Abstract
As an approach to both explore the physical/chemical parameters that drive molecular self-assembly and to generate novel protein oligomers, we have developed a procedure to generate protein dimers from monomeric proteins using computational protein docking and amino acid sequence design. A fast Fourier transform-based docking algorithm was used to generate a model for a dimeric version of the 56-amino-acid β1 domain of streptococcal protein G. Computational amino acid sequence design of 24 residues at the dimer interface resulted in a heterodimer comprised of 12-fold and eightfold variants of the wild-type protein. The designed proteins were expressed, purified, and characterized using analytical ultracentrifugation and heteronuclear NMR techniques. Although the measured dissociation constant was modest (∼300 μM), 2D-[1H,15N]-HSQC NMR spectra of one of the designed proteins in the absence and presence of its binding partner showed clear evidence of specific dimer formation.
Keywords: de novo protein–protein interface, computational protein design, geometric recognition algorithm, protein G, heterodimer, NMR, docking
Molecular self-assembly is the spontaneous association of molecules into stable, structurally well-defined complexes. All major cellular processes depend on the precise, highly specific self-assembly of proteins into functional arrays. Understanding and controlling the physical/chemical parameters that drive protein association is a major goal of protein biochemistry. To date, much progress has been made in this area by analyzing the large body of data collected on naturally occurring protein–protein interfaces (Clackson and Wells 1995; Jones and Thornton 1996; Janin and Seraphin 2003; Janin and Wodak 2003; Nooren and Thornton 2003a,b).
The field of protein design is uniquely positioned to complement these efforts with an inverse approach. That is, instead of analyzing and/or predicting the structures of native complexes, we can explore the physical chemistry of self-assembly through the de novo design of self-assembling protein complexes from previously monomeric proteins. Moreover, the ability to direct a designed protein to bind a target protein in a site-specific manner has potential therapeutic as well as other technological applications.
Computational protein design methods have recently made significant progress toward engineering novel protein–protein interfaces (Kortemme and Baker 2004). For example, Shifman and Mayo used computational methods to generate calmodulin variants with enhanced binding specificity (Shifman and Mayo 2002, 2003); Bolon et al. (2005) re-engineered a protein homodimer into a heterodimer; Havranek and Harbury (2003) generated GCN4-like coiled-coil variants that included the direct consideration of negative design; and Baker, Stoddard, and coworkers generated DNase-inhibitor protein pairs with altered specificity and an artificial endonuclease by fusing two domains from naturally occurring, multidomain endocucleases followed by re-engineering of the newly formed interface (Chevalier et al. 2002; Kortemme et al. 2004).
Here we report the de novo design of a protein–protein heterodimer that was generated by first computationally docking the β1 domain of the streptococcal protein G (GB1) to itself in a structurally specific fashion followed by computational design of the interfacial amino acids so as to drive complex formation (Fig. 1). Wild-type GB1 is a monomeric, 56 amino acid protein domain that has been extensively characterized and used in previous computational protein design studies (Gronenborn et al. 1991; Malakauskas and Mayo 1998).
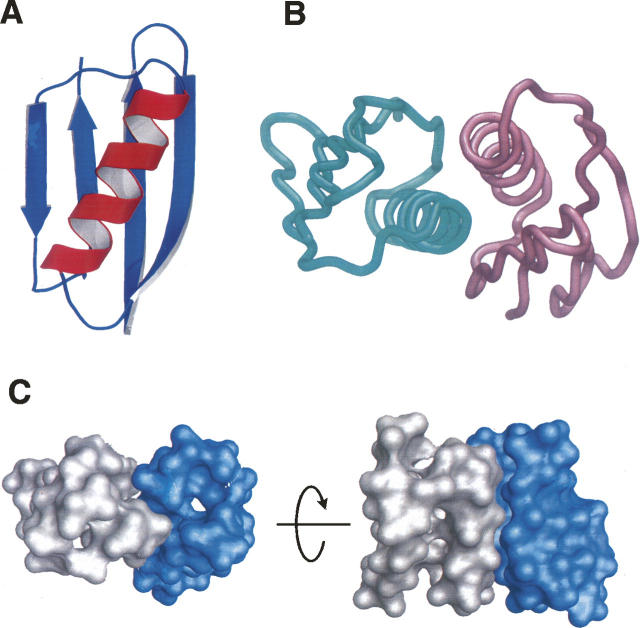
Figure 1.
(A) Ribbon representation of the GB1 monomer structure. (B) Tube representation of the docked GB1 dimer model; (left) GB1A; (right) GB1B. (C) Surface representation of the docked GB1 dimer model showing surface complementarity of the Cβ-truncated monomers used in the docking calculation; (left) GB1A; (right) GB1B.
Results and Discussion
For this study, GB1 dimer orientations were generally restricted to helix-to-helix arrangements. The orientation used for the sequence design calculation (Fig. 1B) corresponds to the docked complex of highest surface complementarity as determined from a rotation-translation search using a surface recognition algorithm and Fourier correlation techniques for evaluating the translational degrees of freedom (Katchalski-Katzir et al. 1992; Gabb et al. 1997). Since the amino acid sequence at the protein–protein interface is determined subsequent to generating the docked orientation, a reduced amino acid side-chain representation was developed by analyzing a series of naturally occurring protein dimers that had their side chains artificially restricted to the Cβ atom. The primary outcome of this analysis was that the effective atomic radii used in the docking procedure were increased from 1.8 Å to 2.15 Å in order to account for the absence of side-chain atoms (Huang et al. 2005).
Docking was carried out first by positioning monomer A, called GB1A, such that its helix axis was along the Y-axis and its β-sheet plane was parallel to the X–Y plane. Monomer B, called GB1B, was then created from monomer A by 180° rotations about the Y- and Z-axes resulting in a head-to-tail, helix-to-helix orientation. This orientation was chosen to direct contacts on mainly helices, as computational designs are more reliable on helices than sheets. The docking search was performed in a 3D grid that was 64 Å on a side using a 0.5 Å step size for the translation search and a 5° increment for the rotation search.
In the second step of the design process, a total of 24 residue positions (13 on GB1A and 11 on GB1B) were considered for sequence optimization using the ORBIT (optimization of rotomers by iterative techniques) suite of protein design programs (Dahiyat and Mayo 1997a). Following the same design principle for designing thermally stable protein variants, residues that are buried in the complex were restricted as hydrophobic residues. This implicitly captures negative design since the created hydrophobic patches destabilize the monomers, coupling dimerization with stability. Of these 24 positions, the 15 “core” positions were restricted to a set of seven hydrophobic amino acids (A, V, L, I, F, Y, and W), and the nine surface positions were restricted to a set of 10 polar amino acids (A, S, T, D, N, H, E, Q, K, and R) resulting in a combinatorial complexity of ∼1021 amino acid sequence combinations. The amino acids at the 88 remaining unoptimized positions were retained as in the wild-type sequence. Since symmetry restraints were not imposed during the docking or sequence design phases, the sequence design step resulted in a pair of protein monomers that had different sets of mutations corresponding to the formation of a heterodimer complex. GB1A is a 12-fold mutant: T16F, T18A, V21E, T25L, K28Y, V29I, K31R, Q32A, Y33L, N35K, D36A, and N37Q. GB1B is an eightfold mutant: A23I, E27A, K28D, K31A, N35A, D40K, Y45A, and D47E. In addition to the largely hydrophobic interface, there are six cross-dimer polar interactions in the structural model. Three of them are side-chain/side-chain interactions: E21A to K40B, D22A to K40B, and Q37A to E47B. The other three are side-chain/backbone interactions: K35A to A45B carbonyl oxygen, D28B to E19A amide hydrogen, and E47B to A36A carbonyl oxygen.
Genes for GB1A and GB1B were constructed by inverse-PCR mutagenesis and separately expressed in Escherichia coli and purified using standard procedures. The expression level for GB1A was similar to that for GB1 (∼40 mg/L), but the yield of GB1B was ∼10-fold lower (∼4 mg/L), consistent with thermal melting temperatures of >100°C and ∼37°C for GB1A and GB1B, respectively (data published previously as part of a stability study) (Barakat et al. 2007). The thermal stability for GB1 is ∼83°C. Our computed energies for GB1A and GB1B in isolation cannot account for the divergent stability observed experimentally. It is unclear as to why GB1A with an exposed hydrophobic patch, which is scored unfavorably in our design algorithm, is hyperthermophilic. GB1B, on the other hand, is likely to be destabilized by the Y45A mutation, as it is known to affect the folding kinetics of the hairpin in protein G (Honda et al. 2000; Kobayashi et al. 2000). In solution at high concentrations (∼1.3 mM), GB1B forms macroscopic fibrils; however, fibril formation does not occur in equimolar solutions of GB1A and GB1B (Shukla et al. 2004).
Sedimentation equilibrium ultracentrifugation suggested a modest dissociation constant of ∼300 μM, but a detailed global analysis was confounded by the non-ideal behavior of GB1B.
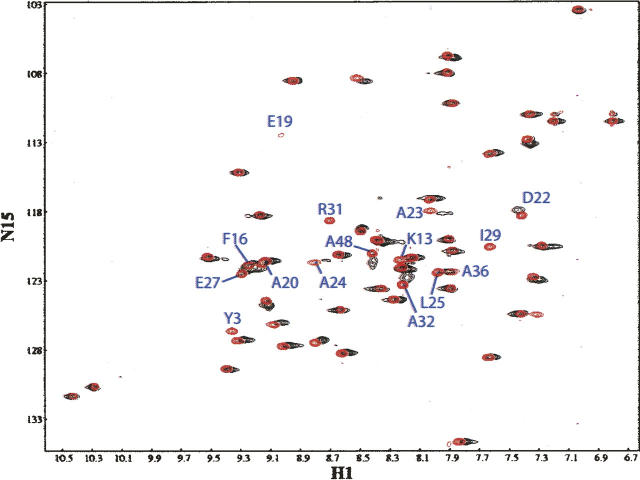
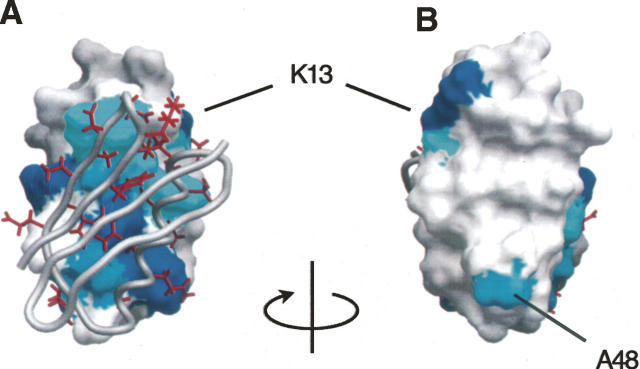
NMR chemical shift perturbation analysis supports complex formation at the designed interface (Fig. 2). Backbone [1H,15N] resonance assignments of free 15N-labeled GB1A were determined using 3D-[1H,15N]-NOESY-HSQC and 3D-[1H,15N]-TOCSY-HSQC. 2D-[1H,15N]-HSQC spectra of 15N-labeled GB1A in the absence and presence of equimolar amounts of unlabeled GB1B show clear signs of specific complex formation with 20 resonances displaying significant perturbation in chemical shift and/or peak intensity (peaks no longer observable: Y3, K13, E19, I29, and R31; peaks with significantly diminished intensity and/or chemical shift perturbation: A20, D22, A23, A24, L25, E27, A32, and A48; peaks with small changes in intensity and/or chemical shift: G14, F16, A26, F30, L33, K35, and A36). With few exceptions (Y3, K13, and A48) the residues corresponding to the perturbed resonances map to the designed dimer interface (Fig. 3). The peaks rendered nonobservable are probably the result of exchange broadening due to fluctuations between two or more states on a microsecond-to-millisecond timescale and imply that the complex may experience a relatively rapid exchange between free and bound states. This explanation appears likely especially in light of the relatively modest binding affinity measured by analytical ultracentrifugation.
Figure 2.
[15N, 1H] HSQC spectra of uniformly enriched 15N-GB1A alone (in red) and in the presence of equimolar quantities of unlabeled GB1B (in black). Example 15N-monomer-A peaks that are nonobservable or exhibit chemical shift perturbations upon complex formation are labeled blue.
Figure 3.
(A) Chemical shift perturbations mapped to the surface of GB1A in the docked model orientation together with GB1B shown as a gray backbone worm on which the interfacial side chains are colored red. (Dark blue) Residues with nonobservable [15N, 1H] HSQC peaks; (lighter blue) those that exhibit chemical shift changes. (B) 180° rotation of A.
Materials and Methods
Assessment of de novo docking parameters
To not bias the docking results with wild-type amino acids, it was necessary to prune all side chains to the Cβ atoms (excluding glycines), and therefore it was not possible to use the geometric recognition algorithm in its original form (Huang et al. 2005). To ascertain optimal discretization values for the proper spacing between the docked models, an extensive analysis was performed on the crystal structures of several natural complexes, the goal of which was to extract from the natural complexes optimized parameters that would provide proper interfacial volume for successful side-chain selection. The accession codes of the 18 PDB files used to extract de novo docking parameters are 1ATN, 1BRS, 1DQJ, 1DZB, 1FCC, 1FDL, 1HRP, 1IGC, 1JHL, 1JTO, 1LPA, 1MLC, 1NCD, 1VFB, 2BTF, 2JEL, 3HFL, and 3HFM.
The distances for each complex that corresponded to the largest correlation were statistically analyzed and resulted in an average value of 2.05 Å ± 0.48 Å. Owing to the relatively large variance in the values for the different complexes, a series of GB1 to GB1 docking calculations were separately conducted with radial distances of 2.00, 2.05, 2.10, 2.15, and 2.20 Å. The resulting docked complexes were analyzed with 3D molecular visualization tools (e.g., GRASP, MOLMOL), and it was concluded that the complex that corresponded to the 2.15 Å radial distance had the best interfacial volume in the set before proceeding with design. The decision was made by visually inspecting the interface for close contacts that often resulted from small radial distances, and by comparing the top ranking dimers on surface complementarity whereby unreasonably large gaps can usually be identified in calculations that do not converge to a specific orientation. The radial distance of 2.15 Å gave a very specific docking orientation with appropriate interface volume and was therefore used to produce the orientation used for design.
Computational protein design
Details of the side-chain selection process performed on interfacial positions including the potential energy function and parameters for solvation, hydrogen bonding, and van der Waals interactions are essentially the same as described previously (Dahiyat and Mayo 1996, 1997a,b; Dahiyat et al. 1997; Gordon et al. 1999). Reclassification of interfacial residues was accomplished with the RESCLASS algorithm (Dahiyat and Mayo 1997a). By comparing monomers in isolation and in the docked orientation, residues reclassified from “surface” to “core” and from “surface” to “boundary” by RESCLASS were considered in the design. Rotamer libraries used during the side-chain selection process were based on the backbone-dependent library of Dunbrack Jr. and Karplus (1993). The Dead-End Elimination theorem (Desmet et al. 1992; Gordon et al. 2003) was used to select for the specific side-chain rotamers that exist in the lowest possible calculated structure (i.e., the global minimum energy conformation).
Protein expression and purification
Synthetic DNA oligonucleotides were used for recursive PCR synthesis of the genes for GB1A and GB1B. One modification to the ORBIT selected sequence was made: Position 28 of GB1A was changed from tryptophan to tyrosine so as to reduce the hydrophobicity of regions that were not fully buried at the interface. The genes were cloned into pET-11a (Novagen), and recombinant protein was expressed by IPTG induction in BL21(DE3) hosts (Invitrogen). The proteins were isolated using a freeze/thaw method (Johnson and Hecht 1994), and purification was accomplished with reverse-phase HPLC using a linear 1% min−1 acetonitrile/water gradient containing 0.1% TFA. The yield of purified protein from expression in rich media was ∼4 mg/L of bacterial growth for GB1B and ∼40 mg/L for GB1A. Labeled GB1A protein, for NMR studies, was prepared with standard M9 minimal media using 15N-ammonium sulfate (2 g/L). Protein purity was verified with standard SDS-PAGE and reverse-phase HPLC, and the correct molecular weight was confirmed by mass spectrometry.
Analytical ultracentrifugation
Sedimentation equilibrium experiments were conducted in a Beckman XL-I Ultima analytical ultracentrifuge equipped with absorbance optics. Runs were carried out at 28,000, 40,000, and 48,000 rpm, at 20°C. Three separate solutions consisting of different concentrations for both free proteins plus the complex were prepared to achieve OD280 readings of ∼0.15, 0.25, and 0.4. The protein concentrations were ∼36 μM, ∼60 μM, and ∼96 μM in a total volume of 110 μL containing 50 mM NaCl and 50 mM NaPi (pH ∼ 6.5). Data were processed using the WinNONLIN software from the National Analytical Ultracentrifuge Facility following standard global nonlinear fitting methods (Lebowitz et al. 2002). The sedimentation equilibrium data were best described by a monomer–dimer self-association model with an estimated K d of 331 μM.
NMR spectroscopy
NMR spectra were collected at 293 K on a Varian UnityPlus 600 MHz spectrometer equipped with an HCN-triple-resonance probe with triple-axis pulse field gradients. Protein concentrations were ∼1.25 mM in 50 mM sodium phosphate (pH ∼ 6.5). Standard 2D-[1H, 15N] HSQC spectra were collected on free 15N-GB1A and 15N-GB1A in a 1:1 stoichiometric complex with unlabeled GB1B and standard 3D-[1H, 15N]-NOESY-HSQC and 3D-[1H, 15N]-TOCSY-HSQC were collected on free 15N-GB1A. Varian data processing software and the program NMRPipe (Delaglio et al. 1995) were used to process the NMR data, and the program NMRView (One Moon Scientific, Inc.) was used to analyze and assign the various spectra.
Electronic supplemental material
The sedimentation equilibrium data and their fits were included in the Supplemental material.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute, the Ralph M. Parsons Foundation, DARPA, the Institute for Collaborative Biotechnologies through grant DAAD19-03-D-0004 from the U.S. Army Research Office, and an IBM Shared University Research Grant.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: John J. Love, Department of Chemistry and Biochemistry, San Diego State University, San Diego, CA 92182, USA; e-mail: jlove@sciences.sdsu.edu; or Stephen L. Mayo, California Institute of Technology, MC 114-96, Pasadena, CA 91125, USA; e-mail: steve@mayo.caltech.edu; fax: (626) 568-0934.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073125207.
References
- Barakat N.H., Barakat, N.H., Carmody, L.J., and Love, J.J. 2007. Exploiting elements of transcriptional machinery to enhance protein stability. J. Mol. Biol. 366: 103–116. doi: 10.1016/j.jmb.2006.10.091. [DOI] [PubMed] [Google Scholar]
- Bolon D.N., Grant, R.A., Baker, T.A., and Sauer, R.T. 2005. Specificity versus stability in computational protein design. Proc. Natl. Acad. Sci. 102: 12724–12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier B.S., Kortemme, T., Chadsey, M.S., Baker, D., Monnat, R.J., and Stoddard, B.L. 2002. Design, activity, and structure of a highly specific artificial endonuclease. Mol. Cell 10: 895–905. [DOI] [PubMed] [Google Scholar]
- Clackson T. and Wells, J.A. 1995. A hot-spot of binding-energy in a hormone–receptor interface. Science 267: 383–386. [DOI] [PubMed] [Google Scholar]
- Dahiyat B.I. and Mayo, S.L. 1996. Protein design automation. Protein Sci. 5: 895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiyat B.I. and Mayo, S.L. 1997a. De novo protein design: Fully automated sequence selection. Science 278: 82–87. [DOI] [PubMed] [Google Scholar]
- Dahiyat B.I. and Mayo, S.L. 1997b. Probing the role of packing specificity in protein design. Proc. Natl. Acad. Sci. 94: 10172–10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiyat B.I., Gordon, D.B., and Mayo, S.L. 1997. Automated design of the surface positions of protein helices. Protein Sci. 6: 1333–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Desmet J., De Maeyer, M., Hazes, B., and Lasters, I. 1992. The dead-end elimination theorem and its use in protein side-chain positioning. Nature 356: 539–542. [DOI] [PubMed] [Google Scholar]
- Dunbrack R.L. and Karplus, M. 1993. Backbone-dependent rotamer library for proteins. Application to side-chain prediction. J. Mol. Biol. 230: 543–574. [DOI] [PubMed] [Google Scholar]
- Gabb H.A., Jackson, R.M., and Sternberg, M.J. 1997. Modelling protein docking using shape complementarity, electrostatics and biochemical information. J. Mol. Biol. 272: 106–120. [DOI] [PubMed] [Google Scholar]
- Gordon D.B., Marshall, S.A., and Mayo, S.L. 1999. Energy functions for protein design. Curr. Opin. Struct. Biol. 9: 509–513. [DOI] [PubMed] [Google Scholar]
- Gordon D.B., Hom, G.K., Mayo, S.L., and Pierce, N.A. 2003. Exact rotamer optimization for protein design. J. Comput. Chem. 24: 232–243. [DOI] [PubMed] [Google Scholar]
- Gronenborn A.M., Filpula, D.R., Essig, N.Z., Achari, A., Whitlow, M., Wingfield, P.T., and Clore, G.M. 1991. A novel, highly stable fold of the immunoglobulin binding domain of streptococcal protein G. Science 253: 657–661. [DOI] [PubMed] [Google Scholar]
- Havranek J.J. and Harbury, P.B. 2003. Automated design of specificity in molecular recognition. Nat. Struct. Biol. 10: 45–52. [DOI] [PubMed] [Google Scholar]
- Honda S., Kobayashi, N., and Munekata, E. 2000. Thermodynamics of a β-hairpin structure: Evidence for cooperative formation of folding nucleus. J. Mol. Biol. 295: 269–278. doi: 10.1006/jmbi.1999.3346. [DOI] [PubMed] [Google Scholar]
- Huang P.S., Love, J.J., and Mayo, S.L. 2005. Adaptation of a fast Fourier transform-based docking algorithm for protein design. J. Comput. Chem. 26: 1222–1232. [DOI] [PubMed] [Google Scholar]
- Janin J. and Seraphin, B. 2003. Genome-wide studies of protein–protein interaction. Curr. Opin. Struct. Biol. 13: 383–388. [DOI] [PubMed] [Google Scholar]
- Janin J. and Wodak, S.J. 2003. Protein modules and protein–protein interaction—Introduction. Adv. Protein Chem. 61: 1–8. [DOI] [PubMed] [Google Scholar]
- Johnson B.H. and Hecht, M.H. 1994. Recombinant proteins can be isolated from E. coli cells by repeated cycles of freezing and thawing. Biotechnology (N. Y.) 12: 1357–1360. [DOI] [PubMed] [Google Scholar]
- Jones S. and Thornton, J.M. 1996. Principles of protein–protein interactions. Proc. Natl. Acad. Sci. 93: 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katchalski-Katzir E., Shariv, I., Eisenstein, M., Friesem, A.A., Aflalo, C., and Vakser, I.A. 1992. Molecular surface recognition: Determination of geometric fit between proteins and their ligands by correlation techniques. Proc. Natl. Acad. Sci. 89: 2195–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N., Honda, S., Yoshii, H., and Munekata, E. 2000. Role of side-chains in the cooperative β-hairpin folding of the short C-terminal fragment derived from streptococcal protein G. Biochemistry 39: 6564–6571. [DOI] [PubMed] [Google Scholar]
- Kortemme T. and Baker, D. 2004. Computational design of protein–protein interactions. Curr. Opin. Chem. Biol. 8: 91–97. [DOI] [PubMed] [Google Scholar]
- Kortemme T., Joachimiak, L.A., Bullock, A.N., Schuler, A.D., Stoddard, B.L., and Baker, D. 2004. Computational redesign of protein–protein interaction specificity. Nat. Struct. Mol. Biol. 11: 371–379. [DOI] [PubMed] [Google Scholar]
- Lebowitz J., Lewis, M.S., and Schuck, P. 2002. Modern analytical ultracentrifugation in protein science: A tutorial review. Protein Sci. 11: 2067–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakauskas S.M. and Mayo, S.L. 1998. Design, structure and stability of a hyperthermophilic protein variant. Nat. Struct. Biol. 5: 470–475. [DOI] [PubMed] [Google Scholar]
- Nooren I.M.A. and Thornton, J.M. 2003a. Diversity of protein–protein interactions. EMBO J. 22: 3486–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nooren I.M.A. and Thornton, J.M. 2003b. Structural characterisation and functional significance of transient protein–protein interactions. J. Mol. Biol. 325: 991–1018. [DOI] [PubMed] [Google Scholar]
- Shifman J.M. and Mayo, S.L. 2002. Modulating calmodulin binding specificity through computational protein design. J. Mol. Biol. 323: 417–423. [DOI] [PubMed] [Google Scholar]
- Shifman J.M. and Mayo, S.L. 2003. Exploring the origins of binding specificity through the computational redesign of calmodulin. Proc. Natl. Acad. Sci. 100: 13274–13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla U.J., Marino, H., Huang, P.S., Mayo, S.L., and Love, J.J. 2004. A designed protein interface that blocks fibril formation. J. Am. Chem. Soc. 126: 13914–13915. [DOI] [PubMed] [Google Scholar]