Abstract
Poxviruses encode immuno-modulatory proteins capable of subverting host defenses. The poxvirus vaccinia expresses a small 14-kDa protein, N1L, that is critical for virulence. We report the crystal structure of N1L, which reveals an unexpected but striking resemblance to host apoptotic regulators of the B cell lymphoma-2 (Bcl-2) family. Although N1L lacks detectable Bcl-2 homology (BH) motifs at the sequence level, we show that N1L binds with high affinity to the BH3 peptides of pro-apoptotic Bcl-2 family proteins in vitro, consistent with a role for N1L in modulating host antiviral defenses.
Keywords: poxvirus, vaccinia virus, virulence, crystal structure, Bcl-2, apoptosis
Poxviruses, such as vaccinia and variola (smallpox), are among the largest animal viruses, carrying a linear double-stranded DNA genome (150–350 kb) with ∼200 distinct genes (Moss 2000). Poxviruses express their own machinery for DNA replication, mRNA transcription, and virion assembly (Moss 2000). They also encode proteins that manipulate host defense mechanisms for efficient viral replication (Johnston and McFadden 2003; Seet et al. 2003; Shchelkunov 2003).
A 14-kDa vaccinia protein, N1L, was initially identified from an attenuated spontaneous deletion mutant (6/2) of vaccinia virus (Kotwal and Moss 1988). N1L is a potent virulence factor, which, when deleted, caused the strongest attenuation observed for any gene that was not essential for growth in culture (Kotwal et al. 1989; Bartlett et al. 2002). Thus, deletion of the N1L gene reduced mortality of intracranially infected mice by a factor of 104 (Kotwal et al. 1989). Furthermore, in the highly attenuated vaccinia Ankara strain, N1L is truncated with a distinct C terminus (Antoine et al. 1998). Although initially described as a secreted “virokine” (Kotwal et al. 1989), N1L is now believed to localize predominantly within the host cell (Bartlett et al. 2002). N1L has 94% sequence identity between vaccinia and variola orthologs (Massung et al. 1993), but appears to be unique to poxviruses (Bartlett et al. 2002).
Understanding the molecular mechanisms of viral immuno-modulatory proteins furthers our insights into the delicate interplay between pathogen and host, illuminates pathways of cellular immunity, and provides new leads for the development of antiviral therapeutics and vaccines. Toward these goals, we report here the crystal structure of N1L, which reveals a compact α-helical architecture characteristic of the Bcl-2 family of host cell apoptotic regulators. In vitro binding studies demonstrate binding to several cellular pro-apoptotic Bcl-2 homology 3 (BH3) domains, suggesting a direct role for N1L in the modulation of host apoptosis.
Results and Discussion
Bcl-2-like structure of vaccinia N1L
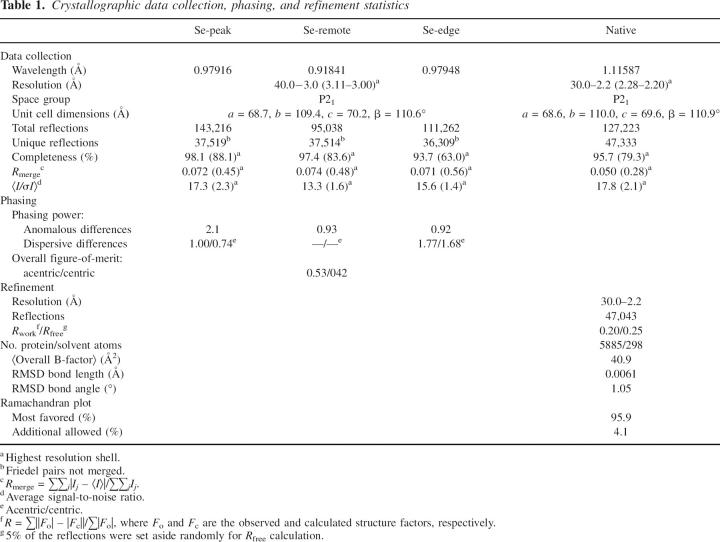
We determined the crystal structure of vaccinia N1L at 2.2 Å resolution (Table 1). The crystals contain six molecules in the asymmetric unit arranged as three symmetric dimers. Conformational heterogeneity occurs in an N-terminal loop (Asn13–Phe24) and at the C terminus (Leu109–Gly115). Otherwise, the six copies are very similar, with RMS main chain deviations of ≤0.7 Å in pairwise comparisons. The refined models include N1L residues 1–115, with three additional residues at the N terminus from the expression vector (Gly−2, Ser−1, and His0); the two C-terminal residues (Glu116 and Lys117) have not been modeled due to poor or absent electron density.
Table 1.
Crystallographic data collection, phasing, and refinement statistics
N1L forms a compact α-helical bundle (Fig. 1A). The N-terminal helix, α1, is connected by a short loop to the α2 with an interhelical angle of ∼80°. The last five residues of α2 (Leu29–Leu33) form a 310 helix followed by a short turn that orients α3 at ∼100° from α2. Three helices, α4, α5, and α6, are nearly antiparallel to each other. The C-terminal helix α6′ (Glu103–Leu113) is contiguous with α6, except that a single 310 helical turn at Glu103 creates a bend, rotating α6′ clockwise by 80° with respect to α6. α6′ is positioned almost perpendicular to the central α5 helix. In the overall organization, the two central α5 and α6 helices are surrounded by two helices (α1 and α2) on one side and two helices (α3 and α4) on the other.
Figure 1.
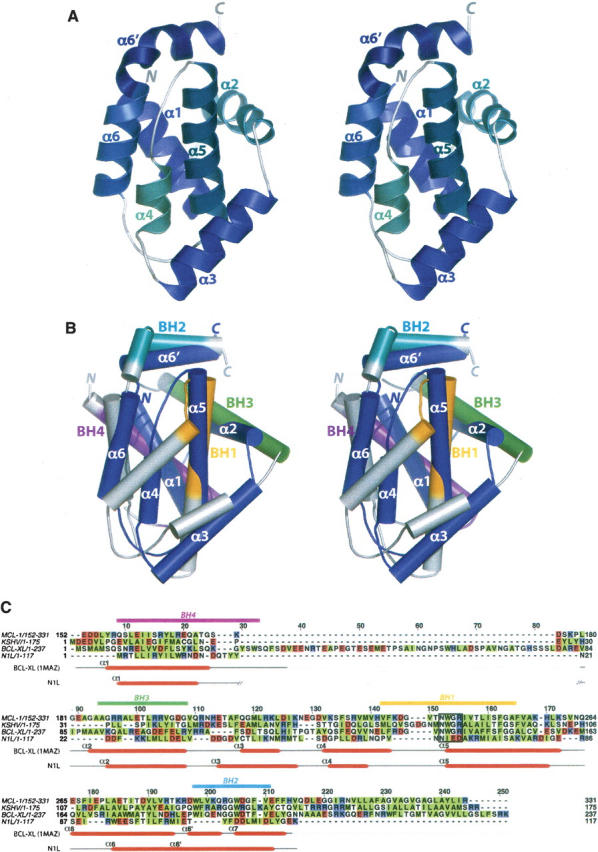
Structure of vaccinia N1L and comparison with Bcl-2 family proteins. (A) Stereo view of the N1L monomer. Helices and termini are labeled. (B) Stereo superposition of N1L (navy) and Bcl-XL (gray; 1MAZ; Muchmore et al. 1996). N1L helices are labeled. Functionally important BH regions of Bcl-XL are colored in magenta (BH4), green (BH3), orange (BH1), and cyan (BH2). (C) Structure-based sequence alignment of N1L with Bcl-2 family members: Mcl-1, mouse myeloid cell leukemia-1 (PDB code 1WSX; Day et al. 2005); KSHV, Kaposi sarcoma virus Bcl-2 homolog (PDB code 1K3K; Huang et al. 2002); human Bcl-XL (PDB code 1MAZ; Muchmore et al. 1996). Hydrophobic residues are highlighted in green, and acidic/basic residues are in red/blue. Secondary structures of N1L and Bcl-XL are indicated below the sequence, and consensus BH motifs are indicated above, with the same color scheme as in B. The highly conserved Bcl-2 signature motif, NWGR, is boxed.
The N1L fold closely resembles that of the Bcl-2 family of cellular apoptotic regulators (Petros et al. 2004), despite a very low sequence identity of 11% (Fig. 1B,C). The DALI server (Holm and Sander 1993) identifies several anti-apoptotic Bcl-2 family proteins as the closest structural neighbors of N1L: mouse myeloid cell leukemia-1 (Mcl-1, PDB code 1WSX; Day et al. 2005; Z-score = 9.6, RMSD = 2.8 Å), Kaposi sarcoma virus Bcl-2 homolog (PDB code 1K3K; Huang et al. 2002; Z = 7.9, RMSD = 3.2 Å), human Bcl-XL (PDB code 1MAZ; Muchmore et al. 1996; Z = 7.0, RMSD = 3.1 Å), Caenorhabditis elegans Bcl-2 protein CED-9 (PDB code 1OHU; Woo et al. 2003; Z = 6.5, RMSD = 3.7 Å), and mouse Bcl-XL (PDB code 1PQ0; Liu et al. 2003; Z = 6.4, RMSD = 4.2 Å). In contrast to the Bcl-2 family members, N1L lacks a C-terminal transmembrane helix (Petros et al. 2004) and in general contains shorter secondary structural elements (Fig. 1B,C). Indeed, vaccinia N1L is the smallest known protein that maintains the Bcl-2-like fold.
Dimeric assembly of N1L
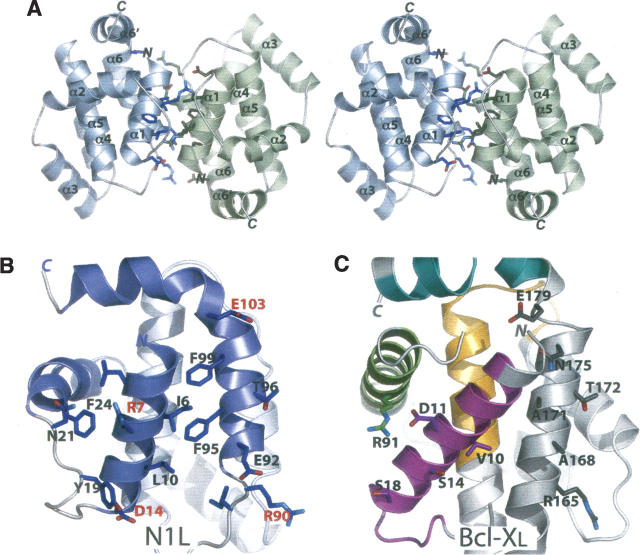
The N1L crystal structure reveals a homodimeric assembly (Fig. 2A) distinct from the monomeric structures reported for host Bcl-2 family members (Petros et al. 2004). Dimerization buries 2100 Å2 of surface, accounting for 30% of the total (6900 Å2) of each subunit. The DCOMPLEX server (Zhou et al. 2005; http://sparks.informatics.iupui.edu/czhang/complex.html) predicts the N1L dimer to be biologically relevant (rather than a crystallization artifact). Gel filtration analysis also suggests that N1L is dimeric in solution at micromolar concentrations (data not shown), consistent with earlier biochemical studies (Bartlett et al. 2002).
Figure 2.
N1L adopts a dimeric structure. (A) Stereo view of the N1L homodimer. The α1 and α6 helices from one N1L monomer (blue) interact in an antiparallel way with equivalent helices in another monomer (green). N and C termini, and helices of each subunit are labeled. (B) Specific α1 and α6 residues at the N1L dimer interface. In the antiparallel N1L dimer, Ile6, Leu10, Phe95, and Phe99 constitute a critical hydrophobic patch whereas Arg7/Asp14 and Arg90/Glu103 pairs form complementary electrostatic surfaces, not present in Bcl-XL. The N1L monomer (blue) in this view is related to the blue monomer in A by 90° about a vertical axis. (C) The same view for Bcl-XL showing analogous residues, which are either not hydrophobic or not complementary in charge. BH1–4 domains are colored in magenta (BH4), green (BH3), orange (BH1), and cyan (BH2), as in Figure 1, B and C.
Molecular contacts at the dimer interface are provided by the α1 and α6 helices (Fig. 2A,B). Bulky hydrophobic α1 residues, Ile6 and Leu10, pack against their counterparts across the dimer interface. In a similar manner, charged α1 residues, Arg7 and Asp14, of one subunit interact with their counter-ions in the second subunit. α6 also provides complementary hydrophobic (Phe95 and Phe99) and charged (Arg90 and Glu103) residues across the dimer interface. Comparable hydrophobic and charged residues are absent in other Bcl-2 family proteins (Fig. 2C). Notably, this antiparallel N1L homodimer is distinct from a recently described Bcl-XL dimer, in which C-terminal halves are swapped between two monomers by formation of a single continuous α5–α6 helix (O'Neill et al. 2006). However, as in the case of the domain-swapped Bcl-XL dimer (O'Neill et al. 2006), the dimer interface of N1L excludes a putative functional face of the molecule, namely the hydrophobic binding groove (Petros et al. 2004) prominent among the Bcl-2 family proteins.
Bcl-2 homology (BH) motifs
The Bcl-2 family of proteins contains at least one of the four “Bcl-2 homology” (BH1–4) regions (Fig. 1B,C) that structurally and functionally support their regulatory roles in apoptosis (Cory and Adams 2002; Danial and Korsmeyer 2004; Petros et al. 2004). Structure-based alignment (Fig. 1C) demonstrates a lack of apparent sequence homology of N1L in regions structurally equivalent to the BH domains. Nevertheless, several key BH-domain interactions appear to be maintained in N1L. For example, an “NIED” sequence found at the beginning of α5 in N1L serves the same structural role as the “NWGR” signature motif of the Bcl-2 BH1 domain. In both N1L and Bcl-2 family proteins, the conserved Asn (Asn65 in N1L and Asn136 in Bcl-XL) at the first position of the motif N-terminally caps the central helix α5. In addition, analogous to the Trp residue at the second position (Trp137 in Bcl-XL), Ile66 of N1L forms hydrophobic contacts with α6/α6′ residues (Tyr105 and Leu109), possibly contributing to the overall structural integrity (Huang et al. 2002). On the other hand, the last two residues in the “NWGR” motif, Gly and Arg, which are crucial for protein–protein interactions among the Bcl-2-related proteins (Sattler et al. 1997), are replaced by Glu67 and Asp68 in N1L.
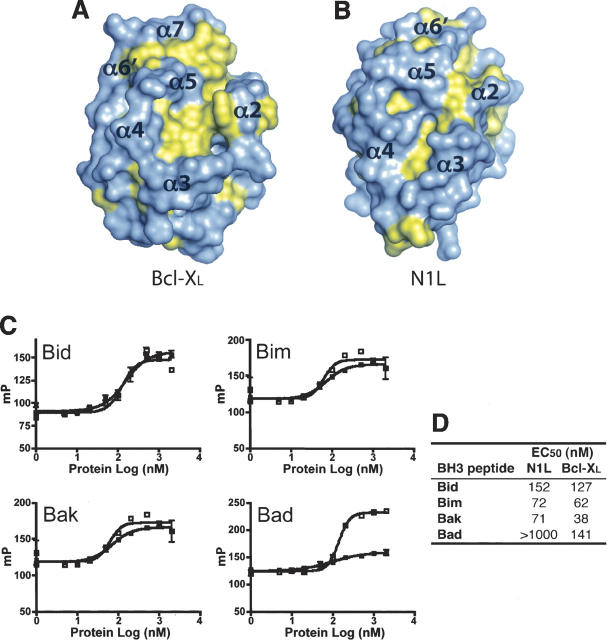
Although N1L lacks consensus BH motifs, its molecular surface contains an elongated hydrophobic patch comparable to that found in the anti-apoptotic Bcl-2 family proteins. In these Bcl-2 proteins, α5 (BH1), α7 (BH2), α2 (BH3), α3, and α4 form a long hydrophobic groove (Fig. 3A) where the BH3 region from another Bcl-2 protein binds to form a heterodimer (Yin et al. 1994; Sattler et al. 1997; Liu et al. 2003). For N1L, a hydrophobic groove is located on the same face of the molecule as in the Bcl-2 family proteins (Fig. 3B), but N1L's groove is narrower and shorter owing to additional charged residues (Glu32, Asp35, Asp38, Glu67, Asp68, and Arg71) and the closer packing of α2 against α5.
Figure 3.
N1L binds to BH3 peptides. (A,B) Hydrophobic surfaces of N1L and Bcl-XL. The solvent-accessible surface of N1L indicates the presence of a small hydrophobic groove on the same face of the molecule as the BH3 binding groove in Bcl-XL. Phe, Trp, Tyr, Met, Ile, Leu, Val, and Ala are colored in yellow. Approximate positions of helices surrounding the groove are indicated on the surfaces. The orientation of each protein is similar to that in Figure 1B. (C) Fluorescence polarization plots (mP = 1000 × fluorescence polarization) of FITC-labeled BH3 domains (Bid, Bim, Bak, and Bad) in the presence of varying concentrations of N1L (▪) or Bcl-XL (□). (D) Tabulated EC50 values from these plots.
N1L binds BH3 peptides in vitro
Heterodimerization between pro- and anti-apoptotic Bcl-2 family proteins is a crucial step in regulating apoptosis and is mediated by the binding of BH3 domains from the pro-apoptotic members to the hydrophobic groove of the anti-apoptotic members (Cory and Adams 2002; Danial and Korsmeyer 2004). We explored potential interactions between N1L and the BH3 domains of pro-apoptotic Bcl-2 members using fluorescence polarization assays (Fig. 3C,D). We found that N1L interacts with peptides comprising the BH3 domains of three different pro-apoptotic Bcl-2 proteins (Bid, Bim, and Bak) with affinities similar to those of the anti-apoptotic Bcl-XL. Curiously, no significant binding was detected between N1L and Bad.
Bcl-2-like proteins in poxviruses
The crystal structure of vaccinia N1L demonstrates the existence of a Bcl-2-like structural fold in the orthopoxviruses. Bcl-2-like proteins have been identified only in fowlpox and canarypox viruses of the avipoxviruses (Afonso et al. 2000; Tulman et al. 2004). Avipoxvirus-encoded Bcl-2 homologs (FPV039 and CNPV058) show sequence homology (∼25% identity and ∼50% similarity) to cellular Bcl-2 proteins and contain recognizable BH1 and BH2 domains as well as a C-terminal transmembrane domain (Afonso et al. 2000; Tulman et al. 2004). Due to the absence of detectable Bcl-2 homologs, most other poxviruses have been assumed to utilize other proteins for controlling host apoptosis (Cuconati and White 2002; Hardwick and Bellows 2003; Taylor and Barry 2006). Vaccinia F1L and myxoma (leporipoxvirus) M11L proteins, for instance, share little sequence homology with Bcl-2 family proteins, yet block apoptosis by inhibiting pro-apoptotic Bak, possibly via their putative BH3-like domain (Wang et al. 2004; Wasilenko et al. 2005; Postigo et al. 2006; Su et al. 2006).
An ortholog search of N1L against other poxviral genomes (http:www.poxvirus.org) yielded a set of uncharacterized proteins from the distantly related nonorthopoxvirus members. Goatpox, sheeppox, and “lumpy skin disease” viruses encode proteins (GTPV_Pellor114, SPPV_A115, and LSDV_WARM144) that share sequence homology (∼20% identity and ∼50% similarity) with the vaccinia N1L. Elucidating the function of these putative orthologs in modulating host immunity will likely provide insights into the molecular basis of host range and virulence across the poxvirus family.
Functional implication and conclusion
Recent studies have demonstrated that N1L targets several components of a multisubunit IκB kinase complex in NF-κB signaling pathways (DiPerna et al. 2004) and reduces cytokine secretion (Zhang et al. 2005). How might this finding be linked to our structural and biochemical observations? One observation that may be pertinent is that a cellular Bcl-2 protein, in addition to regulating mitochondrial-mediated apoptosis, also controls the activation of multiple transcription factors, including NF-κB (Regula et al. 2002; Massaad et al. 2004). Interestingly, the cowpox anti-apoptotic protein CrmA inhibits NF-κB activation by suppressing the caspase-dependent processing of pro-inflammatory cytokines (Ray et al. 1992), suggesting that apoptotic and NF-κB signaling pathways are linked at the molecular level (Bowie et al. 2004). Our identification of a Bcl-2-like protein in vaccinia with the ability to bind BH3 peptides will thus generate testable hypotheses to probe the molecular mechanisms by which N1L counteracts host antiviral defenses.
Materials and methods
N1L expression, purification, and crystallization
The vaccinia N1L coding sequence (Western Reserve strain VACWR028) was PCR amplified and subcloned into the NdeI/BamHI site on the pET15b vector (Novagen). Recombinant N1L protein (117 amino acids), with an N-terminal His6-tag, was expressed in Escherichia coli BL21(DE3) CodonPlus RIL (Strategene) overnight at 15°C by adding isopropyl-β-d-thiogalactopyranoside. Following cell lysis by sonication, the His6-N1L protein was purified through HiTrap Ni2+-chelating and Superdex 200 gel filtration columns (GE Healthcare Bio-Sciences AB). The protein purity was confirmed by SDS-PAGE and peptide mapping mass spectroscopy, and stored in 20 mM Tris-HCl (pH 8), 150 mM NaCl, 4 mM β-mercaptoethanol (β-ME) at −80°C. Seleno-l-methionine (SeMet)-labeled N1L was prepared using minimal M9 medium under metabolic inhibition, as described elsewhere (Van Duyne et al. 1993). Incorporation of seven SeMet residues (including the first Met residue) per N1L molecule into the protein was confirmed by electrospray mass spectrometry analysis.
All crystallization experiments were performed using the hanging-drop vapor diffusion method at 20°C. Two microliters of the N1L or SeMet-N1L (30 mg/mL) were mixed with an equal volume of reservoir containing 5%–10% (w/v) polyethylene glycol 4000, 100 mM Na-K tartrate, 100 mM Tris-HCl (pH 8), and 20 mM β-ME. Monoclinic crystals appeared after 1 wk and continued to grow over a period of 1–2 mo.
Data collection and structure determination
The SeMet and native data sets (Table 1) were collected from a flash-cooled crystal (100 K) at beam lines 9–2 (Stanford Synchrotron Radiation Laboratory, CA) and 12.3.1 (Advanced Light Source, CA), respectively. The cryoprotectant solution consisted of the equilibrated crystallization solution augmented with 30% (v/v) 2-methyl-2,4-pentanediol (MPD). The diffraction data were processed with HKL2000 (Otwinowski and Minor 1997). Forty-two Se sites—seven sites for each of the six molecules in the asymmetric unit—were identified by SHELXD (Schneider and Sheldrick 2002) and refined using SHARP (de La Fortelle and Bricogne 1997). After density modification by SOLOMON (Abrahams and Leslie 1996), maps calculated to 3 Å resolution were used for manual model building using XFIT (McRee 1999) and COOT (Emsley and Cowtan 2004).
The initial model, comprising six copies of residues 1–113, was refined through cycles of model building/fitting and refinement using XFIT (McRee 1999) and CNS (Brünger et al. 1998). Rigid body refinement against the 2.2 Å native data, treating the six molecules as separate rigid groups, was followed by cycles of minimization, simulated annealing, and B-factor refinement, resulting in R work = 0.30 and R free = 0.31. Next, the flexible terminal and loop residues, as well as solvent molecules, were modeled based on 2F o−F c and F o−F c maps, and subjected to further crystallographic refinement without noncrystallographic symmetry restraints, leading to final values of R work = 0.20 and R free = 0.25. The model has excellent stereochemistry as defined by PROCHECK (Laskowski et al. 1993) (Table 1). PDBFIT (McRee 1999), CE (Shindyalov and Bourne 1998), and DALI (Holm and Sander 1993) were used to obtain superposition and RMS deviations of the models. Interhelical angles in the final models were calculated using INTERHLX (K. Yap, University of Toronto). The structure factors and coordinates, comprising six copies of N1L and 282 water and two MPD molecules, have been deposited into the PDB with the accession code 2I39.
Fluorescence polarization assays (FPAs)
Binding of N1L to the BH3 domains of several Bcl-2 family proteins was quantified using fluorescence polarization anisotropy-based peptide binding assays (Zhai et al. 2005). Recombinant human Bcl-XL, lacking the 20-residue C-terminal transmembrane tail, was prepared as previously described (Zhai et al. 2005) and used as a control. Fluorescein isothiocyanate (FITC)-conjugated synthetic peptides comprising the BH3 domains of pro-apoptotic Bcl-2 proteins (BH3-Bid, FITC-aminohexanoyl (Ahx)-EDIIRNIARHLAQVGDSMDR; BH3-Bim, FITC-Ahx-DMRPEIWIAQELRRIGDEFNAYYAR; BH3-Bak, FITC-Ahx-PSSTMGQVGRQLAIIGDDINRRYDS) were prepared at the Burnham Institute's medicinal chemistry core facility, while the FITC-BH3-Bad peptide (NLWAAQRYGRELRRMSD-K[FITC]-FVD) was purchased from Synpep Corporation. Varying concentrations of N1L and Bcl-XL were incubated with 5–15 nM of the FITC-BH3 peptides, and the resulting fluorescence polarization (Analyst TM AD assay Detection system, LJL Biosystem) was used to calculate EC50 values.
Acknowledgments
We thank the Stanford Synchrotron Radiation Laboratory (supported by the Department of Energy, National Institutes of Health, and National Institute of General Medical Sciences) and Advanced Light Source (supported by the U.S. Department of Energy) for use of data collection facilities; L. Bankston, G. Wei, and L.Y. Low for helpful discussions; and S.-J. Cho, C. Bakolitsa, E. Santelli, and Y. Zong for assistance with data analysis. This work was supported by DAMD17-03-2-0038 and NIH U01 AI061139 (R.C.L.), and NIH AI055789 and GM60554 (J.C.R.).
Footnotes
Reprint requests to: Robert C. Liddington, Infectious and Inflammatory Disease Center, Burnham Institute for Medical Research, 10901 North Torrey Pines Road, La Jolla, CA 92037, USA; e-mail: rlidding@burnham.org; fax: (858) 713-9925.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062454707.
References
- Abrahams, J.P. and Leslie, A.G. 1996. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D Biol. Crystallogr. 52: 30–42. [DOI] [PubMed] [Google Scholar]
- Afonso, C.L., Tulman, E.R., Lu, Z., Zsak, L., Kutish, G.F., and Rock, D.L. 2000. The genome of fowlpox virus. J. Virol. 74: 3815–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine, G., Scheiflinger, F., Dorner, F., and Falkner, F.G. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: Comparison with other orthopoxviruses. Virology 244: 365–396. [DOI] [PubMed] [Google Scholar]
- Bartlett, N., Symons, J.A., Tscharke, D.C., and Smith, G.L. 2002. The vaccinia virus N1L protein is an intracellular homodimer that promotes virulence. J. Gen. Virol. 83: 1965–1976. [DOI] [PubMed] [Google Scholar]
- Bowie, A.G., Zhan, J., and Marshall, W.L. 2004. Viral appropriation of apoptotic and NF-κB signaling pathways. J. Cell. Biochem. 91: 1099–1108. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., and Pannu, N.S., et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921. [DOI] [PubMed] [Google Scholar]
- Cory, S. and Adams, J.M. 2002. The Bcl2 family: Regulators of the cellular life-or-death switch. Nat. Rev. Cancer 2: 647–656. [DOI] [PubMed] [Google Scholar]
- Cuconati, A. and White, E. 2002. Viral homologs of BCL-2: Role of apoptosis in the regulation of virus infection. Genes & Dev. 16: 2465–2478. [DOI] [PubMed] [Google Scholar]
- Danial, N.N. and Korsmeyer, S.J. 2004. Cell death: Critical control points. Cell 116: 205–219. [DOI] [PubMed] [Google Scholar]
- Day, C.L., Chen, L., Richardson, S.J., Harrison, P.J., Huang, D.C., and Hinds, M.G. 2005. Solution structure of prosurvival Mcl-1 and characterization of its binding by proapoptotic BH3-only ligands. J. Biol. Chem. 280: 4738–4744. [DOI] [PubMed] [Google Scholar]
- de La Fortelle, E. and Bricogne, G. 1997. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276: 472–494. [DOI] [PubMed] [Google Scholar]
- DiPerna, G., Stack, J., Bowie, A.G., Boyd, A., Kotwal, G., Zhang, Z., Arvikar, S., Latz, E., Fitzgerald, K.A., and Marshall, W.L. 2004. Poxvirus protein N1L targets the I-κB kinase complex, inhibits signaling to NF-κB by the tumor necrosis factor superfamily of receptors, and inhibits NF-κB and IRF3 signaling by toll-like receptors. J. Biol. Chem. 279: 36570–36578. [DOI] [PubMed] [Google Scholar]
- Emsley, P. and Cowtan, K. 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Hardwick, J.M. and Bellows, D.S. 2003. Viral versus cellular BCL-2 proteins. Cell Death Differ. Suppl 1 10: S68–S76. [DOI] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233: 123–138. [DOI] [PubMed] [Google Scholar]
- Huang, Q., Petros, A.M., Virgin, H.W., Fesik, S.W., and Olejniczak, E.T. 2002. Solution structure of a Bcl-2 homolog from Kaposi sarcoma virus. Proc. Natl. Acad. Sci. 99: 3428–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, J.B. and McFadden, G. 2003. Poxvirus immunomodulatory strategies: Current perspectives. J. Virol. 77: 6093–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwal, G.J. and Moss, B. 1988. Vaccinia virus encodes a secretory polypeptide structurally related to complement control proteins. Nature 335: 176–178. [DOI] [PubMed] [Google Scholar]
- Kotwal, G.J., Hugin, A.W., and Moss, B. 1989. Mapping and insertional mutagenesis of a vaccinia virus gene encoding a 13,800-Da secreted protein. Virology 171: 579–587. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26: 283–291. [Google Scholar]
- Liu, X., Dai, S., Zhu, Y., Marrack, P., and Kappler, J.W. 2003. The structure of a Bcl-xL/Bim fragment complex: Implications for Bim function. Immunity 19: 341–352. [DOI] [PubMed] [Google Scholar]
- Massaad, C.A., Portier, B.P., and Taglialatela, G. 2004. Inhibition of transcription factor activity by nuclear compartment-associated Bcl-2. J. Biol. Chem. 279: 54470–54478. [DOI] [PubMed] [Google Scholar]
- Massung, R.F., Esposito, J.J., Liu, L.I., Qi, J., Utterback, T.R., Knight, J.C., Aubin, L., Yuran, T.E., Parsons, J.M., and Loparev, V.N., et al. 1993. Potential virulence determinants in terminal regions of variola smallpox virus genome. Nature 366: 748–751. [DOI] [PubMed] [Google Scholar]
- McRee, D.E. 1999. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125: 156–165. [DOI] [PubMed] [Google Scholar]
- Moss, B. 2000. Poxviridae: The viruses and their replication. In Fields virology, 2nd ed, (eds. D.M. Knipe and P.M. Howley), pp. 2849–2883. Lippincott Williams & Wilkins, Philadelphia.
- Muchmore, S.W., Sattler, M., Liang, H., Meadows, R.P., Harlan, J.E., Yoon, H.S., Nettesheim, D., Chang, B.S., Thompson, C.B., and Wong, S.L., et al. 1996. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature 381: 335–341. [DOI] [PubMed] [Google Scholar]
- O'Neill, J.W., Manion, M.K., Maguire, B., and Hockenbery, D.M. 2006. BCL-XL dimerization by three-dimensional domain swapping. J. Mol. Biol. 356: 367–381. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Petros, A.M., Olejniczak, E.T., and Fesik, S.W. 2004. Structural biology of the Bcl-2 family of proteins. Biochim. Biophys. Acta 1644: 83–94. [DOI] [PubMed] [Google Scholar]
- Postigo, A., Cross, J.R., Downward, J., and Way, M. 2006. Interaction of F1L with the BH3 domain of Bak is responsible for inhibiting vaccinia-induced apoptosis. Cell Death Differ. 13: 1651–1662. [DOI] [PubMed] [Google Scholar]
- Ray, C.A., Black, R.A., Kronheim, S.R., Greenstreet, T.A., Sleath, P.R., Salvesen, G.S., and Pickup, D.J. 1992. Viral inhibition of inflammation: Cowpox virus encodes an inhibitor of the interleukin-1β converting enzyme. Cell 69: 597–604. [DOI] [PubMed] [Google Scholar]
- Regula, K.M., Ens, K., and Kirshenbaum, L.A. 2002. IKKβ is required for Bcl-2-mediated NF-κB activation in ventricular myocytes. J. Biol. Chem. 277: 38676–38682. [DOI] [PubMed] [Google Scholar]
- Sattler, M., Liang, H., Nettesheim, D., Meadows, R.P., Harlan, J.E., Eberstadt, M., Yoon, H.S., Shuker, S.B., Chang, B.S., and Minn, A.J., et al. 1997. Structure of Bcl-xL-Bak peptide complex: Recognition between regulators of apoptosis. Science 275: 983–986. [DOI] [PubMed] [Google Scholar]
- Schneider, T.R. and Sheldrick, G.M. 2002. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58: 1772–1779. [DOI] [PubMed] [Google Scholar]
- Seet, B.T., Johnston, J.B., Brunetti, C.R., Barrett, J.W., Everett, H., Cameron, C., Sypula, J., Nazarian, S.H., Lucas, A., and McFadden, G. 2003. Poxviruses and immune evasion. Annu. Rev. Immunol. 21: 377–423. [DOI] [PubMed] [Google Scholar]
- Shchelkunov, S.N. 2003. Immunomodulatory proteins of orthopoxviruses. Mol. Biol. (Mosk.) 37: 41–53. [PubMed] [Google Scholar]
- Shindyalov, I.N. and Bourne, P.E. 1998. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 11: 739–747. [DOI] [PubMed] [Google Scholar]
- Su, J., Wang, G., Barrett, J.W., Irvine, T.S., Gao, X., and McFadden, G. 2006. Myxoma virus M11L blocks apoptosis through inhibition of conformational activation of Bax at the mitochondria. J. Virol. 80: 1140–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, J.M. and Barry, M. 2006. Near death experiences: Poxvirus regulation of apoptotic death. Virology 344: 139–150. [DOI] [PubMed] [Google Scholar]
- Tulman, E.R., Afonso, C.L., Lu, Z., Zsak, L., Kutish, G.F., and Rock, D.L. 2004. The genome of canarypox virus. J. Virol. 78: 353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne, G.D., Standaert, R.F., Karplus, P.A., Schreiber, S.L., and Clardy, J. 1993. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229: 105–124. [DOI] [PubMed] [Google Scholar]
- Wang, G., Barrett, J.W., Nazarian, S.H., Everett, H., Gao, X., Bleackley, C., Colwill, K., Moran, M.F., and McFadden, G. 2004. Myxoma virus M11L prevents apoptosis through constitutive interaction with Bak. J. Virol. 78: 7097–7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilenko, S.T., Banadyga, L., Bond, D., and Barry, M. 2005. The vaccinia virus F1L protein interacts with the proapoptotic protein Bak and inhibits Bak activation. J. Virol. 79: 14031–14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, J.S., Jung, J.S., Ha, N.C., Shin, J., Kim, K.H., Lee, W., and Oh, B.H. 2003. Unique structural features of a BCL-2 family protein CED-9 and biophysical characterization of CED-9/EGL-1 interactions. Cell Death Differ. 10: 1310–1319. [DOI] [PubMed] [Google Scholar]
- Yin, X.M., Oltvai, Z.N., and Korsmeyer, S.J. 1994. BH1 and BH2 domains of Bcl-2 are required for inhibition of apoptosis and heterodimerization with Bax. Nature 369: 321–323. [DOI] [PubMed] [Google Scholar]
- Zhai, D., Luciano, F., Zhu, X., Guo, B., Satterthwait, A.C., and Reed, J.C. 2005. Humanin binds and nullifies Bid activity by blocking its activation of Bax and Bak. J. Biol. Chem. 280: 15815–15824. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Abrahams, M.R., Hunt, L.A., Suttles, J., Marshall, W., Lahiri, D.K., and Kotwal, G.J. 2005. The vaccinia virus N1L protein influences cytokine secretion in vitro after infection. Ann. N.Y. Acad. Sci. 1056: 69–86. [DOI] [PubMed] [Google Scholar]
- Zhou, H., Zhang, C., Liu, S., and Zhou, Y. 2005. Web-based toolkits for topology prediction of transmembrane helical proteins, fold recognition, structure and binding scoring, folding-kinetics analysis and comparative analysis of domain combinations. Nucleic Acids Res. 33: W193–W197. [DOI] [PMC free article] [PubMed] [Google Scholar]