Abstract
The crystal structures of the natural and recombinant antiviral lectin scytovirin (SVN) were solved by single-wavelength anomalous scattering and refined with data extending to 1.3 Å and 1.0 Å resolution, respectively. A molecule of SVN consists of a single chain 95 amino acids long, with an almost perfect sequence repeat that creates two very similar domains (RMS deviation 0.25 Å for 40 pairs of Cα atoms). The crystal structure differs significantly from a previously published NMR structure of the same protein, with the RMS deviations calculated separately for the N- and C-terminal domains of 5.3 Å and 3.7 Å, respectively, and a very different relationship between the two domains. In addition, the disulfide bonding pattern of the crystal structures differs from that described in the previously published mass spectrometry and NMR studies.
Keywords: lectins, new fold, anomalous scattering, scytovirin
Scytovirin (SVN) is a potent antiviral lectin isolated from the cyanobacterium Scytonema varium (Bokesch et al. 2003) and subsequently produced as a recombinant protein in Escherichia coli (Xiong et al. 2006a). SVN has been reported to exhibit significant activity against human immunodeficiency virus (HIV) (Bokesch et al. 2003; Xiong et al. 2006b). Such potency makes SVN a promising candidate for development as a pharmaceutical agent for prevention of viral infection, specifically in the form of a female-controlled, anti-HIV microbicide.
SVN consists of a single polypeptide chain containing 95 amino acids, with a highly conserved internal repeat. Residues 3–42 (structural domain 1; SD1) and 51–90 (SD2) are 90% identical (36 out of 40), with the remaining three maintaining a similar character and only one significantly different (Fig. 1). Solution structure of SVN obtained by NMR (PDB code 2jmv) (McFeeters et al. 2007) postulated a novel fold for this protein, in which the two halves of the polypeptide create globally similar domains, although large local differences led to the interdomain RMSD of ∼5 Å. It was reported that SVN contained no regular secondary structure, with the main-chain torsion angles (Ramachandran φ/ψ) for only about 1/3 of all residues found in the favored region as defined by MolProbity (Davis et al. 2004), 1/3 in the allowed region, and the remaining 1/3 outside of these two regions.
Figure 1.
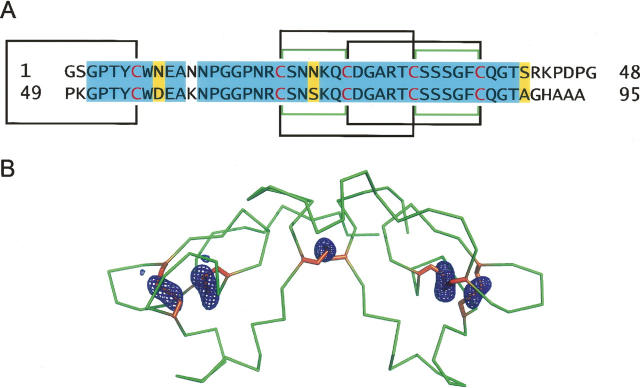
Amino acid sequence and the location of disulfides in scytovirin. (A) The sequence of SVN, with residues in each line belonging to an individual domain. Cysteine residues are red and the disulfide linkages observed in the X-ray structures are marked by black lines, whereas different linkages previously postulated on the basis of mass spectroscopy and NMR data are green. Residues identical in the SD1 and SD2 domains are highlighted in blue, and the similar ones are highlighted in yellow. (B) Anomalous difference Fourier map calculated at 3 Å resolution with data measured at 1.74 Å wavelength, contoured at the 4 σ level. In order to minimize potential bias, the phases were calculated from the refined model of SVN from which all sulfur atoms were removed. Placement of the model was rigid-body optimized in the slightly different unit cell corresponding to these data. The Cα trace of SVN (green) and the side chains of cysteines (yellow) are superimposed on the map.
We present here the high-resolution crystal structure of SVN, which differs in many significant details from its counterpart obtained by solution NMR, although it supports the observation that the fold of SVN is unique. The new structure also deviates from previously published mass spectrometry studies on both native (Bokesch et al. 2003) and recombinant (Xiong et al. 2006a) SVN. Understanding the significance of this new structure for both the mechanism of carbohydrate binding and anti-HIV activity of SVN will necessitate further functional studies.
Results and Discussion
Orthorhombic crystals of both the natural (nonrecombinant) and recombinant SVN belong to space group P212121 and diffract to high resolution. Our attempts to solve the structure using single-wavelength anomalous dispersion (SAD) of cysteine sulfurs and data collected at long wavelength were unsuccessful, although the anomalous difference synthesis calculated a posteriori with these data and refined phases (Fig. 1B) unequivocally identified the positions and covalent connectivity of all sulfur atoms in the structure. The structure was ultimately solved by SAD, with diffraction data measured on a home X-ray source after a brief soak in a solution containing potassium iodide (Dauter et al. 2000). The crystal structure of the nonrecombinant protein was subsequently refined at 1.3 Å resolution, whereas the structure of recombinant SVN was refined to 1.0 Å resolution; both structures utilized data collected on synchrotrons (Table 1). Unless indicated otherwise, the latter structure is discussed below in detail, although there are no meaningful differences between them.
Table 1.
Statistics of data collection and structure refinement
Unlike other lectins with antiviral activity such as cyanovirin or griffithsin, SVN is strictly monomeric, with no indication of oligomerization under any conditions. However, similar to the monomer of cyanovirin, the SVN molecule is highly symmetric with two domains: SD1 and SD2 (Fig. 2A). Residues 1–5 form a tight β hairpin, with an extended chain continuing through residue 11. Pro14 and Gly15 form a tip of the loop with the peptide bond between them assuming two alternative peptide-flipped conformations in the 1.3 Å structure; no such disorder is observed in the 1.0 Å structure. Residues 18–25 form another short loop followed by a helical turn, and 26–32 form another β hairpin. The next β hairpin continues through Cys38, followed by a rather irregular extended chain through Arg43. The interdomain linker includes three prolines (45, 47, and 49), the first two of which are in cis configuration. The linker is very well-ordered, and its configuration is completely unambiguous. The conformation of the SD2 domain is virtually identical to SD1 for residues 51–90 (RMS difference 0.25 Å compared to 3–42). The last four C-terminal residues make extensive contacts with the N-terminal domain, with the terminal carboxylate interacting with the peptide amide and the side chain of Ser2. With the exception of the carbonyl oxygens of prolines 14 and 62 that assume two orientations (only in the 1.3 Å structure), the main chain is fully ordered, including both termini. As discussed previously (McFeeters et al. 2007), the fold of each domain is novel and does not resemble other cysteine-rich lectins, such as wheat-germ agglutinin or hevein.
Figure 2.
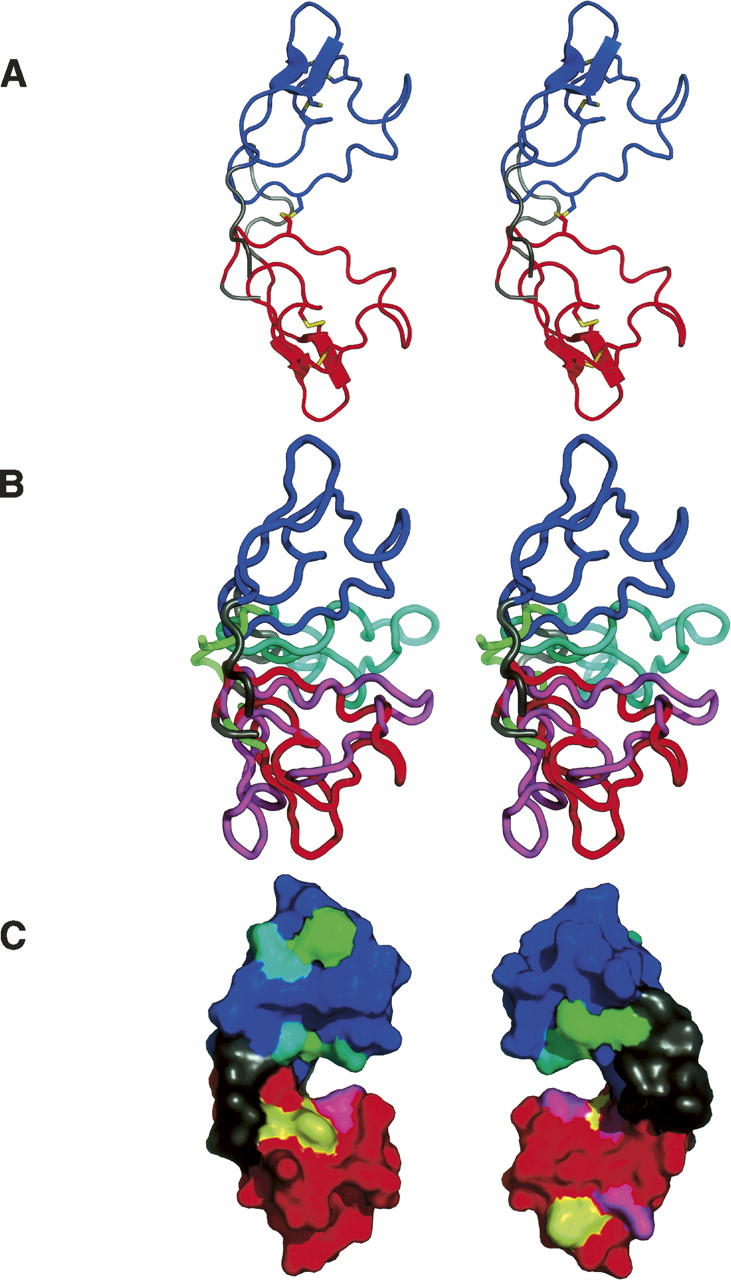
Crystal structure of scytovirin. In all panels, the structure of the 1.3 Å natural SVN is shown with the SD1 domain in red, the SD2 domain in blue, and the remaining terminal and linker regions dark gray. (A) A stereopair showing the overall structure of SVN, with the disulfide linkages shown in yellow. (B) A stereopair showing a superposition of the crystal and NMR structures of SVN. The coordinates of only the SD1 domains were used to superimpose the whole molecules. The NMR structure is shown with the SD1 domain in magenta, the SD2 domain in cyan, and the terminal and linker regions in green. (C) Positions of the proposed carbohydrate-binding residues (McFeeters et al. 2007) in the crystal structure, shown in two views rotated by 180°. The aromatic residues postulated to be involved in carbohydrate binding are shown in yellow for the SD1 domain and green for the SD2 domain. Other carbohydrate-binding residues are shown in magenta for the SD1 domain and cyan for the SD2 domain.
The main-chain torsion angles for all residues are within the favored region of the Ramachandran plot as defined by MolProbity (Davis et al. 2004), whereas 96% are in the most favored and 4% in the additionally allowed region defined by PROCHECK (Laskowski et al. 1993). The limited amount of regular secondary structure includes two β-sheets (residues 31–34 and 38–40, as well as 71–81 and 86–88) and four helical turns (10–12, 23–25, 58–60, and 71–73). However, the intramolecular hydrogen bond network is extensive, with 40 H-bonds formed between the main chain atoms, 38 between the main chain and the side chains, and 10 between side chains only. The two termini are close to each other, with the amide nitrogen of Gly1 making excellent H-bonds with the peptide carbonyls of Cys26 and Arg30, and both the peptide carbonyl and side chain hydroxyl of Ser2 interacting with the C-terminal carboxylate. The extensive involvement of residues 1 and 2 in the stabilization of the structure may explain why their removal leads to the decrease in the anti-HIV activity of the SD1 domain of SVN (Xiong et al. 2006b). The C-terminal strand stabilizes the two-domain structure through several hydrogen bonds between its main chain and the residues in both domains, with a special role of the side chain of His92 that stabilizes the structure through H-bonds with the peptide carbonyl of Thr5 and the carboxylate of Asp75. The only other direct bonds between the SD1 and SD2 domain are the disulfide 7–55 and a putative H-bond between Arg43 and the peptide carbonyl of Thr53. A few hydrophobic interactions complete the interdomain interface.
Surprisingly, the disulfide bonding pattern observed in the crystal structures of SVN (Fig. 1) differs from the pattern reported on the basis of limited proteolysis combined with mass spectrometry (Bokesch et al. 2003; Xiong et al. 2006a) that was utilized in the elucidation of the NMR structures (McFeeters et al. 2007). The disulfides seen in the crystal structures are formed between Cys20–Cys32 and Cys26–Cys38 in SD1, and Cys68–Cys80 and Cys74–Cys86 in SD2. The interdomain disulfide between Cys7 and Cys55 agrees with the previous results. As shown in Figure 1B, the location and covalent connectivity of disulfides is absolutely unambiguous. The distance between the Cα atoms of Cys26 and Cys38 is 5.54 Å, and between Cys20 and Cys32 is 5.93 Å. By contrast, the corresponding distances are 7.18 Å and 4.61 Å between the 20–26 and 32–38 pairs. The observed disulfide pattern is the same in the two high-resolution structures of SVN; thus, the nonrecombinant and recombinant proteins are identical in this respect. This result is particularly surprising in view of the apparently unambiguous assignments of the disulfides in the individual domains of SVN (Xiong et al. 2006b) which supported the originally proposed pattern of disulfide bonds (Bokesch et al. 2003; Xiong et al. 2006a). Additional proteolysis/mass spectrometry studies on a sample of the same material as used for the crystallographic studies also supported the previous assignment of the disulfides (our unpublished data). Furthermore, simultaneous whole-cell anti-HIV assays, performed with the sample of SVN used for crystallography and a recombinant sample prepared previously, indicated that the anti-HIV activity of both samples were indistinguishable from each other with both test samples providing EC50 values of ∼10 nM (data not shown). We are continuing these studies in order to explain the cause of this discrepancy.
The crystal structure of SVN exhibits a number of other significant differences compared to its NMR counterpart. The relationship between the two domains is very different (Fig. 2B), with the two domains separated in the crystal structure, but in close contact in the NMR structure. A large number of NOEs between Cys20 and Gln73 cannot be reconciled with their distance of ∼27 Å observed in the crystal structure. The three substructural elements identified in the NMR structure (see Fig. 3 of McFeeters et al. 2007) differ significantly from their crystallographic counterparts. The top loop (residues 12–17 and 60–65) is a fairly regular β-turn and does not contain the strong twist observed in the NMR structure. The middle turn (residues 22–25 and 70–73) is a regular helical turn, identical in SD1 and SD2. The largest difference between the crystal and NMR structures is in the “bottom knot” (residues 31–39 and 79–87), which in the crystal does not contain a knot at all, but rather two strands of a slightly irregular β-sheet.
Several residues of SVN were postulated to be involved in carbohydrate binding through chemical shift perturbation analysis utilizing the Manα(1 → 2)Manα(1 → 6)Manα(1 → 6)Man tetrasaccharide (McFeeters et al. 2007). In particular, the three aromatic residues in each domain (Tyr6, Trp8, and Phe37 in SD1, and Tyr54, Trp56, and Phe85 in SD2) were proposed to be part of the carbohydrate-binding sites. As seen in Figure 2C, the residues that were so implicated form two separate clusters in each domain, located on both sides of the large interdomain cleft. Asn9 and Asp57, the two nonidentical residues from the SD1 and SD2 domains, both face the cleft and interact through a water molecule. Since the clusters of the putative carbohydrate-binding residues are necessarily identical in the SD1 and SD2 domains, we may only speculate that the difference between the chemical nature of these two side chains might explain the reported differences in oligosaccharide binding of the two domains (Xiong et al. 2006b). However, this assumption needs experimental verification through high-resolution structures of the SVN–carbohydrate complexes.
Materials and Methods
SVN used for structural studies was either purified from Scytonema varium as described before (Bokesch et al. 2003), or was expressed as a recombinant protein in E. coli and purified as described previously (Xiong et al. 2006a). Crystals were obtained by the hanging-drop vapor diffusion method at room temperature. Nonrecombinant SVN crystals were grown in 30% PEG 8000, 0.2 M Li2SO4, 0.1 M Na acetate pH 4.7, protein concentration 30 mg/mL. The recombinant SVN crystals were grown in 1.0 M Na citrate, 0.1 M imidazole, pH 8.0, protein concentration 32 mg/mL. Crystals of both the natural and recombinant SVN are isomorphous in the space group P212121 (Table 1), with a monomer in the asymmetric unit. Several data sets obtained on a home source equipped with a Cu anode, as well as on a synchrotron utilizing 1.75 Å radiation were used in attempts to solve the structure by anomalous scattering of the 10 cysteine sulfurs, but we did not see sufficient anomalous signal and did not succeed in this approach.
X-ray diffraction data for the recombinant crystal used to obtain initial phases were collected with a MAR345 image plate detector mounted on a Rigaku RU-H3R generator operated at 50 kV and 100 mA. Three data sets were collected with synchrotron sources. Data for two crystals of natural SVN were measured with an ADSC Q210 detector on the X9B beamline at NSLS. Data extending to 1.3 Å resolution were collected using the short wavelength of 0.98 Å, whereas a longer wavelength of 1.74 Å was utilized to measure data up to 2.1 Å resolution. Data for recombinant SVN were measured at the SER-CAT beamline 22-ID, at the Advanced Photon Source (APS), on a MAR 300CCD detector. Prior to data collection crystals were briefly soaked in a crystallization buffer containing additional 10% ethylene glycol, whereas the crystals of the recombinant SVN were soaked in a buffer containing the mother liquor, 10 mM KI, and 20% glycerol. The crystals were rapidly cooled in a nitrogen stream (temperature 100 K). All data were processed and scaled using the HKL2000 package (Otwinowski and Minor 1997). Diffraction data from the iodide-soaked and nonrecombinant SVN crystals were measured in a single rotation pass. However, the atomic resolution data from the recombinant SVN crystal were collected in three passes: first low-resolution (30–2.5 Å), then medium-resolution (5–1.6 Å), and finally a high-resolution pass (3.0–1.0 Å), in order to adequately measure both the weak and strong intensities. The effective exposure differed by about six in each pass and all measured intensities were scaled and merged together. The merging statistics suggested a possibility of some radiation damage inflicted on the crystal during the high-resolution pass consisting of 200 images of 0.5° width and 1 sec exposure to the full beam of the undulator X-ray source. The statistics of all diffraction data are presented in Table 1.
The structure was solved using the single-wavelength anomalous diffraction (SAD) approach applied to the data collected with the copper-source wavelength of 1.54 Å from the crystal soaked in cryo-solution containing 1 M potassium iodide. The data were interpreted by XPREP (Bruker AXS), which indicated that the anomalous signal-to-noise ratio <ΔF ano>/<σ(F)> was larger than 1.3 for resolution ranges extending to 2.0 Å. SHELXD (Schneider and Sheldrick 2002) provided a convincing solution of the iodide substructure of 12 ions with E obs/E calc correlation of 37%. A run of SHELXE (Sheldrick 2002) with the use of native 1.3 Å data indicated a large contrast between two enantiomers (pseudo-free CC 73.4 vs. 50.3%). The phases produced by SHELXE were input to ARP/wARP (Perrakis et al. 1999) in the warpNtrace mode, which after 50 cycles built 90 residues, all docked in sequence. However, the missing residues (Gly1, a stretch Pro45-Asp46-Pro47, and Ala95) had very clear electron density and were easily modeled using Coot (Emsley and Cowtan 2004).
The structures of the nonrecombinant and recombinant SVN were refined independently of each other with the program REFMAC5 (Murshudov et al. 1997) and rebuilt with Coot (Emsley and Cowtan 2004). The results of the refinements are summarized in Table 1. The coordinates were compared with the program ALIGN (Cohen 1997). Since no average NMR structure of SVN was deposited in the PDB, the coordinates of the first model in the 20-model assembly were used for all comparisons. The coordinates and structure factors have been deposited in the PDB with accession codes 2qt4 and 2qsk for the 1.3 and 1.0 Å data sets, respectively.
Acknowledgments
We acknowledge the use of beamline 22-ID of the Southeast Regional Collaborative Access Team (SER-CAT), located at the Advanced Photon Source, Argonne National Laboratory. Use of the APS was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S Government. This research was also supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and in part by the Intramural AIDS Targeted Antiviral Program of the Office of the Director of the National Institutes of Health to A.W.
Footnotes
Reprint requests to: Alexander Wlodawer, Macromolecular Crystallography Laboratory, NCI, Frederick, MD 21702-1201, USA; e-mail: wlodawer@ncifcrf.gov; fax: (301) 846-6322.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073157507.
References
- Bokesch H.R., O'Keefe, B.R., McKee, T.C., Pannell, L.K., Patterson, G.M., Gardella, R.S., Sowder, R.C., Turpin, J., Watson, K., Buckheit Jr, R.W., et al. 2003. A potent novel anti-HIV protein from the cultured cyanobacterium Scytonema varium . Biochemistry 42: 2578–2584. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. 1992. The free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature 355: 472–474. [DOI] [PubMed] [Google Scholar]
- Cohen G.E. 1997. ALIGN: A program to superimpose protein coordinates, accounting for insertions and deletions. J. Appl. Crystallogr. 30: 1160–1161. [Google Scholar]
- Dauter Z., Dauter, M., and Rajashankar, K.R. 2000. Novel approach to phasing proteins: Derivatization by short cryo soaking with halides. Acta Crystallogr. D56: 232–237. [DOI] [PubMed] [Google Scholar]
- Davis I.W., Murray, L.W., Richardson, J.S., and Richardson, D.C. 2004. MOLPROBITY: Structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32: W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P. and Cowtan, K. 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: Program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26: 283–291. [Google Scholar]
- McFeeters R.L., Xiong, C., O'Keefe, B.R., Bokesch, H.R., McMahon, J.B., Ratner, D.M., Castelli, R., Seeberger, P.H., and Byrd, R.A. 2007. The novel fold of scytovirin reveals a new twist for antiviral entry inhibitors. J. Mol. Biol. 369: 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin, A.A., and Dodson, E.J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D53: 240–255. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Morris, R., and Lamzin, V.S. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6: 458–463. [DOI] [PubMed] [Google Scholar]
- Schneider T.R. and Sheldrick, G.M. 2002. Substructure solution with SHELXD. Acta Crystallogr. D58: 1772–1779. [DOI] [PubMed] [Google Scholar]
- Sheldrick G.M. 2002. Macromolecular phasing with SHELXE. Z. Kristallogr. 217: 644–650. doi: 10.1524/zkri.217.12.644.20662. [DOI] [Google Scholar]
- Xiong C., O'Keefe, B.R., Botos, I., Wlodawer, A., and McMahon, J.B. 2006a. Overexpression and purification of scytovirin, a potent, novel anti-HIV protein from the cultured cyanobacterium Scytonema varium . Protein Expr. Purif. 46: 233–239. [DOI] [PubMed] [Google Scholar]
- Xiong C., O'Keefe, B.R., Byrd, R.A., and McMahon, J.B. 2006b. Potent anti-HIV activity of scytovirin domain 1 peptide. Peptides 27: 1668–1675. [DOI] [PubMed] [Google Scholar]