Abstract
Follicular lymphoma (FL) is a B cell non-Hodgkin’s lymphoma (NHL) that frequently displays a t(14;18) translocation. Clonal evolution and histological transformation of FL is frequently associated with the accumulation of secondary genetic alterations. It has been demonstrated that the BCL-6 gene can be altered by chromosomal rearrangements and by mutations clustering in its 5′ noncoding region in a significant fraction of FL and diffuse large cell lymphoma (DLCL). To elucidate the role of the BCL-6 gene alterations in the histological transformation and clonal progression of FL, we analyzed serial biopsy specimens from 12 patients with FL. Two cases of FL showed no histological alteration in the second biopsy, and 10 cases of FL showed morphological transformation to DLCL in the second biopsy. Southern blot analysis was used to detect rearrangement of the BCL-6 gene, polymerase chain reaction-single strand conformation polymorphism and sequence analysis were performed for identification of mutations in the 5′ noncoding region of the BCL-6 gene, and immunohistochemical analysis was applied to reveal the BCL-6 protein expression. No BCL-6 gene rearrangement was detected in any of the samples, but a total of 58 mutations were found in the 5′ noncoding region of the BCL-6 gene in seven cases. In five cases, both the FL and the clonally related FL or DLCL, and in two cases only the DLCL samples were mutated. The mutations were identical in multiple biopsy specimens of FL that did not show morphological transformation. In six patients where FL cells underwent morphological transformation, considerable intraclonal sequence heterogeneity was observed, indicating an ongoing type of somatic mutation. Based on the pattern of shared and nonshared mutations, the genealogical relationship of neoplastic clones could be established. In all of these cases, the histological transformation of FL was associated with the emergence of a subpopulation marked by new sites of mutations in the BCL-6 5′ noncoding sequences. In three of these six cases, the histological transformation is also associated with the reduced expression of the BCL-6 protein. These findings demonstrate that mutation of the 5′ noncoding region of the BCL-6 gene developed in the clonal evolution of FL, and at different time points in the lymphoma evolution different clonotypes dominate.
Follicular lymphoma (FL) is a B cell non-Hodgkin’s lymphoma (NHL) that is believed to originate from germinal center (GC) cells. 1 Generally, FL starts as a relatively indolent malignancy; however, it has a tendency to transform into high-grade aggressive diffuse large cell lymphoma (DLCL) during the later stage of the disease. 2-4 Approximately 85% of FLs are associated with the t(14;18) translocation, which juxtaposes the BCL-2 oncogene with the immunoglobulin (Ig) heavy chain (H) gene. 5,6 In the clonal evolution and histological transformation of the FL, the neoplastic cells retain the t(14;18) translocation and frequently acquire additional genetic abnormalities at cytogenetic 7-11 and molecular levels. 12-16
The BCL-6 gene is located on chromosome 3q27 and encodes a POX/Zinc finger protein, which functions as a sequence-specific DNA-binding transcription repressor. 17 The BCL-6 protein is expressed in B cells within GC and is involved in the control of GC formation and T-cell-dependent antigen responses. 17,18 Several lines of evidence suggest that structural alterations of the regulatory region of the BCL-6 gene are involved in lymphomagenesis. Chromosomal rearrangements affecting the BCL-6 gene cluster within a 4-kb region spanning the promoter and first noncoding exon are associated with ∼40% of DLCL and ∼10% of FL. 19-22 Recent studies also indicate that the BCL-6 gene may be altered by somatic point mutations clustering within the 5′ noncoding regions of the gene in malignancies, which display a GC phenotype and hypermutated Ig variable (V) genes. 23 Approximately 70% of DLCL, 45% of FL, 58% of AIDS-related NHL, 39% of Burkitt’s lymphoma, and 44% of posttransplant lymphoproliferative disorders carry BCL-6 genes mutated in the 5′ noncoding region. 24-27 These mutations are of somatic origin, frequently biallelic, and found in cases displaying either normal or rearranged BCL-6 alleles, indicating their independence of chromosomal translocation. 24,26
To further characterize the nature of BCL-6 gene mutations in FL and to gain insight into the role of the BCL-6 gene in lymphoma progression, we have performed longitudinal analysis of the BCL-6 gene organization in sequential biopsy specimens from patients with FL that showed ho histological alterations in subsequent biopsy specimens or that underwent morphological transformation to DLCL. Our results indicate ongoing somatic mutations of the BCL-6 5′ noncoding sequences in the majority of FLs and DLCLs, and provide evidence that histological transformation of the FL may be associated with the emergence of subclones marked by divergent BCL-6 clonotypes.
Materials and Methods
Pathological Samples and DNA Extraction
Sequential biopsy samples of 12 patients with FL observed at Stanford University Medical Center and University Medical School of Pécs were selected for this study based on the availability of frozen tissue for the molecular analyses. Diagnoses were based on histopathological, immunophenotypic, and immunogenotypic analyses. 15 All lymphoma samples were classified according to the Revised European-American Lymphoma Classification proposed by the International Lymphoma Study Group. 1 The histology of the first lymph node biopsy in six patients (Cases 5–8, 10, and 11) was FL, provisional cytologic grade I, in four patients (Cases 1, 2, 9, and 12) was FL, provisional cytological grade II, and in two patients (Cases 3 and 4) was FL, provisional cytological grade III. In two cases (Cases 11 and 12), the histology of the second biopsy was identical with the first biopsy. In Cases1–10, histology of the second biopsy was classified as DLCL (Table 1) ▶ . All samples included in this study displayed monoclonal IgH gene rearrangement and t(14;18) translocation. In each case, the subsequent biopsy samples showed identical Ig heavy chain gene rearrangements and identical breakpoint sequences of the t(14;18) translocation, indicating common clonal origin of the tumor samples of the first and the second biopsy. Genomic DNAs were extracted from cryopreserved tissue samples using the salting out technique. 28
Table 1.
Summary of the BCL-6 Gene Rearrangement, PCR-SSCP Analysis of the BCL-6 5′ Noncoding Regions and Expression of the BCL-6 Protein in 12 Cases of Paired FL and Subsequent FL or DLCL Samples
| Case | Sample | Date of biopsy | Histology | BCL-6 rearrangement | BCL-6 5′ Noncoding region | BCL-6 expression (%) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| XbaI | BamHI | E1.10 | E1.11 | E1.12 | E1.13 | |||||
| 1 | A | 1991 | FL-II | G | G | WT | M | M | WT | 100 |
| B | 1992 | DLCL | G | G | WT | M | WT | WT | 100 | |
| 2 | A | 1986 | FL-II | G | G | WT | M | WT | WT | 100 |
| B | 1992 | DLCL | G | G | WT | M | WT | WT | 100 | |
| 3 | A | 1989 | FL-III | G | G | WT | M | WT | M | 100 |
| B | 1990 | DLCL | G | G | WT | M | WT | M | 100 | |
| 4 | A | 1988 | FL-III | G | G | WT | WT | WT | WT | 100 |
| B | 1989 | DLCL | G | G | WT | WT | WT | WT | 100 | |
| 5 | A | 1985 | FL-I | G | G | WT | M | WT | WT | 100 |
| B | 1988 | DLCL | G | G | WT | M | WT | WT | 20 | |
| 6 | A | 1987 | FL-I | G | G | WT | WT | WT | WT | 100 |
| B | 1988 | DLCL | G | G | WT | M | M | WT | 10 | |
| 7 | A | 1983 | FL-I | G | G | WT | WT | WT | WT | 100 |
| B | 1990 | DLCL | G | G | M | M | WT | WT | 10 | |
| 8 | A | 1984 | FL-I | G | G | WT | WT | WT | WT | 100 |
| B | 1986 | DLCL | G | G | WT | WT | WT | WT | 100 | |
| 9 | A | 1989 | FL-II | G | G | WT | WT | WT | WT | 100 |
| B | 1990 | DLCL | G | G | WT | WT | WT | WT | 100 | |
| 10 | A | 1986 | FL-I | G | G | WT | WT | WT | WT | ND |
| B | 1987 | DLCL | G | G | WT | WT | WT | WT | ND | |
| 11 | A | 1998 | FL-I | G | G | WT | M | WT | WT | 100 |
| B | 1999 | FL-I | G | G | WT | M | WT | WT | 100 | |
| 12 | A | 1997 | FL-II | G | G | WT | WT | WT | WT | 100 |
| B | 1999 | FL-II | G | G | WT | WT | WT | WT | 100 |
A, first biopsy sample; B, second biopsy sample; FL-I, follicular lymphoma—cytological grade I; FL-II, follicular lymphoma—cytological grade II; FL-III, follicular lymphoma—cytological grade III; DLCL, diffuse large cell lymphoma; G, germline configuration; M, mutations identified in PCR-SSCP fragments; ND, not detected; WT, wild-type.
Southern Blot Analysis of BCL-6 Gene Rearrangement
The presence of BCL-6 gene rearrangement was analyzed by the 4.0-kb Sac fragment BCL-6 probe. 19 Five-microgram aliquots of genomic DNA were digested with the BamHI or XbaI restriction endonucleases according to the manufacturer’s instructions (Boehringer Mannheim GmbH, Mannheim, Germany), electrophoresed in 0.8 or 1% agarose gels, denatured with alkali, neutralized, and transferred to nitrocellulose filters (Schleicher & Schuell, Keene, NH). The filters were hybridized in 50% formamide/3× standard citrate (SSC) buffer at 37°C to DNA probes that had been 32P-labeled using the Random Primed DNA Labeling Kit (Boehringer Mannheim) according to the manufacturer’s instructions. The filters were washed in 0.2 × SSC/0.5% sodium dodecyl sulfate at 60°C for 2 hours and then autoradiographed at −70°C for 16 to 48 hours, as described before. 29
Polymerase Chain Reaction-Single Strand Conformation Polymorphism (PCR-SSCP) Analysis of the 5′ Noncoding Region of the BCL-6 Gene
PCR-SSCP analysis of BCL-6 5′ noncoding regions was performed on four partially overlapping PCR fragments (E1.10, E.1.11, E1.12, and E1.13) spanning 998 bp located downstream of the first BCL-6 noncoding exon. 24,26 The selection of this region for the mutational analysis of BLC-6 was based on evidence that ∼45% of FLs and ∼70% of de novo DLCLs carry somatic mutations in these sequences. 24,26
DNA Sequencing
PCR products encompassing fragments E1.10, E1.11, E1.12, and E1.13 were cloned into the pCR vector using the TA Cloning Kit (Invitrogen Corp., San Diego, CA). In samples subjected to DNA sequencing, six independent subclones were analyzed. DNA sequencing was performed directly from a small-scale plasmid preparation using the Sequenase version 2.0 (United States Biochemical, Cleveland, OH) system according to the manufacturer’s instructions. DNA sequences were analyzed using the MacVector version 4.5 (Eastern Kodak Co., New Haven, CT) software and the GenBank data base.
Analysis of the Sequence Polymorphism of the 5′ Noncoding Region of the BCL-6 Gene
To analyze whether sequence alterations of the 5′ noncoding region of the BCL-6 gene shared by the FL and DLCL samples of Cases 3 and 5 are associated with somatic mutation or sequence polymorphism, normal tissue samples of these two patients were amplified by the primers specific for the E1.13 and E1.11 fragments, respectively. The PCR products were cloned, sequenced, and compared with the corresponding BCL-6 5′ noncoding sequences of the tumor samples.
Immunohistochemistry
The expression of the BCL-6 protein was detected by using the mAb PG-B6 directed against the amino-terminal portion of the human BCL-6 gene product. 30 Immunostaining for BCL-6 was performed on frozen sections by the alkaline phosphatase-anti-alkaline phosphatase method. 31
Results
Analysis of BCL-6 Gene Rearrangement
Genomic DNAs from paired samples of FL and subsequent FL or DLCL were digested with BamHI and XbaI restriction endonucleases and hybridized with radiolabeled BCL-6 probe. None of the FL or transformed DLCL samples of the 12 patients displayed BCL-6 gene rearrangement in either the BamHI- or XbaI-digested DNAs (Table 1) ▶ .
PCR-SSCP Analysis of the 5′ Noncoding Sequences of the BCL-6 Gene
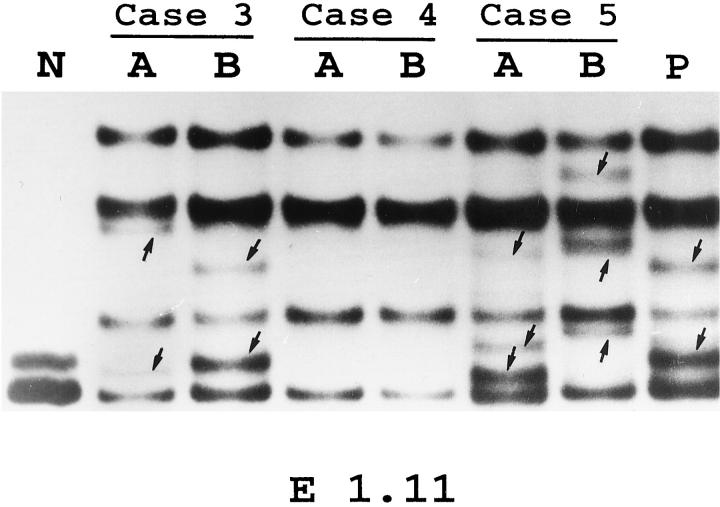
In each case, the samples of the first and second biopsies were evaluated in parallel. The 24 samples from the 12 patients displayed a total of 16 PCR-SSCP variants. Representative examples of the PCR-SSCP analyses are shown in Figure 1 ▶ . In five cases (Cases 4, 8–10, and 12), neither first nor second biopsy samples showed altered migration patterns. In two cases (Cases 6 and 7) only the second sample, and in five cases (Cases 1–3, 5, and 11) both the first and subsequent samples, showed altered electrophoretic migration patterns. In those cases, where both the first and the second biopsy samples showed altered PCR-SSCP migration, the patterns were identical in one case (Case 11) and different in six cases (Cases 1–3 and 5–7).
Figure 1.
PCR-SSCP analysis of the 5′ noncoding regions of BCL-6 gene in paired serial biopsy specimens of FL (A) and transformed DLCL (B). Representative results obtained for PCR-amplified fragments from the E1.11 regions of BCL-6 gene in Cases 3–5 are shown. A positive control (P), represented by a tumor sample known to harbor BCL-6 mutations, as well as sample that was not denatured before electrophoresis (N) are shown. Arrows identify abnormally migrating bands.
Nucleotide Sequence Analysis of the BCL-6 5′ Noncoding Sequences
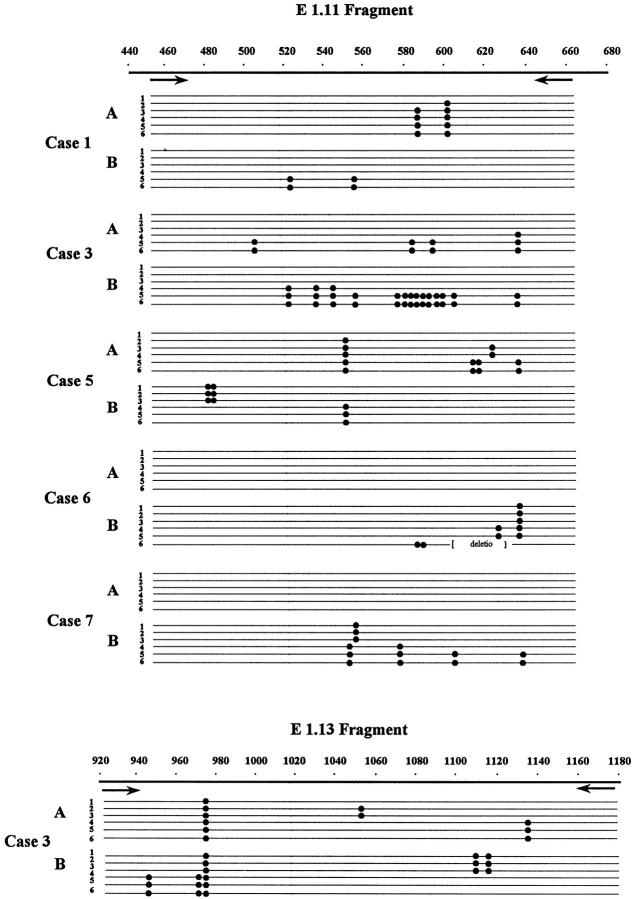
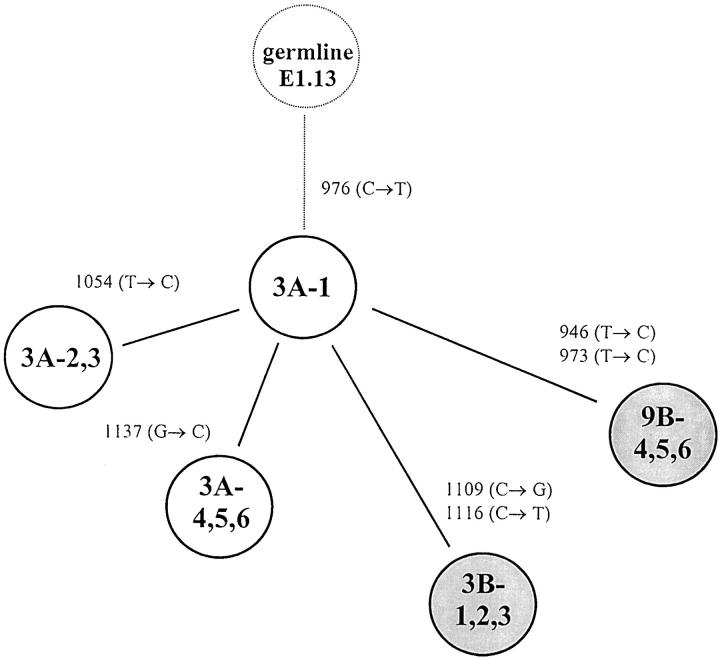
To confirm and characterize the mutations affecting the BCL-6 5′ noncoding region, we cloned and sequenced those pairs of PCR products in which any of the samples showed altered migration pattern in the PCR-SSCP analysis. In these cases, DNAs were PCR-amplified under the same conditions as described for the SSCP, but the radioactive nucleotide was omitted. In each sample, six independent bacterial isolates were analyzed and compared with germline sequences of the BCL-6 5′ noncoding region. For each tumor sample studied, PCR fragments that appeared abnormal by SSCP analysis were found to contain mutations. The mutations found are shown in Table 2 ▶ . In 14 samples of the seven cases analyzed, a total of 58 alterations were detected. The mutations observed included single basepair substitutions and a 31-bp stretch deletion. All mutations found were distributed in one or two clusters of clonally related BCL-6 5′ noncoding sequences, indicating mono- or biallelic distribution of the mutations. In three samples (5B, 6B, and 7B), both alleles carried mutations. In two cases (Cases 6 and 7) only the DLCL samples, and in five cases (Cases 1–3, 5, and 11) both the FL and subsequent FL or DLCL samples were mutated. In six samples (1A, 3A, 3B, 5A, 6B, and 7B), the nucleotide sequences showed evidence of intraclonal heterogeneity, ie, clones of a given sample differed from the others in one or more nucleotides. To distinguish intraclonal heterogeneity from possible Taq error, we considered sequence heterogeneity as intraclonal divergence provided nucleic acid differences are found in more than one clones. Sequences with intraclonal heterogeneity are demonstrated in Figure 2 ▶ . In those four cases (Cases 1–3 and 5) where both FL and DLCL samples were mutated, mutations were different in the subsequent biopsy samples; however, in Cases 3 and 5 a single shared mutation was detected. From the pattern of shared and unique mutations, assuming that shared mutations represent single events and not independent mutations, genealogical trees of the evolution of tumor cells could be constructed. A representative example of a genealogical tree is shown in Figure 3 ▶ .
Table 2.
Distributions of Mutations within the BCL-6 5′ Noncoding Sequences in Seven Cases of Paired FL and Subsequent FL or DLCL Samples
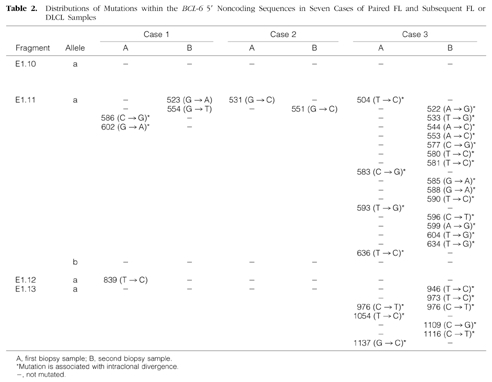
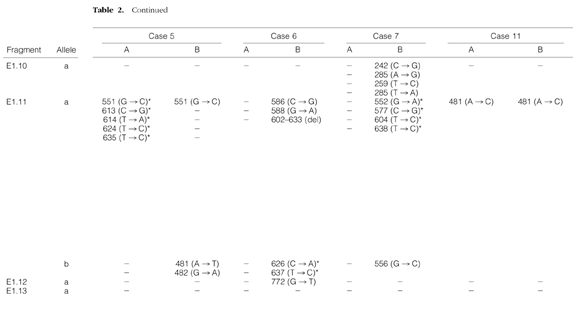
Figure 2.
Schematic representation of the BCL-6 5′ noncoding region diversification and sequence intraclonal heterogeneity in FL (A) and transformed DLCL (B) samples. Each sequence is represented as a horizontal line. The first nucleotide of the BCL-6 cDNA is designated as position +1. The arrows represent the positions of the primers. Within each sequence, nucleotide differences from the BCL-6 cDNA sequences are indicated by dots. The type and exact positions of the mutations are specified in Table 2 ▶ .
Figure 3.
Probable clonal relationship of FL and transformed DLCL clones in Case 3. The genealogical tree is constructed from the pattern of shared and unique mutations of E1.13 PCR fragment of the BCL-6 5′ noncoding sequences (Table 2 ▶ and Figure 2 ▶ ), assuming that shared mutations represent single events. The FL clones (A) are drawn as open circles and DLCL clones (B) are drawn as shaded circles. The number of mutations that separate each branch is given.
Analysis of the Sequence Polymorphism of the 5′ Noncoding Region of the BCL-6 Gene
In Cases 3 and 5, shared mutations of the 5′ noncoding region of the BCL-6 gene were detected in FL and subsequent DLCL samples at positions 976 (C → T) and 551 (G → C), respectively. To determine whether shared mutations are associated with sequence polymorphism or somatic mutation, we have analyzed the BCL-6 5′ noncoding region sequences in normal tissue samples of these two patients. The sequence analysis of the normal tissue samples revealed no alterations in the BCL-6 5′ noncoding sequences. Thus, these findings are consistent with our presumption that nucleotide sequence alterations shared by FL and DLCL samples in Cases 3 and 5 arose by somatic mutation.
Expression of BCL-6 Gene
Expression of the BCL-6 gene was detected by immunoreactivity of tumor cells by anti-BCL-6 mAb. The staining pattern on frozen tissue sections was nuclear. The percentage of the BCL-6-positive cells in FL and subsequent FL or DLCL samples is summarized in Table 1 ▶ . In all of the FL and in six of the DLCL samples, 100% of the tumor cells expressed the BCL-6 protein. In three transformed DLCL samples (Cases 5–7), the number of the BCL-6-positive cells reduced to 10 to 20%.
Discussion
We have analyzed sequential biopsy samples from 12 patients with FL that showed no histological transformation in subsequent biopsy specimens or underwent histological transformation to DLCL for structural alterations of the BCL-6 gene. We found no rearrangement of the BCL-6 gene in any of the samples, but multiple mutations of the 5′ noncoding sequences of the BCL-6 gene were detected in seven cases. In those cases where FL underwent histological transformation, these mutations generated considerable intraclonal divergence of the neoplastic clones in both FL and transformed DLCL. The histological transformation of FL was associated with the emergence of new subpopulations representing divergent clonotypes of the BCL-6 5′ noncoding sequences. These findings suggest that genetic diversity of the 5′ noncoding sequences of the BCL-6 gene is generated in the FL or DLCL clones and histological transformation is associated with clonotypic shift of the original FL clone.
Somatic mutation of the 5′ noncoding region of the BCL-6 gene has been described in B cell NHLs of the GC/post-GC origin. 23-25,32 These mutations are always found in a 3.5-kb region spanning the first noncoding exon and clustering in the 5′ region of the first intron, which may occur in the absence of physical linkage to antigen receptor loci. In the present study, we have revealed further characteristics of these mutations showing that mutation of the 5′ noncoding region of the BCL-6 gene is associated with considerable intraclonal heterogeneity in FL and transformed DLCL cells. This sequence heterogeneity within the tumor clones indicates that the tumor cells are still under the influence of the mutation mechanism after neoplastic transformation. Although intraclonal heterogeneity of the BCL-6 5′ noncoding sequences has not been reported in NHLs, several lines of evidence suggested such a possibility. In the majority of FLs, the PCR-SSCP analysis of the BCL-6 5′ noncoding sequences showed multiple variant fragments, suggesting subclonal variations of the neoplastic clone. 24 Furthermore, Migliazza et al 24 found additional mutations in subcloned products, which were interpreted as a possible consequence of Taq polymerase error. Although some of the nucleotide substitutions found in our study may be attributed to the infidelity of Taq polymerase, this explanation is unlikely to account for more than a few of the mutations. The ongoing mutation rate of approximately 0.3% in the BCL-6 5′ noncoding region of NHLs analyzed in this study is an order of magnitude greater than our Taq error rate of 0.03%, which was calculated from the analysis of unmutated BCL-6 5′ noncoding sequences in our laboratory. In addition to the high frequency, many of the uncommon mutations were shared by different clones of a given BCL-6 5′ noncoding sequence, which also indicates that they are most likely of lymphoma cell origin.
The follow-up sequence analysis of the 5′ noncoding region of the BCL-6 gene revealed considerable differences between sequences of FL and subsequent DLCL samples. These nucleotide differences ranged from two to 18 nucleotides. Because there were identical Ig gene rearrangements and identical t(14;18) breakpoints of the subsequent biopsy samples, mutated BCL-6 5′ noncoding sequences derived from a single neoplastic B cell clone in each case. The common clonal origin of the FL and DLCL cells is further supported by the findings that in two cases, single shared mutations were detected in the subsequent biopsy samples. Any nucleotide alterations not shared by both FL and DLCL cells necessarily accumulated during clonal evolution or histological transformation of the neoplastic cells. The extent and nature of BCL-6 mutations detected in this study is most similar to the hypermutation mechanism of IgV genes detected in the clonal evolution and histological transformation of FL. 33 This observation is also supported by our previous findings. The IgH gene mutational analysis performed in Case 5 showed similar type of intraclonal heterogeneity and clonal selection in the histological transformation of FL as BCL-6 5′ noncoding sequences in this study. 34
The functional significance of the intraclonal divergence of the BCL-6 5′ noncoding sequences and the pathogenic role of the intensive clonal selection of the mutated sequences in histological transformation of FL are unknown. Although functional characterization of BCL-6 is still in its early stages, it is thought that the BCL-6 5′ noncoding region contains important regulatory elements. Initial studies on tumor-derived BCL-6 alleles indicate that few mutations can significantly deregulate BCL-6 expression, whereas others are apparently functionally irrelevant or associated with silent alleles. 35 In three cases with FL that underwent histological transformation, the 100% of BCL-6 immunoreactivity of the FL cells reduced to 10 to 20% reactivity in the transformed DLCL sample. Although this finding suggest that the development of additional mutations in the BCL-6 5′ noncoding region may deregulate the expression of the BCL-6 gene, further functional analysis of the BCL-6 gene is required to demonstrate whether mutations detected in the DLCL samples cause biological and/or functional alterations of the tumor cells.
The intensive clonal selection of the mutated BLC-6 5′ noncoding sequences in histological transformation of FL suggests that they may have been selected based on their functional role. As a result of ongoing somatic mutation, the tumor cell population becomes heterogeneous, and a mutational variant having selective growth advantage compared to those of parental clones gives rise to the DLCL cell population. Functional selection of the neoplastic clones in clonal evolution and histological transformation of FLs has also been suggested by the mutational analysis of IgVH genes. 34,36-38 The pattern and distribution of these mutations suggest that antigen selection may play an important role in the clonal evolution and histological transformation of FL. Although the nature of antigens that appear to function through the Ig receptor expressed by NHLs is not known, self-antigens have been considered as potential candidates. 36 In this regard, it would be of particular interest to know the type of selective force that influences the BCL-6 gene in clonal selection and histological transformation of FL.
Ongoing somatic mutation of the BCL-6 5′ noncoding sequences and clonal selection of these mutations have also been detected in normal GC B cells. 32,39 The pattern of mutations is not clearly different in normal and malignant B cells, which suggests that mutations may not have a pathological effect. Furthermore, in our study ongoing mutations have been detected in the majority of neoplastic clones both before and after transformation of FL. These results may indicate that mutations of the BCL-6 5′ noncoding sequences represent genetic instability of the neoplastic cells. Because the majority of FLs, which eventually will transform to DLCL, shows mutations in the BCL-6 5′ noncoding region, it is possible that these mutations are simply a reflection of an increased genetic instability, which, in turn, may result in alterations of other oncogenes or tumor suppressor genes responsible for histological transformation of FL.
In conclusion, this study provides the first evidence that somatic mutation of the 5′ noncoding sequences of the BCL-6 gene may occur after neoplastic transformation of FL and DLCL, and demonstrates that histological transformation of FL is associated with clonal selection of the mutated clones. However, it remains to be determined whether these mutations are responsible for histological transformation of FLs or are simply a reflection of increased genetic instability.
Footnotes
Address reprint requests to András Matolcsy, M.D., Ph.D., Department of Pathology, University Medical School of Pécs, H-7624 Pécs, Szigeti út 12, Hungary. E-mail: amatolc@pathology.pote.hu.
Supported by grants from the Hungarian Ministry of Culture and Education FKFP 0931/97, OTKA T023588 OTKA TO25782, and ETT 365/96 (to A. M.) and by grant CA34233 from the National Institutes of Health (to R. A. W.).
References
- 1.Harris NL, Jaffe ES, Stein H, Banks PM, Chan JK, Cleary ML, Delsol G, Wolf-Peeters C, Falini B, Gatter KC, Grogan TM, Isaacson PG, Knowles DM, Mason DY, Muller-Hermelink HK, Pileri SA, Piris MA, Ralfkiaer E, Warnke RA: A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood 1994, 84:1361-1392 [PubMed] [Google Scholar]
- 2.Garvin AJ, Simon RM, Osborne CK, Merrill J, Young RC, Berard CW: An autopsy study of histologic progression in non-Hodgkin’s lymphomas: 192 cases from the National Cancer Institute. Cancer 1983, 52:393-398 [DOI] [PubMed] [Google Scholar]
- 3.Oviatt DL, Cousar JB, Collins RD, Flexner JM, Stein RS: Malignant lymphomas of follicular center cell origin in humans. V. Incidence, clinical features, and prognostic implications of transformation of small cleaved cell nodular lymphoma. Cancer 1984, 53:1109-1114 [DOI] [PubMed] [Google Scholar]
- 4.Ersbøll J, Schultz HB, Pedersen-Bjergaard J, Nissen NI: Follicular low-grade non-Hodgkin’s lymphoma: long-term outcome with or without tumor progression. Eur J Haematol 1989, 42:155-163 [DOI] [PubMed] [Google Scholar]
- 5.Cleary ML, Sklar J: Nucleotide sequence of a t(14;18) chromosomal breakpoint in follicular lymphoma and demonstration of a breakpoint-cluster region near a transcriptionally active locus on chromosome 18. Proc Natl Acad Sci USA 1985, 82:7439-7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleary ML, Galili N, Sklar J: Detection of a second t(14;18) breakpoint cluster region in human follicular lymphomas. J Exp Med 1986, 164:315-320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richardson ME, Chen QG, Filippa DA, Offit K, Hampton A, Koduru PR, Jhanwar SC, Lieberman PH, Clarkson BD, Chaganti RSK: Intermediate- to high-grade histology of lymphomas carrying t(14;18) is associated with additional nonrandom chromosome changes. Blood 1987, 70:444-447 [PubMed] [Google Scholar]
- 8.Armitage JO, Sanger WG, Weisenburger DD, Harrington DS, Linder J, Bierman PJ, Vose JM, Purtilo DT: Correlation of secondary cytogenetic abnormalities with histologic appearance in non-Hodgkin’s lymphomas bearing t(14;18)(q32;q21). J Natl Cancer Inst 1988, 80:576-580 [DOI] [PubMed] [Google Scholar]
- 9.Whang-Peng J, Knutsen T, Jaffe ES, Steinberg SM, Raffeld M, Zhao WP, Duffey P, Condron K, Yano T, Longo DL: Sequential analysis of 43 patients with non-Hodgkin’s lymphoma: clinical correlations with cytogenetic, histologic, immunophenotyping, and molecular studies. Blood 1995, 85:203-216 [PubMed] [Google Scholar]
- 10.Bernell P, Jacobsson B, Liliemark J, Hjalmar V, Arvidsson I, Hast R: Gain of chromosome 7 marks the progression from indolent to aggressive follicle centre lymphoma and is a common finding in patients with diffuse large B-cell lymphoma: a study by FISH. Br J Haematol 1998, 101:487-491 [DOI] [PubMed] [Google Scholar]
- 11.Elenitoba-Johnson KSJ, Gascoyne RD, Lim MS, Chhanabai M, Jaffe ES, Raffeld M: Homozygous deletions at chromosome 9p21 involving p16 and p15 are associated with histologic progression in follicle center lymphoma. Blood 1998, 91:4677-4685 [PubMed] [Google Scholar]
- 12.Yano T, Jaffe ES, Longo DL, Raffeld M: MYC rearrangements in histologically progressed follicular lymphomas. Blood 1992, 80:758-767 [PubMed] [Google Scholar]
- 13.Lo Coco F, Gaidano G, Louie DC, Offit K, Chaganti RSK, Dalla-Favera R: p53 mutations are associated with histologic transformation of follicular lymphoma. Blood 1993, 82:2289–2295 [PubMed]
- 14.Sander CA, Yano T, Clark HM, Harris C, Longo DL, Jaffe ES, Raffeld M: p53 mutation is associated with progression in follicular lymphomas. Blood 1993, 82:1994-2004 [PubMed] [Google Scholar]
- 15.Matolcsy A, Casali P, Warnke RA, Knowles DM: Morphologic transformation of follicular lymphoma is associated with somatic mutation of the translocated bcl-2 gene. Blood 1996, 88:3937-3944 [PubMed] [Google Scholar]
- 16.Pinyol M, Cobo F, Bea S, Jares P, Nayach I, Fernandez PL, Montserrat E, Cardesa A, Campo E: p16INK4a gene inactivation by deletions, mutations, and hypermethylation is associated with transformed and aggressive variants of non-Hodgkin’s lymphomas. Blood 1998, 91:2977-2984 [PubMed] [Google Scholar]
- 17.Chang C-C, Ye BH, Chaganti RSK, Dalla-Favera R: BCL6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc Natl Acad Sci USA 1996, 93:6947-6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye BH, Cattoretti G, Shen Q, Zhang J, Hawe N, de Waard R, Leung C, Nouri-Shirazi M, Orazi A, Chaganti RSK, Rothman P, Stall AM, Pandolfi PP, Dalla-Favera R: The BCL-6 proto-oncogene controls germinal-centre formation, and Th2-type inflammation. Nat Genet 1997, 16:161-170 [DOI] [PubMed] [Google Scholar]
- 19.Ye BH, Lista F, Lo Coco F, Knowles DM, Offit K, Chaganti RSK, Dalla-Favera R: Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science 1993, 262:747–749 [DOI] [PubMed]
- 20.Lo Coco F, Ye BH, Lista F, Corradini P, Offit K, Knowles DM, Chaganti RSK, Dalla-Favera R: Rearrangements of BCL6 gene in diffuse large cell non-Hodgkin’s lymphoma. Blood 1994, 83:1757–1759 [PubMed]
- 21.Bastard C, Deweindt C, Kerckaert JP, Lenormand B, Rossi A, Pezzella F, Fruchart C, Duval C, Monconduit M, Tilly H: LAZ3 rearrangements in non-Hodgkin’s lymphoma: correlation with histology, immunophenotype, karyotype, and clinical outcome in 217 patients. Blood 1994, 83:2423-2427 [PubMed] [Google Scholar]
- 22.Muramatsu M, Akasaka T, Kadowaki N, Ohno H, Yamabe H, Edamura S, Dor S, Mori T, Okuma M, Fukuhara S: Rearrangement of the BCL6 gene in B-cell lymphoid neoplasms: comparison with lymphomas associated with BCL2 rearrangement. Br J Haematol 1996, 93:911-920 [DOI] [PubMed] [Google Scholar]
- 23.Peng H-Z, Du M-Q, Koulis A, Aiello A, Dogan A, Pan L-X, Isaacson P: Nonimmunglobulin gene hypermutation in germinal center B cells. Blood 1999, 93:2167–2172 [PubMed]
- 24.Migliazza A, Martinotti S, Chen W, Fusco C, Ye BH, Knowles DM, Offit K, Chaganti RSK, Dalla-Favera R: Frequent somatic hypermutation of the 5′ noncoding region of the BCL6 gene in B-cell lymphoma. Proc Natl Acad Sci USA 1995, 92:12520-12524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Capello D, Carbone A, Pastore C, Gloghini A, Saglio G, Gaidano G: Point mutations of the BCL-6 gene in Burkitt’s lymphoma. Br J Haematol 1997, 99:168-170 [DOI] [PubMed] [Google Scholar]
- 26.Gaidano G, Carbone A, Pastore C, Capello D, Migliazza A, Gloghini A, Roncella S, Ferrarini M, Saglio G, Dalla-Favera R: Frequent mutation of the 5′ noncoding region of the BCL-6 gene in acquired immunodeficiency syndrome-related non-Hodgkin’s lymphomas. Blood 1997, 89:3755-3762 [PubMed] [Google Scholar]
- 27.Cesarman E, Chadburn A, Liu Y-F, Migliazza A, Dalla-Favera R, Knowles DM: BCL-6 gene mutations in posttransplantation lymphoproliferative disorders predict response to therapy and clinical outcome. Blood 1998, 92:2294-2302 [PubMed] [Google Scholar]
- 28.Miller SA, Dykes DD, Polesky HF: A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acid Res 1988, 16:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matolcsy A, Chadburn A, Knowles DM: De novo CD5-positive, and Richter’s syndrome-associated diffuse large B cell lymphomas are genotypically distinct. Am J Pathol 1995, 147:207-216 [PMC free article] [PubMed] [Google Scholar]
- 30.Flenghi L, Ye BH, Fizzotti M, Bigerna B, Cattoretti G, Venturi S, Pacini R, Pileri S, Lo Coco P, Pescarmona E, Pelicci P-G, Dalla-Favera R, Falini B: A specific monoclonal antibody (GP-B6) detects expression of the BCL-6 protein in germinal center B cells. Am J Pathol 1995, 147:405–411 [PMC free article] [PubMed]
- 31.Cordell JL, Falini B, Erber WN, Ghosh AK, Abdulaziz Z, MacDonald S, Pulord KAF, Stein H, Mason DY: Immunoenzymatic labeling of monoclonal antibodies using immune complexes of alkaline phosphatase and monoclonal anti-alkaline phosphatase (APAAP complexes). J Histochem Cytochem 1984, 32:219-229 [DOI] [PubMed] [Google Scholar]
- 32.Pasqualucci L, Migliazza A, Fracchiolla N, William C, Neri A, Baldini L, Chaganti RSK, Klein U, Küppers R, Rajewsky K, Dalla-Favera R: BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc Natl Acad Sci USA 1998, 95:11816-11821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zelenetz AD, Chen TT, Levy R: Histologic transformation of follicular lymphoma to diffuse lymphoma represents tumor progression by a single malignant B cell. J Exp Med 1991, 173:197-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matolcsy A, Schatter EJ, Knowles DM, Casali P: Clonal evolution of B cells in transformation from low- to high-grade lymphoma. Eur J Immunol 1999, 29:1253-1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Migliazza A, Ye BH, Fracchiolla N, Neri A, Baldini L, Chaganti RSK, Dalla-Favera R: Mutations of the 5′ non-coding region of BCL-6 are common in B cell malignancies and can deregulate BCL-6 expression. Blood 1997, 90(suppl 1):177a(abstr.) [Google Scholar]
- 36.Friedman DF, Cho EA, Goldman J, Carmack CE, Besa EC, Hardy RR, Silberstein EL: The role of clonal selection in the pathogenesis of an autoreactive human B cell lymphoma. J Exp Med 1991, 174:525-537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bahler DW, Levy R: Clonal evolution of a follicular lymphoma: evidence for antigen selection. Proc Natl Acad Sci USA 1992, 89:6770-6774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu D, Hawkins RE, Hamblin TJ, Stevenson FK: Clonal history of a human follicular lymphoma as revealed in the immunoglobulin variable region genes. Br J Haematol 1994, 86:505-512 [DOI] [PubMed] [Google Scholar]
- 39.Shen HM, Peters A, Baron B, Zhu X, Storb U: Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science 1998, 280:1750-1752 [DOI] [PubMed] [Google Scholar]