Abstract
To clarify the effects of humanizing a murine antibody on its specificity and affinity for its target, we examined the interaction between hen egg white lysozyme (HEL) and its antibody, HyHEL-10 variable domain fragment (Fv). We selected a human antibody framework sequence with high homology, grafted sequences of six complementarity-determining regions of murine HyHEL-10 onto the framework, and investigated the interactions between the mutant Fvs and HEL. Isothermal titration calorimetry indicated that the humanization led to 10-fold reduced affinity of the antibody for its target, due to an unfavorable entropy change. Two mutations together into the interface of the variable domains, however, led to complete recovery of antibody affinity and specificity for the target, due to reduction of the unfavorable entropy change. X-ray crystallography of the complex of humanized antibodies, including two mutants, with HEL demonstrated that the complexes had almost identical structures and also paratope and epitope residues were almost conserved, except for complementary association of variable domains. We conclude that adjustment of the interfacial structures of variable domains can contribute to the reversal of losses of affinity or specificity caused by humanization of murine antibodies, suggesting that appropriate association of variable domains is critical for humanization of murine antibodies without loss of function.
Keywords: antibody, protein–protein interaction, humanization, CDR grafting, X-ray crystallography, thermodynamics
Extensive studies from various viewpoints have revealed that complementarity-determining regions (CDRs) in variable domains of antibodies play a critical role in antigen specificity and affinity, with their shape complementarities to the antigens (Mariuzza and Pojak 1993; Braden and Poljak 1995; Davies and Cohen 1996; Padlan 1996). Therefore, several approaches to antibody engineering based on CDRs have been reported (Jirholt et al. 1998; Soderlind et al. 2000; Ewert et al. 2004; Nishibori et al. 2006). In particular, to reduce immune response against murine antibodies in human hosts, transplantation of a set of CDRs from murine antibodies to appropriate scaffolds of some human antibodies has been attempted, so-called “antibody humanization” or “CDR grafting” (Verhoeyen et al. 1988), and some humanized antibodies have been used in clinical trials and, indeed, have been utilized for internal or external medicine (Stochwin and Holmes 2003; Stern and Herrmann 2005).
Some studies have indicated, however, that grafting of the six CDRs of murine antibodies onto appropriate human frameworks often resulted in reduced affinity or specificity for the target antigen (Jones et al. 1986; Riechmann et al. 1988; Verhoeyen et al. 1988). Although the CDR-grafting method is widely used for humanizing murine antibodies, there are few general strategies for recovery of the affinity of humanized antibodies for their targets (Queen et al. 1989; Co et al. 1991, 1992; Foote and Winter 1992; Ohtomo et al. 1995), requiring several trial-and-error approaches.
To address how antibodies can recognize their target antigens with high specificity and affinity, both structural and thermodynamic information is required (Sturtevant 1994). X-ray crystal analysis can clarify structural aspects of the complementarity of the interactions (Braden and Poljak 1995; Davies and Cohen 1996; Padlan 1996), and isothermal titration calorimetry can provide useful information for quantitative assessment of the energetic contribution of residues to the interaction (Lemmon and Ladbury 1994; Schwarz et al. 1995; Torigoe et al. 1995; Xavier et al. 1997; Furukawa et al. 1999). Thus, the combination of these two approaches should be especially valuable in providing insight into structural and thermodynamic consequences of grafting CDRs onto selected framework regions. However, only a few studies have precisely analyzed structural changes due to humanization (Holmes et al. 1998, 2001), and there has been no report of thermodynamic consequences of humanizing a murine antibody.
We have focused on the interaction between hen egg white lysozyme (HEL) and its monoclonal murine antibody, HyHEL-10 (Kam-Morgan et al. 1993; Ueda et al. 1993; Tsumoto et al. 1994a; Pons et al. 1999), whose structural features have been analyzed by X-ray crystallography in Fab–HEL (Padlan et al. 1989) and Fv–HEL (Kondo et al. 1999) complexes. A bacterial expression system of the HyHEL-10 Fv fragment, which consists of the associated variable domains of an antibody, has been established (Ueda et al. 1996; Merk et al. 1999), and the corresponding Fv–HEL interactions have been investigated by using mutant Fv fragments (Tsumoto et al. 1994a, 1995, 1996). The combination of thermodynamic data with structural results should be a powerful tool for the precise description of the mutant Fv–HEL interactions (Shiroishi et al. 2001; Yokota et al. 2003).
Here, using isothermal titration calorimetry and X-ray crystallography, we report the effects of humanization and mutations into two interfacial residues on the high specificity and affinity of murine HyHEL-10 (mHyHEL-10) Fv. Mutations at two residues in the VH–VL interface of humanized HyHEL-10 (hHyHEL-10) led to almost complete recovery to the murine type from thermodynamic and structural viewpoints. On the basis of our thermodynamic and structural results, we discuss what causes the reduction of affinity via humanization and how the function of humanized antibody can be recovered.
Results
Humanization of HyHEL-10
To humanize the HyHEL-10 variable domains, some frameworks for grafting the murine CDRs were screened with a homology-based approach. Finally, human VH region (GenBank accession number AAA52875) and VL region (PIR accession number S40362) were selected on the basis of a homology modeling (Supplemental Fig. S1). The amino acid sequence of the humanized VH displayed 76% identity to that of the parental murine VH, while that of humanized VL displayed 78% identity to that of the parental murine VL.
Preparation of hHyHEL-10 Fv
The humanized HyHEL-10 (hHyHEL-10) Fv was expressed in Escherichia coli and purified from the culture supernatant. The hHyHEL-10 Fv were highly purified by affinity chromatography using HEL-Sepharose and gel filtration using a Sephacryl S-200 HR (>95%).
Thermodynamic analysis of the interaction between lysozyme and humanized Fv fragment
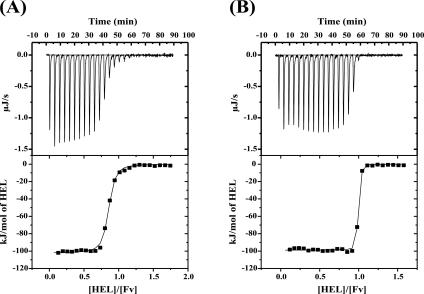
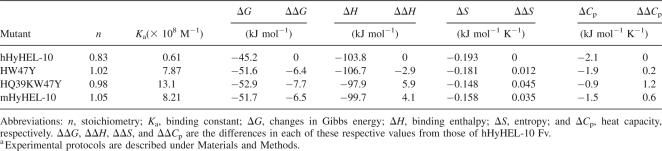
To investigate the interaction between HEL and hHyHEL-10 Fv, thermodynamic analysis was performed by ITC (Fig. 1). We carried out ITC at four different temperatures under otherwise identical conditions. Thermodynamic parameters (30°C and pH 7.2) calculated from the titration curves are summarized in Table 1, and the temperature dependence of enthalpy changes due to binding is shown in Figure 2.
Figure 1.
Titration calorimetry of the interaction between the HyHEL-10 Fv fragment and HEL. (A, B) (Top) Typical calorimetric titration of hHyHEL-10 mutant Fv fragment (5 μM) with 50 μM HEL at pH 7.2 and 30°C; (bottom) integration plot of the data calculated from the raw data. The solid line corresponds to the best-fit curve obtained by least-squares deconvolution. (A) hHyHEL-10; (B) HQ39KW47Y mutant.
Table 1.
Thermodynamic parameters of the interactions between Fv and HEL at 30°C and pH 7.2 in phosphate buffera
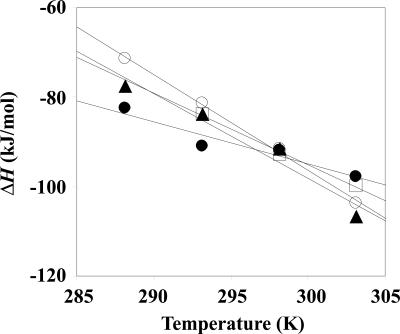
Figure 2.
Temperature dependence of the enthalpy change of the interaction between HyHEL-10 Fv and HEL: open circles, hHyHEL-10; solid triangles, HW47Y; solid circles, HQ39KW47Y; open squares, mHyHEL-10.
The binding constant (K a) of hHyHEL-10 Fv for HEL was ∼10-fold smaller than that of the parental murine Fv fragment for HEL. Although the binding enthalpy (ΔH) of the humanized Fv–HEL interaction was slightly increased by 4.1 kJ mol−1 at 30°C relative to the murine Fv–HEL interaction, the negative entropy change of interaction was increased, resulting in the reduced affinity for HEL. The heat capacity change (ΔC p) estimated from the temperature dependence of enthalpy change was −2.1 kJ mol−1 K−1.
Preparation of hHyHEL-10 mutant Fvs
Critical contribution of appropriate associations between VH and VL chains has been suggested in several antigen–antibody systems (Takahashi et al. 1994; Khalifa et al. 2000; Hugo et al. 2003); we have recently observed that changes in specificity of the antibody were correlated well with the changes in the relative orientations of VH, VL, and HEL (Kumagai et al. 2003). To recover the affinity reduced via humanization, we focused on some residues of VH at the VH–VL interface, i.e., Gln39 and Trp47 of hHyHEL-10; the corresponding residues in murine HyHEL-10 are Lys39 and Tyr47, respectively. We then constructed three mutants, HQ39K, HW47Y, and HQ39KW47Y. These mutants were expressed in E. coli and purified from the culture supernatant. Two mutants (HW47Y and HQ39KW47Y) were highly purified by affinity chromatography using HEL-Sepharose and gel filtration using a Sephacryl S-200 HR (>95%). However, the HQ39K mutant was separated into the VH and VL chains during gel filtration, and so this mutant was not used for the following analyses.
Thermodynamic analysis of the interaction between lysozyme and mutated humanized Fv fragments
To investigate the interaction between HEL and mutated hHyHEL-10 Fvs, thermodynamic analysis was performed by ITC. Thermodynamic parameters are summarized in Table 1.
HW47Y mutant Fv had almost identical affinity for lysozyme relative to the murine Fv, indicating that the increase in the negative entropy change was compensated by the increase in the negative enthalpy change. On the other hand, the binding constant of the HW47Y mutant for HEL was ∼10-fold higher than that of humanized Fv, which resulted from the favorable changes in both binding enthalpy and entropy. Furthermore, to alter the structure at the interface between VH and VL, the double mutant (i.e., HQ39KW47Y) was prepared and characterized. The affinity constant of the HQ39KW47Y double-mutant Fv was slightly larger than that of parental murine and the HW47Y mutant Fvs. HQ39KW47Y mutant behaved differently than HW47Y mutant: The decrease in the negative enthalpy change for HQ39KW47Y mutant was compensated for by the decrease in the negative entropy change, resulting in high affinity for HEL. The heat capacity changes for the HW47Y and HQ39KW47Y mutant were estimated to be −1.9 and −0.9 kJ mol−1 K−1, respectively.
Crystal structures of mutant Fv–HEL complexes
To precisely describe the mutant Fv–HEL interactions from a structural viewpoint, we determined the crystal structures of the three mutant–HEL complexes (Fig. 3). Crystallographic data are summarized in Supplemental Table S2. The maximum resolution of the X-ray data used in the refinements ranged from 1.9 to 2.4 Å, and the R factors of the refined structures were 0.214–0.221.
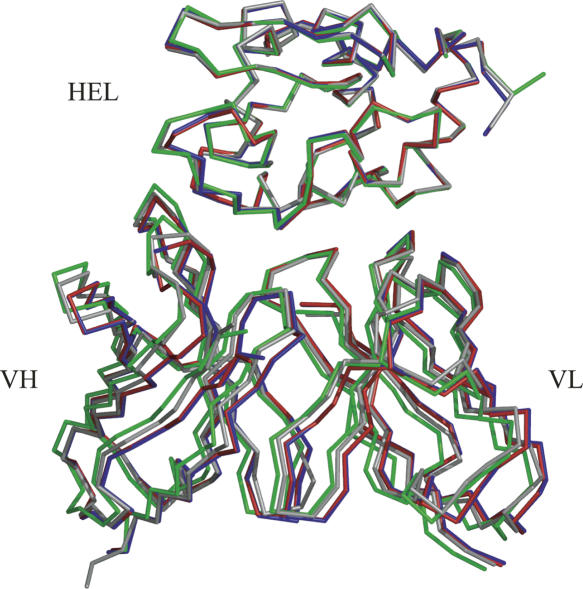
Figure 3.
Overall structure of the hHyHEL-10 Fv–HEL and mutant–HEL complexes. The structure of the three humanized Fv–HEL complexes, whose Cα coordinates of HEL are superimposed on the Cα coordinates of HEL complexed with mHyHEL-10, is superposed on the structure of Fv (gray). Red, hHyHEL-10 Fv–HEL complex; blue, HW47Y Fv–HEL complex; green, HQ39KW47Y Fv–HEL complex. Generated with PyMOL (DeLano Scientific).
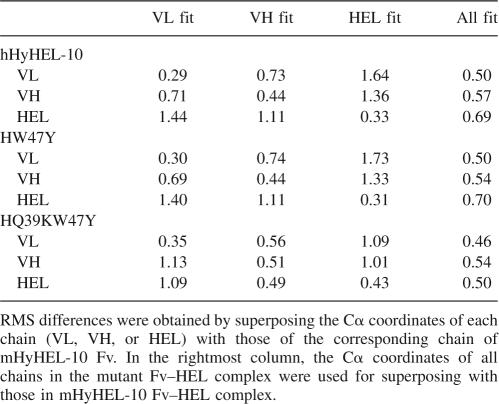
The mutant Fv–HEL complexes were superimposed onto the wild-type Fv–HEL complex (Kondo et al. 1999). The root-mean-square deviations (RMSDs) between the Cα atoms of the mutant Fv structures and those of the wild-type Fv structure are summarized in Table 2. The overall structures of the HyHEL-10 mutant Fv–HEL complexes are similar to that of the HyHEL-10 wild-type Fv–HEL complex (Table 2, column “All fit”).
Table 2.
RMS differences in the Cα atom of each chain (Å)
No major changes in the relative orientations of VL, VH, and HEL were observed in the hHyHEL-10 Fv–HEL and mutant Fv–HEL complexes (Table 2, column “All fit”). However, the RMSDs of VL and VH chains in the hHyHEL-10 complex were 1.64 and 1.36 Å, respectively, when HEL of the hHyHEL-10 Fv–HEL complex was superposed on that of the mHyHEL-10 complex. When each chain of the HW47Y mutant complex was superimposed onto the corresponding chain of the mHyHEL-10–HEL complex, the RMSDs were almost the same as in the case of the hHyHEL-10 complex. On the other hand, the RMSDs of the VL and VH chains in the HQ39KW47Y mutant were decreased (1.09 and 1.01 Å, respectively), indicating that the relative orientations of VL, VH, and HEL were altered due to the double mutation.
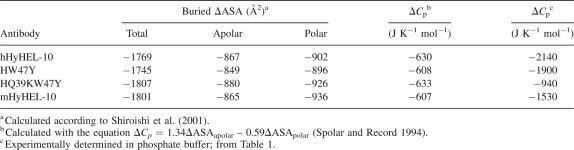
The calculated contact area of the mHyHEL-10 Fv–HEL complexes was 1801 Å2 (Kondo et al. 1999). Those of hHyHEL-10 and mutant complexes were 1769, 1745, and 1807 Å2 for the hHyHEL-10, HW47Y mutant, and HQ39KW47Y mutant, respectively (Table 3).
Table 3.
Heat capacity changes of the interactions between HyHEL-10 and HEL
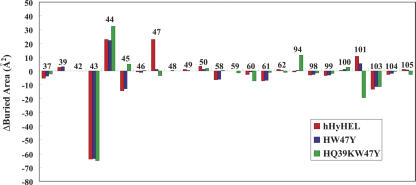
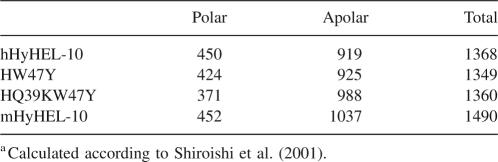
The contact area of VH–VL interfaces of the murine complex was 1490 Å2; those of hHyHEL-10, HW47Y, and HQ39KW47Y complexes were 1368, 1349, and 1360 Å2, respectively. The decreases in the contact areas were mainly caused by the interactions around Lys43 and Leu45 in the H chain (Fig. 4). Note that large changes in apolar areas of the interfaces have been observed due to the humanization (hHyHEL-10 and HW47Y mutant) (Table 4).
Figure 4.
Change in the buried areas of the VH–VL interface in hHyHEL-10 mutants relative to mHyHEL-10.
Table 4.
Interfacial areas between VH and VL (Å2)a
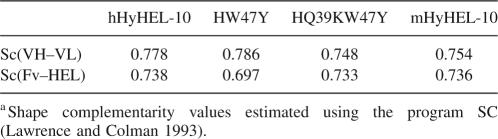
Shape-complementarity (Sc) values of mHyHEL-10, hHyHEL-10, and the two mutant complexes have been calculated using the program Sc (Table 5) (Lawrence and Colman 1993). The results showed that no major changes in shape complementarities were observed in either the antigen–antibody or VH–VL interfaces.
Table 5.
Shape complementarity valuesa
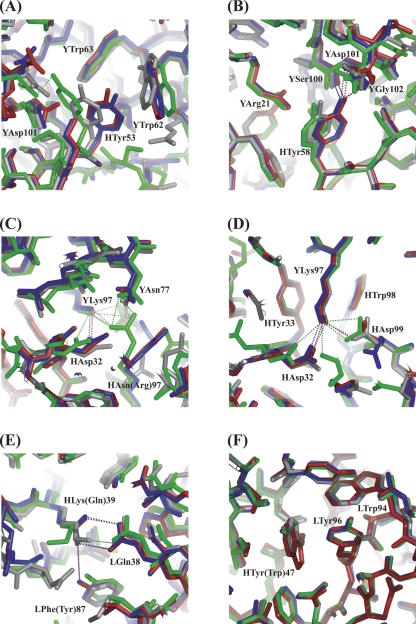
Local structural changes in the antigen–antibody interfaces due to humanization have been localized around Tyr53, Tyr58, Arg97, and Asp99 in the H chain (Fig. 5A–D); on the other hand, those in the VH–VL interfaces were localized around the mutated sites: i.e., sites 39 and 47 in the VH chain (Fig. 5E,F).
Figure 5.
Comparison of local structures in mHyHEL-10-HEL and hHyHEL-10-HEL complexes (hydrogen bonds and salt bridges are depicted as dotted lines). (A–D) Local structural changes in the antigen–antibody interfaces due to humanization of mHyHEL-10: (A) region around site 53 of VH; (B) region around site 58 of VH; (C) region around site 97 of VH; (D) region around site 99 of VH. (E, F) Comparison of local structures around the mutated sites: (E) region around site 39 of VH; (F) region around site 47 of VH. Red, hHyHEL-10 Fv–HEL complex; blue, HW47Y Fv–HEL complex; green, HQ39KW47Y Fv–HEL complex; gray, mHyHEL-10 Fv–HEL complex.
Discussion
To address the effects of grafting CDRs of a murine antibody onto the selected framework region of a human antibody, i.e., humanization of a murine antibody, on the specificity and affinity of the antibody for its target, we examined the interaction between hen egg white lysozyme (HEL) and its murine antibody HyHEL-10 (mHyHEL-10) variable domain fragment. We selected a human antibody framework sequence with high homology, grafted sequences of six complementarity-determining regions of murine HyHEL-10 onto the selected human framework, and investigated the interactions between the mutant Fvs and HEL. In the following subsections, we discuss and correlate our thermodynamic and structural findings.
Thermodynamic analysis of hHyHEL-10 Fv–HEL interactions
Humanization of the antibody increased the negative enthalpy change for the hHyHEL-10 Fv–HEL interaction (−ΔH became more negative) compared to that for the wild-type mHyHEL-10–HEL interaction; however, humanization decreased the affinity constant (K a) more than 10-fold (Table 1). These results indicate that the entropy loss increased due to humanization of mHyHEL-10. In order to improve the affinity of the antibody for its target, we then focused on the residues at sites 39 and 47, which are located in the VH–VL interface. Substitution of one Trp residue at site 47 of the VH chain with Tyr led to almost complete recovery of the affinity via an increase in the negative enthalpy change. Note that the mutation at site 47 reduced the entropy loss. One additional mutation at site 39 of the VH chain led to significant reduction of the negative enthalpy and entropy changes, resulting in further improvement of the affinity for the target antigen, HEL. These results indicate the notable reduction of the entropy loss due to the second mutation in the VH–VL interface has led to the complete recovery of the affinity for the target.
Comparison of the hHyHEL-10 Fv–HEL and mutant hHyHEL-10 Fv–HEL structures with the mHyHEL-10 Fv–HEL structure
The overall structure of the hHyHEL-10 Fv–HEL complex is similar to the structure of the mHyHEL-10 Fv–HEL complex (Fig. 3), and almost all of the noncovalent bonds have been found in the hHyHEL-10 complex. The present structural data indicated that no major changes in the interfacial interactions have been introduced due to humanization and mutations into the interfacial residues. The area of HEL covered by the hHyHEL-10 Fv, however, decreased by 32 Å2 in comparison with the mHyHEL-10 Fv–HEL complex (Table 3); this decrease was due to a decrease in the polar contact areas. Although no significant changes in these areas have been observed, recovery of the polar contact area could be achieved via the two mutations.
The area of the VH–VL interface of the hHyHEL-10 Fv–HEL complex decreased by 122 Å2 in comparison with that of the murine antibody, due to significant reductions at sites 43 and 45 in the VH chain (Fig. 4) in spite of the slight increase at site 47. The structural changes are in part compensated for by mutation at site 47 of VH and completely compensated for by further mutation at site 39 of VH. The relative orientation of VH, VL, and HEL has been changed due to humanization and the two mutations into the interfacial residues of hHyHEL-10 Fv.
Note that the Lys residue is rarely observed at site 39 in murine antibodies, whereas Gln is frequently observed (Vargas-Madrazo and Paz-Garcia 2003). Recently, we and another group have suggested that the Lys residue at this site is critical for association of the antibody mHyHEL-10 with its target HEL (Masuda et al. 2006). Induced fitting of antibodies via changes in association of variable domains was first proposed by Bhat et al. (1990). Critical contribution of appropriate associations between VH and VL chains has been suggested in several antigen–antibody systems (Takahashi et al. 1994; Khalifa et al. 2000; Hugo et al. 2003; Kumagai et al. 2003); thus, we conclude that some particular residues in VH–VL interfaces should be taken into account for engineering humanized antibodies.
The most useful tie between the thermodynamic and structural data is based on the relationship between the change in heat capacity and the change in the surface area that is exposed to solvent on going from one equilibrium state to another (Morton and Ladbury 1996). For example, Spolar and Record successfully related the thermodynamic and structural changes on formation of a protein–DNA complex (Spolar and Record 1994). Thus, from the temperature dependency of the enthalpy changes (Fig. 2), we estimated the heat capacity changes (ΔC p) of the interactions between the antibodies and the antigen (Table 1). We calculated the ΔC p values on the basis of structural data and Equation 1:
reported by Spolar and Record (1994) (Table 3). Our data include large discrepancies between the calculated values and the experimentally determined values in the cases of mHyHEL-10, hHyHEL-10, HW47Y mutant. Such discrepancies were first pointed out by Morton and Ladbury (1996). Possible explanations for such discrepancies are restriction of water molecules in the interface (Dunitz 1994; Morton and Ladbury 1996) and recognition-coupled structural changes due to induced fitting (Spolar and Record 1994; Myszka et al. 2000). Precisely estimating the contribution of water molecules and structural changes is difficult; however, significant contributions of larger negative ΔC p values may originate from structural changes due to mutations, since the changes in relative orientations of VH, VL, and HEL due to humanization seem to be correlated with increase in negative ΔC p values. Again, note that the negative ΔC p value was significantly reduced via two mutations into the interfacial residues of hHyHEL-10.
Conclusion
Structural and thermodynamic consequences of humanizing a murine antibody
Humanization of anti-HEL murine antibody HyHEL-10 led to 10-fold reduced affinity of the antibody for its target, due to an unfavorable entropy change. Structural analyses clearly indicate that all interfacial interactions between Fv and HEL were conserved; relative orientations of VH, VL, and HEL have been changed. Two mutations into the interface of the variable domains, however, led to complete recovery of the antibody's affinity and specificity for the target, due to reduction of an unfavorable entropy change. Structural analyses suggest that the HQ39KW47Y mutant complex has a tertiary structure almost identical to that of the mHyHEL-10–HEL complex, except for some interactions of the Fv–HEL interface. From these results, we conclude that appropriate association of variable domains is critical for humanization of murine antibodies without loss of function. The mutations of humanized antibodies in the framework regions of mHyHEL-10 Fv at characteristic sites 39 and 47 at VH–VL interfaces may change the relative orientations of VH, VL, and HEL and improve affinity or specificity. The residues focused on in this study are completely buried in the VH–VL interface; thus, the present strategy, i.e., structural adjustment of VH–VL interfaces, may be favorable for functional improvement of the humanized antibodies since the mutations at the interface may cause little antigenicity of the engineered antibodies. Fine-tuning of affinity or specificity of humanized antibodies might be successfully performed by careful design of variable domain interactions.
Materials and Methods
Materials
All enzymes for genetic engineering were obtained from Takara Shuzo Co., Ltd., Toyobo Biologics, Roche Diagnostics, and New England Biolabs. Isopropyl β-d-(−)-thiogalactopyranoside (IPTG) was obtained from Wako Fine Chemicals, Inc. All other reagents were of biochemical-research grade. The antigen, HEL (Seikagaku-Kogyo Co.), was purified by ion-exchange chromatography on SP–Sepharose FF (Amersham-Biosciences), followed by gel filtration on Superdex 75 pg (Amersham-Biosciences) equilibrated with phosphate-buffered saline (PBS). The eluate was dissolved in water at a concentration of 0.54 mM prior to use.
Humanization of HyHEL-10
The amino acid sequences of the humanized HyHEL-10 VH and VL were constructed by a CDR-grafting method. BLASTP (Altschul et al. 1990) was used to search for the human sequences most similar to the HyHEL-10 variable domains. Some human sequences were chosen to graft the murine CDRs, and the homology modeling was performed using the grafted variable domain fragments. The genes encoding the humanized VH and VL were separately synthesized from nine oligonucleotides by using an overlap-extension PCR method previously described (Sato et al. 1994; Mehta et al. 1997). The oligonucleotides, ranging from 77 to 97 bp with overlapping regions of 20 bp, were designed using codons that exist with high frequency in E. coli; the designed oligonucleotides are summarized in Supplemental Table S1. The oligonucleotides were assembled, and the full-length gene was amplified by PCR. A second PCR was performed using the DNA fragments amplified by the first PCR as templates and the two external primers. The second PCR product was digested with NcoI and SacII and cloned into a pRA vector containing T7 promoter (Asano et al. 2006). The resulting vectors were designated pRA-hVH and pRA-hVL. The correctness of the humanized versions was confirmed by DNA sequencing (ABI 3100 Avant Genetic Analyzer, Applied Biosystems).
Construction of expression vectors of hHyHEL-10 and mutant Fvs
To produce the Fv fragment, a vector for coexpressing both VH and VL chains was constructed. Each chain was fused to pelB signal peptide for secretory expression in E. coli. In brief, the genes encoding both PelB and VL were amplified by PCR using pRA-hVL as a template and the primers as follows. The nucleotide primers were 5′-ACTAGTTATTTCAAGGAGACAGTCATAATG-3′ and 5′-GGATCCGCTATTAATGGTGGTGATGATGGTG-3′, respectively (SpeI and BamHI sites are underlined). The PCR product was digested with SpeI and BamHI and ligated into the same sites of pRA-hVH to give the expression vector pRA-hHL (the SpeI and BamHI sites are downstream of the hVH gene).
Site-directed mutagenesis
Site-directed mutagenesis was accomplished by PCR mutagenesis. The oligonucleotide primer pairs for mutations at sites 39 and 47 of VH were 5′-CTGGATTCGCAAACCGCCGGGCAAAGGT-3′, 5′-ACCTTTGCCCGGCGGTTTGCGAATCCAG-3′, 5′-CCGCCGGGCAAAGGTCTGGAATATATTGGCTATGTGA-3′, and 5′-TCACATAGCCAATATATTCCAGACCTTTGCCCGGCGG-3′, respectively (mutated sites are underlined).
Expression and purification of hHyHEL-10 Fv
E. coli BL21 (DE3) was transformed with the expression vector described above, and transformants were grown at 28°C in 2× YT broth containing 100 μg/mL ampicillin for 10 h. To induce the expression of Fv fragment, IPTG was added to a final concentration of 1 mM, and the culture was incubated overnight at 28°C. The supernatant was separated from the culture by centrifugation at 6000g for 15 min at 4°C and salted out with ammonium sulfate at 80% saturation. The precipitates were collected by centrifugation at 7000g for 30 min at 4°C and dissolved in PBS. The protein solution was dialyzed against the PBS overnight and then centrifuged at 7000g for 20 min. After the precipitates were removed, the supernatant was loaded onto a HEL-Sepharose column that had been previously equilibrated with the same buffer, and in which HEL was immobilized to CNBr-activated Sepharose 4B (GE Healthcare). The column was washed with the same buffer, 200 mM NaCl/100 mM Tris-HCl (pH 8.5), and then the adsorbed protein was eluted with 100 mM glycine-HCl (pH 2.0). The eluate was quickly neutralized with 200 mM NaCl/1 M Tris-HCl (pH 7.5). The fraction containing Fv was loaded onto a Sephacryl S-200 HR equilibrated with 200 mM NaCl/50 mM phosphate buffer (pH 7.2).
Isothermal titration calorimetry
Thermodynamic parameters between HEL and Fv were determined by isothermal titration calorimetry (ITC) using a VP-ITC calorimeter (MicroCal) under conditions as follows. The Fv fragment at a concentration of 5 μM in 200 mM NaCl/50 mM phosphate buffer (pH 7.2) in a calorimeter cell was titrated with the HEL at a concentration of 50 μM in the same buffer at four temperatures (20°C, 25°C, 30°C, and 35°C). The HEL solution was injected 25 times in portions of 10 μL during a period of 20 s. Thermogram data were analyzed with the program Origin version 5 (MicroCal).
The enthalpy change (ΔH) and association constant (K a) for the Fv–HEL interaction were obtained from the titration curve. The Gibbs free energy change (ΔG = –RT ln K a) and the entropy change [ΔS = (–ΔG + ΔH)/T] for the binding were calculated from ΔH and K a. The heat capacity change (ΔCp) was estimated from the temperature dependency of the enthalpy change.
Crystallization of the hHyHEL-10 Fv–HEL complexes
The hHyHEL-10 Fv and HEL solutions were mixed in a molar ratio of 1:1.2 and then dialyzed against 10 mM Tris-HCl (pH 8.0). Finally, the Fv–HEL complex solution was concentrated to 31 mg mL−1. Initial screening for crystallization conditions was performed by the sparse matrix method at 20°C by using Crystal Screen and Crystal Screen 2 kits (Hampton Research). A crystal of the wild-type Fv–HEL complex was grown by vapor diffusion using the hanging-drop method against a reservoir containing 100 mM MES (pH 6.4), 10 mM ZnSO4, and 28%–31% (w/v) polyethylene glycol monomethyl ester 550. Crystals of the mutant Fv–HEL complexes were also obtained under similar conditions.
Data collection and structural determination
Data for wild-type Fv–HEL complex were collected at the Photon Factory (Tsukuba, Japan) beamline BL-5A, using an ADSC Quantum 315 CCD detector, and were processed using the program HKL2000 (Otwinowski and Minor 1997). Data for mutant Fv–HEL complexes were collected at the Photon Factory beamline NW12A, using an ADSC Quantum 210 CCD detector, and were also processed using the program HKL2000. Crystallographic data and statistics of the data collection are summarized in Supplemental Table S2. Initially, the structure of the HW47Y mutant Fv–HEL complex was determined by a molecular replacement (MR) method with the program MOLREP (Vagin and Teplyakov 2000) in the CCP4 suite (Collaborative Computational Project Number 4 1994). The search model for MR was derived from the structure of parental murine Fv–HEL complex. The structures of the other Fv–HEL complexes (wild type and HQ39KW47Y) were determined by a MR method using the structure of HW47Y mutant Fv–HEL complex as a search model. Refinements of the structures of the Fv–HEL complexes were carried out using the programs O (Jones et al. 1991) and CNS (Brünger et al. 1998). The atomic coordinates and structural factors for each Fv–HEL complex have been deposited in the Protein Data Bank (ID codes hHyHEL-10, HW47Y, and HQ39KW47Y for 2EKS, 2EIZ, and 2YSS, respectively).
Estimation of protein concentration
The concentration of HEL was estimated taking A 1% 280 = 26.5 (Imoto et al. 1972). The concentrations of the hHyHEL-10 Fv fragments were estimated taking A 1% 280 = 22.9 for the wild type and A 1% 280 = 21.2 for the HW47Y and HQ39KW47Y mutants.
Electronic supplementary material
The supplementary material contains two tables and one figure. Supplemental Table S1 presents the primers of overlap-extension PCR for synthesis of hHyHEL-10 genes. Supplemental Table S2 shows the crystallographic data for Fv–HEL complexes. Supplemental Figure S1 shows the amino acid sequences of mHyHEL-10 and hHyHEL-10 selected.
Acknowledgments
We thank N. Sakabe, S. Wakatsuki, M. Suzuki, and N. Igarashi of the Photon Factory for their kind help with the data collection. This work was supported in part by Grants-in-Aid for General Research from the Japan Society for the Promotion of Science. Additional support was provided through Grants-in-Aid for Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Izumi Kumagai, Department of Biomolecular Engineering, Graduate School of Engineering, Tohoku University, Aoba-yama 6-6-11-606, Aoba-ku, Sendai 980-8579, Japan; e-mail: kmiz@kuma.che.tohoku.ac.jp; fax: +81-22-795-6164.
Abbreviations: VH, variable region of immunoglobulin heavy chain; VL, variable region of immunoglobulin light chain; Fv, fragment of immunoglobulin variable regions; mHyHEL-10, murine HyHEL-10; hHyHEL-10, humanized HyHEL-10; Fab, antigen binding fragment of immunoglobulin; HW47Y mutant, mutant hHyHEL-10 Fv in which Trp47 of the VH chain is substituted with Tyr; HQ39K mutant, mutant hHyHEL-10 Fv in which Gln39 of the VH chain is substituted with Lys; ITC, isothermal titration calorimetry.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073156708.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Asano, R., Sone, Y., Makabe, K., Tsumoto, K., Hayashi, H., Katayose, Y., Unno, M., Kudo, T., Kumagai, I. Humanization of the bispecific epidermal growth factor receptor × CD3 diabody and its efficacy as a potential clinical reagent. Clin. Cancer Res. 2006;12:4036–4042. doi: 10.1158/1078-0432.CCR-06-0059. [DOI] [PubMed] [Google Scholar]
- Bhat, T.N., Bentley, G.A., Fischmann, T.O., Boulot, G., Poljak, R.J. Small rearrangements in structures of Fv and Fab fragments of antibody D1.3 on antigen binding. Nature. 1990;347:483–485. doi: 10.1038/347483a0. [DOI] [PubMed] [Google Scholar]
- Braden, B.C., Poljak, R.J. Structural features of the reactions between antibodies and protein antigens. FASEB J. 1995;9:9–16. doi: 10.1096/fasebj.9.1.7821765. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Co, M.S., Deschamps, M., Whitley, R.J., Queen, C. Humanized antibodies for antiviral therapy. Proc. Natl. Acad. Sci. 1991;88:2869–2873. doi: 10.1073/pnas.88.7.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Co, M.S., Avdalovic, N.M., Caron, P.C., Avdalovic, M.V., Scheinberg, D.A., Queen, C. Chimeric and humanized antibodies with specificity for the CD33 antigen. J. Immunol. 1992;48:1149–1154. [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Davies, D.R., Cohen, G.H. Interactions of protein antigens with antibodies. Proc. Natl. Acad. Sci. 1996;93:7–12. doi: 10.1073/pnas.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunitz, J.D. The entropic cost of bound water in crystals and biomolecules. Science. 1994;264:670. doi: 10.1126/science.264.5159.670. [DOI] [PubMed] [Google Scholar]
- Ewert, S., Honegger, A., Plückthun, A. Stability improvement of antibodies for extracellular and intracellular applications: CDR grafting to stable frameworks and structure-based framework engineering. Methods. 2004;34:184–199. doi: 10.1016/j.ymeth.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Foote, J., Winter, G. Antibody framework residues affecting the conformation of the hypervariable loops. J. Mol. Biol. 1992;224:487–499. doi: 10.1016/0022-2836(92)91010-m. [DOI] [PubMed] [Google Scholar]
- Furukawa, K., Akasako-Furukawa, A., Shirai, H., Nakamura, H., Azuma, T. Junctional amino acids determine the maturation pathway of an antibody. Immunity. 1999;11:329–338. doi: 10.1016/s1074-7613(00)80108-9. [DOI] [PubMed] [Google Scholar]
- Holmes, M.A., Buss, T.N., Foote, J. Conformational correction mechanisms aiding antigen recognition by a humanized antibody. J. Exp. Med. 1998;187:479–485. doi: 10.1084/jem.187.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, M.A., Buss, T.N., Foote, J. Structural effects of framework mutations on a humanized anti-lysozyme antibody. J. Immunol. 2001;167:296–301. doi: 10.4049/jimmunol.167.1.296. [DOI] [PubMed] [Google Scholar]
- Hugo, N., Weidenhaupt, M., Beukes, M., Xu, B., Janson, J.C., Vernet, T., Altschuh, D. VL position 34 is a key determinant for the engineering of stable antibodies with fast dissociation rates. Protein Eng. 2003;16:381–386. doi: 10.1093/protein/gzg042. [DOI] [PubMed] [Google Scholar]
- Imoto, T., Johnson, T.M., North, A.C.T., Phillips, D.C., Rupley, J.A. Vertebrate lysozymes. In: Boyer P., editor. The enzymes. Vol. 7. Academic Press; San Diego, CA: 1972. pp. 665–868. [Google Scholar]
- Jirholt, P., Ohlin, M., Borrebaeck, C.A., Soderlind, E. Exploiting sequence space: Shuffling in vivo formed complementarity determining regions into a master framework. Gene. 1998;215:471–476. doi: 10.1016/s0378-1119(98)00317-5. [DOI] [PubMed] [Google Scholar]
- Jones, P.T., Dear, P.H., Foote, J., Neuberger, M.S., Winter, G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature. 1986;321:522–525. doi: 10.1038/321522a0. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kam-Morgan, L.N., Smith-Gill, S.J., Taylor, M.G., Zhang, L., Wilson, A.C., Kirsch, J.F. High-resolution mapping of the HyHEL-10 epitope of chicken lysozyme by site-directed mutagenesis. Proc. Natl. Acad. Sci. 1993;90:3958–3962. doi: 10.1073/pnas.90.9.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalifa, M.B., Weidenhaupt, M., Choulier, L., Chatellier, J., Rauffer-Bruyere, N., Altschuh, D., Vernet, T. Effects on interaction kinetics of mutations at the VH-VL interface of Fabs depend on the structural context. J. Mol. Recognit. 2000;13:127–139. doi: 10.1002/1099-1352(200005/06)13:3<127::AID-JMR495>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Kondo, H., Shiroishi, M., Matsushima, M., Tsumoto, K., Kumagai, I. Crystal structure of anti-Hen egg white lysozyme antibody (HyHEL-10) Fv-antigen complex. Local structural changes in the protein antigen and water-mediated interactions of Fv-antigen and light chain–heavy chain interfaces. J. Biol. Chem. 1999;274:27623–27631. doi: 10.1074/jbc.274.39.27623. [DOI] [PubMed] [Google Scholar]
- Kumagai, I., Nishimiya, Y., Kondo, H., Tsumoto, K. Structural consequences of target epitope-directed functional alteration of an antibody. The case of anti-hen lysozyme antibody, HyHEL-10. J. Biol. Chem. 2003;278:24929–24936. doi: 10.1074/jbc.M301149200. [DOI] [PubMed] [Google Scholar]
- Lawrence, M.C., Colman, P.M. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- Lemmon, M.A., Ladbury, J.E. Thermodynamic studies of tyrosyl-phosphopeptide binding to the SH2 domain of p56lck. Biochemistry. 1994;33:5070–5076. doi: 10.1021/bi00183a010. [DOI] [PubMed] [Google Scholar]
- Mariuzza, R.A., Pojak, R.J. The basics of binding: Mechanisms of antigen recognition and mimicry by antibodies. Curr. Opin. Immunol. 1993;5:50–55. doi: 10.1016/0952-7915(93)90080-c. [DOI] [PubMed] [Google Scholar]
- Masuda, K., Sakamoto, K., Kojima, M., Aburatani, T., Ueda, T., Ueda, H. The role of interface framework residues in determining antibody V(H)/V(L) interaction strength and antigen-binding affinity. FEBS J. 2006;273:2184–2194. doi: 10.1111/j.1742-4658.2006.05232.x. [DOI] [PubMed] [Google Scholar]
- Mehta, D.V., DiGate, R.J., Banville, D.L., Guiles, R.D. Optimized gene synthesis, high level expression, isotopic enrichment, and refolding of human interleukin-5. Protein Expr. Purif. 1997;11:86–94. doi: 10.1006/prep.1997.0785. [DOI] [PubMed] [Google Scholar]
- Merk, H., Stiege, W., Tsumoto, K., Kumagai, I., Erdmann, V.A. Cell-free expression of two single-chain monoclonal antibodies against lysozyme: Effect of domain arrangement on the expression. J. Biochem. 1999;125:328–333. doi: 10.1093/oxfordjournals.jbchem.a022290. [DOI] [PubMed] [Google Scholar]
- Morton, C.J., Ladbury, J.E. Water-mediated protein-DNA interactions: The relationship of thermodynamics to structural detail. Protein Sci. 1996;5:2115–2118. doi: 10.1002/pro.5560051018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myszka, D.G., Sweet, R.W., Hensley, P., Brigham-Burke, M., Kwong, P.D., Hendrickson, W.A., Wyatt, R., Sodroski, J., Doyle, M.L. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. 2000;97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibori, N., Horiuchi, H., Furusawa, S., Matsuda, H. Humanization of chicken monoclonal antibody using phage-display system. Mol. Immunol. 2006;43:634–642. doi: 10.1016/j.molimm.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Ohtomo, T., Tsuchiya, M., Sato, K., Shimizu, K., Moriuchi, S., Miyao, Y., Akimoto, T., Akamatsu, K., Hayakawa, T., Ohsugi, Y. Humanization of mouse ONS-M21 antibody with the aid of hybrid variable regions. Mol. Immunol. 1995;32:407–416. doi: 10.1016/0161-5890(95)00017-9. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z., Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Padlan, E.A. X-ray crystallography of antibodies. Adv. Protein Chem. 1996;49:57–133. doi: 10.1016/s0065-3233(08)60488-x. [DOI] [PubMed] [Google Scholar]
- Padlan, E.A., Silverton, E.W., Sheriff, S., Cohen, G.H., Smith-Gill, S.J., Davies, D.R. Structure of an antibody–antigen complex: Crystal structure of the HyHEL-10 Fab–lysozyme complex. Proc. Natl. Acad. Sci. 1989;86:5938–5942. doi: 10.1073/pnas.86.15.5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons, J., Rajpal, A., Kirsch, J.F. Energetic analysis of an antigen/antibody interface: Alanine scanning mutagenesis and double mutant cycles on the HyHEL-10/lysozyme interaction. Protein Sci. 1999;8:958–968. doi: 10.1110/ps.8.5.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen, C., Schneider, W.P., Selick, H.E., Payne, P.W., Landolfi, N.F., Duncan, J.F., Avdalovic, N.M., Levitt, M., Junghans, R.P., Waldmann, T.A. A humanized antibody that binds to the interleukin 2 receptor. Proc. Natl. Acad. Sci. 1989;86:10029–10033. doi: 10.1073/pnas.86.24.10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, L., Clark, M., Waldmann, H., Winter, G. Reshaping human antibodies for therapy. Nature. 1988;332:323–327. doi: 10.1038/332323a0. [DOI] [PubMed] [Google Scholar]
- Sato, K., Tsuchiya, M., Saldanha, J., Koishihara, Y., Ohsugi, Y., Kishimoto, T., Bendig, M.M. Humanization of a mouse anti-human interleukin-6 receptor antibody comparing two methods for selecting human framework regions. Mol. Immunol. 1994;31:371–381. doi: 10.1016/0161-5890(94)90115-5. [DOI] [PubMed] [Google Scholar]
- Schwarz, F.P., Tello, D., Goldbaum, F.A., Mariuzza, R.A., Poljak, R.J. Thermodynamics of antigen–antibody binding using specific anti-lysozyme antibodies. Eur. J. Biochem. 1995;228:388–394. [PubMed] [Google Scholar]
- Shiroishi, M., Yokota, A., Tsumoto, K., Kondo, H., Nishimiya, Y., Horii, K., Matsushima, M., Ogasahara, K., Yutani, K., Kumagai, I. Structural evidence for entropic contribution of salt bridge formation to a protein antigen–antibody interaction: The case of hen lysozyme-HyHEL-10 Fv complex. J. Biol. Chem. 2001;276:23042–23050. doi: 10.1074/jbc.M100480200. [DOI] [PubMed] [Google Scholar]
- Soderlind, E., Strandberg, L., Jirholt, P., Kobayashi, N., Alexeiva, V., Aberg, A.M., Nilsson, A., Jansson, B., Ohlin, M., Wingren, C., et al. Recombining germline-derived CDR sequences for creating diverse single-framework antibody libraries. Nat. Biotechnol. 2000;18:852–856. doi: 10.1038/78458. [DOI] [PubMed] [Google Scholar]
- Spolar, R.S., Record, M.T. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- Stern, M., Herrmann, R. Overview of monoclonal antibodies in cancer therapy: Present and promise. Crit. Rev. Oncol. Hematol. 2005;54:11–29. doi: 10.1016/j.critrevonc.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Stochwin, L.H., Holmes, S. The role of therapeutic antibodies in drug discovery. Biochem. Soc. Trans. 2003;31:433–436. doi: 10.1042/bst0310433. [DOI] [PubMed] [Google Scholar]
- Sturtevant, J.M. The thermodynamic effects of protein mutations. Curr. Opin. Struct. Biol. 1994;4:69–78. [Google Scholar]
- Takahashi, H., Tamura, H., Simba, N., Shimada, I., Arata, Y. Role of the domain-domain interaction in the construction of the antigen combining site. A comparative study by 1H–15N shift correlation NMR spectroscopy of the Fv and Fab fragments of anti-dansyl mouse monoclonal antibody. J. Mol. Biol. 1994;243:494–503. doi: 10.1006/jmbi.1994.1675. [DOI] [PubMed] [Google Scholar]
- Torigoe, H., Nakayama, T., Imazato, M., Shimada, I., Arata, Y., Sarai, A. The affinity maturation of anti-4-hydroxy-3-nitrophenylacetyl mouse monoclonal antibody. A calorimetric study of the antigen–antibody interaction. J. Biol. Chem. 1995;270:22218–22222. doi: 10.1074/jbc.270.38.22218. [DOI] [PubMed] [Google Scholar]
- Tsumoto, K., Nakaoki, Y., Ueda, Y., Ogasahara, K., Yutani, K., Watanabe, K., Kumagai, I. Effect of the order of antibody variable regions on the expression of the single-chain HyHEL10 Fv fragment in E. coli and the thermodynamic analysis of its antigen-binding properties. Biochem. Biophys. Res. Commun. 1994a;201:546–551. doi: 10.1006/bbrc.1994.1736. [DOI] [PubMed] [Google Scholar]
- Tsumoto, K., Ueda, Y., Maenaka, K., Watanabe, K., Ogasahara, K., Yutani, K., Kumagai, I. Contribution to antibody–antigen interaction of structurally perturbed antigenic residues upon antibody binding. J. Biol. Chem. 1994b;269:28777–28782. [PubMed] [Google Scholar]
- Tsumoto, K., Ogasahara, K., Ueda, Y., Watanabe, K., Yutani, K., Kumagai, I. Role of Tyr residues in the contact region of anti-lysozyme monoclonal antibody HyHEL10 for antigen binding. J. Biol. Chem. 1995;270:18551–18557. doi: 10.1074/jbc.270.31.18551. [DOI] [PubMed] [Google Scholar]
- Tsumoto, K., Ogasahara, K., Ueda, Y., Watanabe, K., Yutani, K., Kumagai, I. Role of salt bridge formation in antigen–antibody interaction. Entropic contribution to the complex between hen egg white lysozyme and its monoclonal antibody HyHEL10. J. Biol. Chem. 1996;271:32612–32616. doi: 10.1074/jbc.271.51.32612. [DOI] [PubMed] [Google Scholar]
- Ueda, Y., Tsumoto, K., Watanabe, K., Kumagai, I. Synthesis and expression of a DNA encoding the Fv domain of an anti-lysozyme monoclonal antibody, HyHEL10, in Streptomyces lividans . Gene. 1993;129:129–134. doi: 10.1016/0378-1119(93)90708-b. [DOI] [PubMed] [Google Scholar]
- Ueda, H., Tsumoto, K., Kubota, K., Suzuki, E., Nagamune, T., Nishimura, H., Schueler, P.A., Winter, G., Kumagai, I., Mohoney, W.C. Open sandwich ELISA: A novel immunoassay based on the interchain interaction of antibody variable region. Nat. Biotechnol. 1996;14:1714–1718. doi: 10.1038/nbt1296-1714. [DOI] [PubMed] [Google Scholar]
- Vagin, A., Teplyakov, A. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- Vargas-Madrazo, E., Paz-Garcia, E. An improved model of association for VH-VL immunoglobulin domains: Asymmetries between VH and VL in the packing of some interface residues. J. Mol. Recognit. 2003;16:113–120. doi: 10.1002/jmr.613. [DOI] [PubMed] [Google Scholar]
- Verhoeyen, M., Milstein, C., Winter, G. Reshaping human antibodies: Grafting an antilysozyme activity. Science. 1988;239:1534–1536. doi: 10.1126/science.2451287. [DOI] [PubMed] [Google Scholar]
- Xavier, K.A., Shick, K.A., Smith-Gil, S.J., Willson, R.C. Involvement of water molecules in the association of monoclonal antibody HyHEL-5 with bobwhite quail lysozyme. Biophys. J. 1997;73:2116–2125. doi: 10.1016/S0006-3495(97)78242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota, A., Tsumoto, K., Shiroishi, M., Kondo, H., Kumagai, I. The role of hydrogen bonding via interfacial water molecules in antigen–antibody complexation. The HyHEL-10-HEL interaction. J. Biol. Chem. 2003;278:5410–5418. doi: 10.1074/jbc.M210182200. [DOI] [PubMed] [Google Scholar]