophylactic intranasal treatment with fragments of 1,3-β-glucanase olive pollen allergen prevents airway inflammation in a murine model of type I allergy. Int. Arch. Allergy Immunol. 2006;139:175–180. doi: 10.1159/000091162. [DOI] [PubMed] [Google Scholar]
- Quiocho, F.A. Carbohydrate-binding proteins: Tertiary structures and protein–sugar interactions. Annu. Rev. Biochem. 1986;55:287–315. doi: 10.1146/annurev.bi.55.070186.001443. [DOI] [PubMed] [Google Scholar]
- Radauer, C., Breiteneder, H. Pollen allergens are restricted to few protein families and show distinct patterns of species distribution. J. Allergy Clin. Immunol. 2006;117:141–147. doi: 10.1016/j.jaci.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Rodríguez, R., Villalba, M., Batanero, E., González, E.M., Monsalve, R.I., Huecas, S., Tejera, M.L., Ledesma, A. Allergenic diversity of the olive pollen. Allergy. 2002;S57:6–16. doi: 10.1034/j.1398-9995.2002.057s71006.x. [DOI] [PubMed] [Google Scholar]
- Rodríguez, R., Villalba, M., Batanero, E., Palomares, O., Salamanca, G. Emerging pollen allergens. Biomed. Pharmacother. 2007;61:1–7. doi: 10.1016/j.biopha.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Seavey, B.R., Farr, E.A., Westler, W.M., Markley, J.L. A relational database for sequence-specific protein NMR data. J. Biomol. NMR. 1991;1:217–236. doi: 10.1007/BF01875516. [DOI] [PubMed] [Google Scholar]
- Treviño, M.A., García-Mayoral, M.F., Barral, P., Villalba, M., Santoro, J., Rico, M., Rodríguez, R., Bruix, M. NMR solution structure of Ole e 6, a major allergen from olive tree pollen. J. Biol. Chem. 2004;279:39035–39041. doi: 10.1074/jbc.M406045200. [DOI] [PubMed] [Google Scholar]
- Wallevik, K. SS-interchanged and oxidized isomers of bovine serum albumin separated by isoelectric focusing. Biochim. Biophys. Acta. 1976;420:42–56. doi: 10.1016/0005-2795(76)90343-3. [DOI] [PubMed] [Google Scholar]
- Yamasaki, K., Kigawa, T., Inoue, M., Yamasaki, T., Yabuki, T., Aoki, M., Seki, E., Matsuda, T., Tomo, Y., Terada, T., et al. Solution structure of the major DNA-binding domain of Arabidopsis thaliana ethylene-insensitive3-like3. J. Mol. Biol. 2005;348:253–264. doi: 10.1016/j.jmb.2005.02.065. [DOI] [PubMed] [Google Scholar]