Abstract
The crystal structure of thioredoxin (AaTrx) from the acetic acid bacterium Acetobacter aceti was determined at 1 Å resolution. This is currently the highest resolution crystal structure available for any thioredoxin. Thioredoxins facilitate thiol-disulfide exchange, a process that is expected to be slow at the low pH values encountered in the A. aceti cytoplasm. Despite the apparent need to function at low pH, neither the active site nor the surface charge distribution of AaTrx is notably different from that of Escherichia coli thioredoxin. Apparently the ancestral thioredoxin was sufficiently stable for use in A. aceti or the need to interact with multiple targets constrained the variation of surface residues. The AaTrx structure presented here provides a clear view of all ionizable protein moieties and waters, a first step in understanding how thiol-disulfide exchange might occur in a low pH cytoplasm, and is a basis for biophysical studies of the mechanism of acid-mediated unfolding. The high resolution of this structure should be useful for computational studies of thioredoxin function, protein structure and dynamics, and side-chain ionization.
Keywords: acidophile, thioredoxin, thiol-disulfide exchange, crystal structure
Acetobacter aceti is a Gram-negative α-proteobacterium that oxidizes ethanol to acetic acid, a trait useful in vinegar production throughout recorded human history (Bergey et al. 1984). The discovery and early study of the acetic acid bacteria by Pasteur was facilitated by their resistance to ethanol and acetic acid (Pasteur 1868). Acetic acid is a potent microbicide by virtue of its ability to penetrate cell membranes, causing cytoplasmic acidification and proton gradient collapse in susceptible cells. A. aceti survives by tolerating an acidic cytoplasm; as the external pH drops, the cytoplasmic pH falls from around 6 to 4 during mid-logarithmic growth (Menzel and Gottschalk 1985). Acetic acid resistance strategies include an acetate transporter driven by the proton gradient (Matsushita et al. 2005; Nakano et al. 2006), at least one citric acid cycle component (Fukaya et al. 1990), and potentially several proteins induced during acid adaptation (Steiner and Sauer 2001, 2003). We have shown that several cytoplasmic A. aceti proteins are intrinsically acid resistant (Constantine et al. 2006; Francois and Kappock 2006; Francois et al. 2006). Maintaining the proper function of metabolism over a broad pH range is another striking ability of A. aceti that has received less attention.
Thiol-disulfide exchange is a reaction that is particularly slow at low pH. The reactivity of the incoming thiolate nucleophile depends strongly on its pK a (Whitesides et al. 1977). A sharp decrease in exchange rate with decreasing pH is observed for small molecule thiol-disulfide exchange reactions, to below ∼1 M−1 s−1 at pH <7 (Pleasants et al. 1989).
Thioredoxin (Trx) is a small monomeric protein that functions in a variety of cytoplasmic redox roles in bacteria (Kumar et al. 2004). A Cys-X-X-Cys motif in Trx is maintained in a reduced dithiol state by Trx reductases (Holmgren 1989). Thiol-disulfide exchange is a two-step reaction: second-order formation of a heterodisulfide is followed by a rapid intramolecular displacement of the departing thiolate. The active site environment of Escherichia coli thioredoxin A (EcTrx) accelerates disulfide exchange with respect to small-molecule analogs by lowering the pK a of the attacking nucleophile Cys32 (Kallis and Holmgren 1980), raising the pK a of its buried partner Cys35 (LeMaster 1996), and stabilizing the heterodisulfide intermediate (Wynn et al. 1995). These effects are thought to be due to the local electrostatic and dielectric environment created by the surrounding residues, including a buried, protonated Asp26 (Wilson et al. 1995; Wynn et al. 1995; Jeng and Dyson 1996; LeMaster 1996; Chivers et al. 1997; Dyson et al. 1997; Dillet et al. 1998). Similar perturbations are observed by placing small-molecule thiols in nonaqueous solvents (Singh and Whitesides 1990).
A key role of Trx is to reduce class I ribonucleotide reductase (RNR) (Laurent et al. 1964). RNR, in turn, catalyzes reduction of NDPs (or NTPs) to dNDPs (or dNTPs), a potentially rate-limiting process in DNA replication (Thelander and Reichard 1979; Howell et al. 1993). If Trx is unavailable, glutaredoxin can reduce RNR, but heterodisulfide formation is still required (Holmgren 1976). Clearly, any hindrance in the ability to effect thiol-disulfide exchange would be deleterious to A. aceti survival. The kinetic problem caused by sluggish thiol-disulfide exchange is only exacerbated in periplasmic proteins that function as protein disulfide synthases and isomerases, some of which are Trx orthologs (Ritz and Beckwith 2001).
EcTrx is a familiar subject of protein structure (Holmgren et al. 1975; Katti et al. 1990; Jeng et al. 1994; Bhutani and Udgaonkar 2003) and folding studies (Kelley et al. 1986; Ladbury et al. 1993; Maier et al. 1999; Huber et al. 2005). EcTrx is even acid resistant (Hiraoki et al. 1988). Similarly, A. aceti Trx (AaTrx) should be a good candidate for biophysical studies of the mechanism of acid-mediated unfolding of A. aceti proteins. This is important, as the A. aceti proteins previously studied in this laboratory are all multisubunit enzymes with an unfortunate tendency to unfold irreversibly (Constantine et al. 2006; Francois and Kappock 2006; Francois et al. 2006).
Here we present the cloning, purification, and high-resolution structure of AaTrx as a first step in understanding how electron flow by thiol-disulfide exchange occurs at low pH and as a basis for biophysical studies of acid-mediated unfolding.
Results and Discussion
Cloning, isolation, and characterization of AaTrx
A draft genome sequence of the industrial vinegar strain A. aceti 1023 contains an open reading frame with 57% identity to EcTrx. The inferred protein sequence of AaTrx is shown in Figure 1, aligned with the EcTrx sequence. The gene was cloned, and AaTrx (108 amino acids, 11.6 kDa) was expressed in E. coli as a partly soluble protein. Exploiting the serendipitous thermostability of A. aceti proteins, host cell proteins were efficiently removed from AaTrx by a heat-denaturation step. After ammonium sulfate fractionation and size-exclusion chromatography, a mixture of protein bands migrating between 7 and 11 kDa was obtained on SDS-PAGE. Urea/SDS-PAGE showed a single band, migrating at ∼15 kDa (data not shown). Electrospray ionization mass spectrometry (ESI-MS) indicated removal of the starting methionine residue, with an observed m/z = 11,553 ± 3 Da (11,556.3 Da expected for [M-Met]). Analytical gel filtration showed a single peak with an apparent molecular weight of 11 kD, consistent with a monomeric protein.
Figure 1.
Sequence alignment of AaTrx and EcTrx. Identical residues are highlighted.
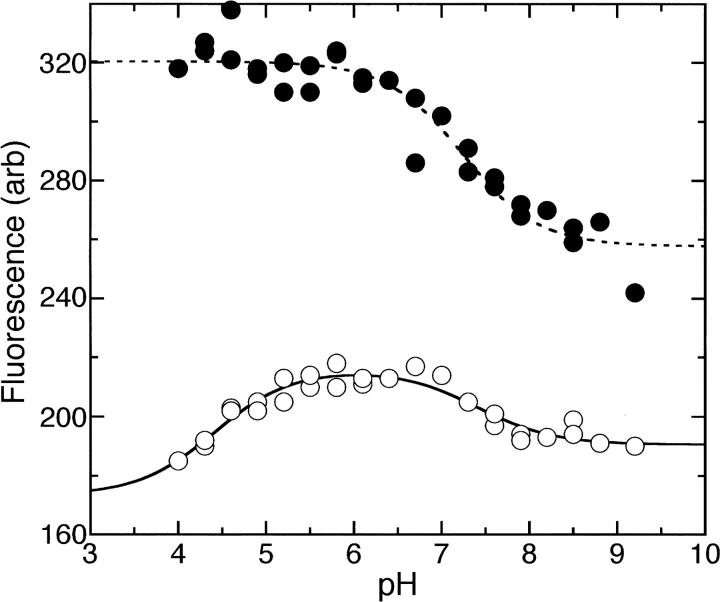
The fluorescence properties of oxidized and reduced AaTrx were measured as a function of pH in an experiment similar to those reported for EcTrx (Holmgren 1972; Dyson et al. 1997). The pH-dependent fluorescence changes in EcTrx report on the ionization states of residues near Trp28, including Asp26, and possibly Cys32 (Dyson et al. 1997). As observed for EcTrx, the reduced form of AaTrx shows a much more dramatic change in fluorescence intensity with pH than does the oxidized form (Fig. 2). Fitting the alkaline side of the reduced AaTrx titration curve gives an apparent pK a of 7.24, compared with the apparent pK a of 6.75 determined for EcTrx (Holmgren 1972).
Figure 2.
Fluorescence emission intensity at 350 nm as a function of pH for reduced AaTrx (•) and oxidized AaTrx (○). The dotted line is a fit to the equation y = [F1(10-pH) + F2(10-pKa)]/[10-pH + 10-pKa]; pK a = 7.2 ± 0.1, F1 = 320 ± 2, F2 = 258 ± 4. The solid line is a fit to the equation y = [F1(10−2pH) + F2(10-pKa1-pH) + F3(10-pKa1-pKa2)]/[10−2pH + 10-pKa1-pH + 10-pKa1-pKa2]; pK a1 = 4.4 ± 0.2, pK a2 = 7.4 ± 0.2, F1 = 173 ± 9, F2 = 216 ± 2, F3 = 191 ± 2.
Crystal structure determination
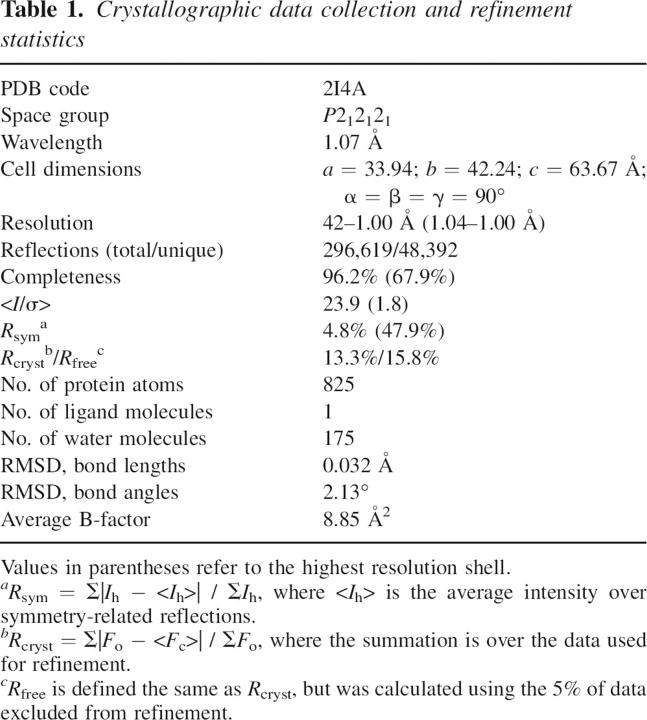
The crystal structure of AaTrx was determined by molecular replacement with a data set extending to 1 Å resolution. All amino acid residues present in the purified protein were readily modeled into the electron density, and alternative conformations were modeled for the side chains of Thr4, Ser20, Leu24, Met37, Ile45, and Leu99. The high resolution of the data allowed loosening of the geometric restraints in Refmac; 95.6% of residues are in the most favored regions of the Ramachandran plot, while the remaining 4.4% are in the additional allowed regions. Data collection and refinement statistics are given in Table 1, and a representative region of electron density is shown in Figure 3A. Coordinates and structure factors have been deposited in the RCSB Protein Data Bank (PDB) with accession code 2I4A.
Table 1.
Crystallographic data collection and refinement statistics
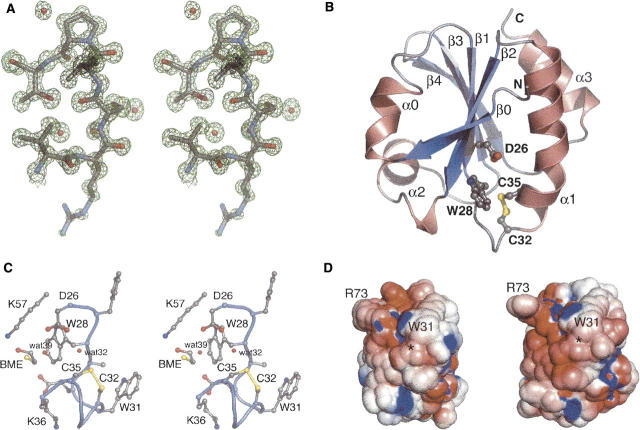
Figure 3.
(A) Stereo view of a representative portion of the σA-weighted 2F o-F c map calculated using the final refined structure. The map is contoured at 1.5 σ. Pro76, a cis proline, is shown at top. The side chain of Arg73, shown at the bottom, is solvent exposed and somewhat disordered. (B) Ribbon diagram of AaTrx. Helices and strands are numbered according to the Trx fold (Martin 1995) and correspond to residues 4–8 (β0), 9–15 (α0), 22–27 (β1), 33–49 (α1), 53–59 (β2), 60–70 (α2, interrupted by Pro64), 77–82 (β3), 85–91 (β4), and 96–105 (α3). Selected residues are shown in ball-and-stick representation. The termini are labeled with N and C, respectively. (C) Stereo view of a ball-and-stick representation of selected residues near the AaTrx active site. (D) Electrostatic rendering of the solvent-accessible surface of AaTrx (left) and EcTrx (right), computed at pH 7 with (red) −5 kT/e and (blue) +5 kT/e. Locations of Trp31 and Arg73 are located for orientation. The location of the Cys32–Cys35 disulfide is marked with an asterisk.
Protein fold and notable residues
Like other Trx forms (Martin 1995), AaTrx consists of a five-stranded β-sheet surrounded by four α-helices (Fig. 3B). The structure of AaTrx closely resembles that of oxidized EcTrx (PDB code 2TRX, RMSD 0.79 Å over 105 Cα atoms). The most notable differences are in residues 1–4; in AaTrx, these residues pack more closely against the rest of the protein, with the carbonyl oxygen of His3 forming the first hydrogen bond of the β-sheet. This arrangement allows the side chain of Asp2 to occupy the space filled by Asp43 in EcTrx (Gly43 in AaTrx). Conformational differences also exist in the loop region comprising residues 50–52. Both of these regional differences are near crystal contacts and may be attributable to crystal packing. All key EcTrx active site residues are conserved in AaTrx, including Trp28, Cys32, Cys35, and Asp26, Lys36, and Lys57 (Fig. 3C).
AaTrx is present in the crystal in the oxidized form. Cys32 and Cys35 form a disulfide bond with a sulfur–sulfur distance of 2.31 Å; this distance is longer than the 2.09 Å measured in EcTrx. To explore whether this long disulfide bond length represents a mixture of oxidized and reduced forms, we calculated a 2F o-F c omit map using phases from a structure refined without residues 28–36. Examining the electron density at increasing contour levels from 1 to 10 σ showed only a single position for each sulfur atom. Although AaTrx was stored and crystallized in the presence of 15 mM 2-mercaptoethanol (BME), the disulfide apparently reoxidized during crystal growth or data collection. Refinement of the AaTrx structure against a lower-resolution (1.2 Å) data set collected earlier from the same crystal (see Materials and Methods) also showed well-defined sulfur positions, with a disulfide bond distance of 2.21 Å, suggesting that oxidiation of the disulfide occurred prior to data collection (data not shown).
Near the disulfide, clear electron density for a molecule of BME was observed. The hydrocarbon portion of BME packs against the side chain of Trp28, and the hydroxyl group forms a hydrogen bond with the side chain of Lys57. The BME sulfur is 7.3 Å from the Cys35 sulfur, precluding formation of a mixed disulfide bond in this conformation. However, rotation about the Cys35 Cα–Cβ bond, and rotation about the BME C–C bond would bring the sulfurs within 2.3 Å of each other. Therefore, a subtle conformational change could accommodate the formation of a mixed disulfide between protein and reducing agent. The BME binding site is on the “shielded side” of the disulfide (Eklund et al. 1984) rather than on the accessible, hydrophobic side thought to be important for substrate binding. Examination of the proposed model of a Trx–Trx reductase complex (Sandalova et al. 2001) suggests that this shielded side might be accessible to the physiological reductant Trx reductase.
A protonated Asp26 residue is implicated in proton transfer to and from Cys35 (Chivers and Raines 1997). AaTrx Asp26 should be fully protonated at the crystallization pH of 4.6 (Chivers et al. 1997). In the AaTrx crystal structure, the side chain of Asp26 forms hydrogen bonds with two water molecules (waters 32 and 39); water 39 forms additional hydrogen bonds with the BME sulfhydryl and the Cys35 carbonyl, while water 32 forms two hydrogen bonds to the Cys35 sulfhydryl and the Phe27 carbonyl. Water 32 is well positioned to participate in proton transfer between the Asp26 and Cys35 side chains.
Adaptation to low pH
The acidic A. aceti cytoplasm appears to exert selective pressure to increase the acid stability of its cytoplasmic components. Such adaptation is evident in the structures of its proteins. The surfaces of the two other A. aceti proteins with known structures are quite different from their E. coli counterparts. The surface of A. aceti citrate synthase is highly decorated with basic side chains, and has many fewer uncompensated negative charges than E. coli citrate synthase (Francois et al. 2006). PurE from A. aceti has a distribution of positive and negative charges on its surface, distinguishing it from the acidic surface of E. coli PurE (Constantine et al. 2006). In contrast, the surface charge distributions of AaTrx and EcTrx are very similar (Fig. 3D). Comparing the two structures, nearly all differences in charged surface residues are offset by compensating differences elsewhere on the protein surface. This lack of apparent surface adaptation in AaTrx may not be surprising since EcTrx is acid-stable (Hiraoki et al. 1988); therefore, Trx may not have been subject to selective pressure to further improve acid stability in a low pH cytoplasm. In contrast, there may have been unusually strong selective pressure not to alter protein surface residues, given the many different targets of Trx in the cell.
It might be expected that the active site of Trx from an acidophile would be perturbed to maintain Cys32 as a reactive thiolate at lower pH. However, as measured by the fluorescence of Trp28, there is no downward shift in the apparent pK as of AaTrx active site residues relative to EcTrx. Consistent with this result, there are no major structural differences apparent in the active site regions of oxidized AaTrx and EcTrx. These observations suggest that alterations in residues near the AaTrx active site were not needed for low pH adaptation, or might have disrupted the essential Asp26-Cys35-Cys32 proton transfer system.
The Trx redox potential, a pH-dependent thermodynamic quantity, is critical for its proper physiological functions. At and just below neutrality, the midpoint potential of EcTrx has a slope of –59 mV per unit pH, as is expected for a proton-coupled electron transfer (Setterdahl et al. 2003). As the internal pH of A. aceti varies, the disulfide reduction potential of AaTrx and its targets may vary in parallel, minimizing differences in reduction potential. It is also possible that the local pH and dielectric environment within AaTrx-target protein complexes facilitates thiol-disulfide exchange at low solution pH. Subtle structural differences not immediately apparent from the crystal structure may affect the AaTrx active site. Future work will examine how the reduction potential of AaTrx varies over the range of pH's encountered in the A. aceti cytoplasm.
Materials and methods
AaTrx cloning
Shotgun sequencing of the industrial vinegar strain A. aceti 1023 to fourfold coverage (S.W. Clifton, R.K. Wilson, and T.J. Kappock, unpubl.) revealed a single open-reading frame (Contig0.22_19289_19618_AAC0295) with 57% identity to EcTrx. The nucleotide sequence was deposited in GenBank with accession number DQ869238. This gene was amplified by PCR with Vent DNA polymerase (New England Biolabs), A. aceti strain 1023 genomic DNA (3 ng), and oligodeoxynucleotide primers 1041 (5′-AACCAGCATATGAGTGAACATACGC) and 1042 (5′-ACCTGAATTCTAATTACTGAGCGCTTTCTAC) from IDT, and was cloned into the NdeI and EcoRI sites of pET23a (Novagen) to generate plasmid pJK295. DNA sequencing revealed the expected sequence.
AaTrx expression and purification
E. coli BL21(DE3) cells harboring pJK295 were propagated on solid LB medium containing 0.1 mg/L ampicillin (LB/Amp). A starter culture grown overnight at 37°C was used to inoculate a production culture (1 L LB/Amp) at a 1:20 dilution. After the culture reached an optical density of 0.6 at 600 nm, isopropylthio-β-galactopyranoside (IPTG) was added to a final concentration of 0.4 mM. Cells were grown another 4 h, harvested by centrifugation, and either used immediately or stored at −80°C. All subsequent steps were performed at 4°C unless noted. Cells (typically 7 g per L of culture) were resuspended in 5 vol of TM buffer (0.05 M Tris-HCl at pH 8.0, and 15 mM BME) and disrupted by three cycles of sonication. After removing debris by centrifugation at 27,000g for 20 min (“fast spin”), the supernatant was placed in a hybridization oven at 70°C for 20 min, then chilled on ice. After a fast spin to remove solids, the supernatant was adjusted to 30% saturation by the addition of solid ammonium sulfate (176 g/L) with stirring over 30 min. After stirring another 30 min, solids were removed by a fast spin and the supernatant was adjusted to 70% saturation by the addition of solid ammonium sulfate (270 g/L) with stirring over 30 min. After stirring another 30 min, solids were collected by a fast spin, dissolved in a minimal volume (∼15 mL) of TM buffer, and applied to Sephadex G50 (3.5 × 12 cm) equilibrated and eluted with TM buffer. Fractions containing AaTrx were identified by SDS-PAGE, pooled (∼70 mL), and concentrated to <15 mL (>4 mg/mL) by ultrafiltration (Amicon Centriprep YM10; Millipore). Single-use aliquots were frozen and stored at −80°C until used. Typically, 4 mg of pure AaTrx was obtained from a 1 L culture.
Analytical methods
Denaturing urea gel electrophoresis (urea/SDS-PAGE) was performed using 20% polyacrylamide gels (37.5:1 acrylamide:bis-acrylamide) containing 0.125 M Tris-HCl (pH 8.8), 0.1% (w/v) SDS, and 8 M urea. Brief heating at 37°C was required to dissolve the urea. These gels were polymerized, run, and stained (Coomassie Blue) like typical SDS-PAGE gels (Ausubel et al. 2000).
ESI-MS was performed by infusing AaTrx (50 μg) via an in-line C18 guard column (MS Scientific) into a Micromass Q-TOF Ultima quadrupole MS. The mobile phase was acetonitrile:water:trifluoroacetic acid (80:20:0.1), and the accelerating potential was 10 eV.
Analytical gel filtration was carried out at 5°C on a Tosoh TSK-GEL G2000SWXL column (0.78 × 30 cm) in 20 mM Tris (pH 8.0), 100 mM NaCl with a flow rate of 4 mL/min. AaTrx (120 μg) was loaded in 15 μL. The column was calibrated with thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), equine myoglobin (17 kDa), and vitamin B12 (1.3 kDa). The volume versus log(molecular weight) method was used to construct a calibration curve.
Fluorescence measurements were carried out on a Cary Eclipse fluorimeter equipped with a thermostatted cell holder at 22°C using a 1-cm fluorimeter cell that contained 2.0 mL of solution. The excitation wavelength was 280 nm (5 nm slitwidth), and emission spectra were collected from 290–450 nm. Reduced and oxidized AaTrx were prepared by incubating the protein for at least 20 min with 50 mM of either dithiothreitol or its oxidized form, trans-4,5-dihydroxy-1,2-dithiane, respectively. In each case, no change in emission spectrum was observed at DTT concentrations between 20 and 75 mM after incubations of 3–60 min. Fluorescence measurements were performed at varied pH by diluting oxidized or reduced AaTrx (5 μL, 0.9 mg/mL) into 2 mL of a solution containing 100 mM KCl plus an acetic acid/MES/Tris constant ionic strength triple buffer (Ellis and Morrison 1982). Emission intensities at 340 nm were plotted as a function of pH and apparent pK as were obtained from fits to forms of the Henderson-Hasselbalch equation.
Crystallization of AaTrx
Crystals were grown at room temperature by the hanging-drop vapor diffusion method. Drops consisted of 1 μL of protein in buffer TM and 1 μL of reservoir solution; reservoir volumes were 500 μL. Crystals initially grew in 100 mM sodium acetate (pH 4.6–5), 50–100 mM ammonium acetate, and 30% (w/v) PEG 4000. More reliable nucleation and better morphology were achieved by using crystals grown in these conditions for microseeding into fresh drops with reservoir solutions containing 100 mM sodium acetate (pH 4.6), 100 mM ammonium acetate, and 24% (w/v) PEG 4000. Prior to data collection, crystals were briefly soaked in a cryoprotectant solution (100 mM sodium acetate at pH 4.6, 100 mM ammonium acetate, 25% [w/v] PEG 4000, and 30% [v/v] ethylene glycol), and then frozen in liquid nitrogen. The data set described here was collected from a crystal that was allowed to grow for 1 mo after seeding.
Data collection and processing
Crystallographic data were collected at the Advanced Light Source beamline 4.2.2, using a 2θ-offset. A data set extending to 1.2 Å was collected from a single crystal (data not shown); this crystal was then stored in liquid nitrogen and used later to collect an additional data set that extended to 1.0 Å resolution. The 1.0 Å data set was used for structure determination and refinement. Data were indexed, integrated, and scaled using d*TREK (Pflugrath 1999); the crystal was indexed with a primitive orthorhombic lattice, and examination of systematic absences allowed assignment of the space group as P212121. Intensities were then converted to structure factor amplitudes using the Truncate (French and Wilson 1978) program in CCP4i (Collaborative Computational Project No. 4 1994; Potterton et al. 2003), and 5% of the data were set aside as the test set for R free calculations.
Structure determination, model building, and refinement
The structure of AaTrx was determined by molecular replacement in MOLREP (Vagin and Teplyakov 1997). The search model consisted of one monomer of oxidized EcTrx (PDB code 2TRX) with side chains trimmed to minimally conserved shapes using SEAMAN (Kleywegt and Jones 1996). Rigid body refinement, simulated annealing, and grouped B-factor refinement were then carried out in CNS (Brünger et al. 1998) using data to 2 Å. This yielded an R-factor of 33.5% (R free 41.0%). Rebuilding was carried out and side chains were mutated to correspond to the A. aceti sequence, using the program O (Jones et al. 1991). Additional rounds of refinement and water picking were carried out using REFMAC 5 (Vagin et al. 2004) with ARP/wARP (Perrakis et al. 1997) using data to 1.05 Å and allowing anisotropic B factor refinement. The Cys32–Cys35 disulfide was refined in the oxidized form with a target bond length of 2.03 Å. Hydrogens added with the program MOLPROBITY (Davis et al. 2004) were used in later refinement rounds. In the final refinement, data to 1.00 Å were used. The refined model has an R-factor of 13.3% (R free 15.8%). Residues in the model are numbered beginning after the starting methionine and correspond to the residue numbering in EcTrx.
Computational analysis and figure preparation
Electrostatic calculations were performed at pH 7.0 using PDB2PQR (Dolinsky et al. 2004) and APBS (Baker et al. 2001). Figures were rendered with PyMOL (DeLano 2002).
Acknowledgments
We thank C. Nelson and J. Nix for assistance with crystal screening and data collection and processing, and J. Swensson for initial crystallization experiments. We thank P. Chivers and J. Jez for comments on the manuscript. This research was supported by grants from the NSF (CAREER MCB 0347250) and Herman Frasch Foundation (531-HF02) to T.J.K. and a Washington University Summer Scholars Program in Biology and Biomedical Research to B.Z.H. Mass spectrometry was provided by the Washington University Mass Spectrometry Resource, supported by the NIH (P41RR0954).
Footnotes
Reprint requests to: T. Joseph Kappock, Department of Chemistry, Washington University in Saint Louis, St. Louis Missouri 63130, USA; e-mail: kappock@wustl.edu; fax: (314) 935-4481.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062519707.
Abbreviations: AaTrx, Acetobacter aceti thioredoxin; Amp, ampicillin; BME, 2-mercaptoethanol; DTT, dithiothreitol; EcTrx, E. coli thioredoxin A; ESI-MS, electrospray ionization mass spectrometry; LB, Luria-Bertani medium; RMSD, root-mean-square deviation; Trx, thioredoxin.
References
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.D., Smith, J.A., and Struhl, K. 2000. Current protocols in molecular biology. John Wiley & Sons, New York.
- Baker, N.A., Sept, D., Joseph, S., Holst, M.J., and McCammon, J.A. 2001. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. 98: 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergey, D.H., Krieg, N.R., and Holt, J.G. 1984. Bergey's manual of systematic bacteriology. Williams & Wilkins, Baltimore, MD.
- Bhutani, N. and Udgaonkar, J.B. 2003. Folding subdomains of thioredoxin characterized by native-state hydrogen exchange. Protein Sci. 12: 1719–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, M., and Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921. [DOI] [PubMed] [Google Scholar]
- Chivers, P.T. and Raines, R.T. 1997. General acid/base catalysis in the active site of Escherichia coli thioredoxin. Biochemistry 36: 15810–15816. [DOI] [PubMed] [Google Scholar]
- Chivers, P.T., Prehoda, K.E., Volkman, B.F., Kim, B.M., Markley, J.L., and Raines, R.T. 1997. Microscopic pKa values of Escherichia coli thioredoxin. Biochemistry 36: 14985–14991. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4. 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50: 760–763. [DOI] [PubMed] [Google Scholar]
- Constantine, C.Z., Starks, C.M., Mill, C.P., Ransome, A.E., Karpowicz, S.J., Francois, J.A., Goodman, R.A., and Kappock, T.J. 2006. Biochemical and structural studies of N 5-carboxyaminoimidazole ribonucleotide mutase (PurE) from the acidophilic bacterium Acetobacter aceti . Biochemistry 45: 8193–8208. [DOI] [PubMed] [Google Scholar]
- Davis, I.W., Murray, L.W., Richardson, J.S., and Richardson, D.C. 2004. MOLPROBITY: Structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32: W615–W619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano, W.L. 2002. The PyMOL users manual. San Carlos, CA.
- Dillet, V., Dyson, H.J., and Bashford, D. 1998. Calculations of electrostatic interactions and pKas in the active site of Escherichia coli thioredoxin. Biochemistry 37: 10298–10306. [DOI] [PubMed] [Google Scholar]
- Dolinsky, T.J., Nielsen, J.E., McCammon, J.A., and Baker, N.A. 2004. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32: W665–W667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson, H.J., Jeng, M.F., Tennant, L.L., Slaby, I., Lindell, M., Cui, D.S., Kuprin, S., and Holmgren, A. 1997. Effects of buried charged groups on cysteine thiol ionization and reactivity in Escherichia coli thioredoxin: Structural and functional characterization of mutants of Asp 26 and Lys 57. Biochemistry 36: 2622–2636. [DOI] [PubMed] [Google Scholar]
- Eklund, H., Cambillau, C., Sjöberg, B.M., Holmgren, A., Jörnvall, H., Höög, J.O., and Brändén, C.I. 1984. Conformational and functional similarities between glutaredoxin and thioredoxins. EMBO J. 3: 1443–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, K.J. and Morrison, J.F. 1982. Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol. 87: 405–426. [DOI] [PubMed] [Google Scholar]
- Francois, J.A. and Kappock, T.J. 2006. Alanine racemase from the acidophile Acetobacter aceti . Protein Expr. Purif. doi:10.1016/j.pep.2006.05.016. [DOI] [PubMed]
- Francois, J.A., Starks, C.M., Sivanuntakorn, S., Jiang, H., Ransome, A.E., Nam, J.-W., Constantine, C.Z., and Kappock, T.J. 2006. Structure of a NADH-insensitive hexameric citrate synthase that resists acid inactivation. Biochemistry 45: 13487–13499. [DOI] [PubMed] [Google Scholar]
- French, G.S. and Wilson, K.S. 1978. On the treatment of negative intensity observations. Acta Crystallogr. A 34: 517–525. [Google Scholar]
- Fukaya, M., Takemura, H., Okumura, H., Kawamura, Y., Horinouchi, S., and Beppu, T. 1990. Cloning of genes responsible for acetic acid resistance in Acetobacter aceti . J. Bacteriol. 172: 2096–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoki, T., Brown, S.B., Stevenson, K.J., and Vogel, H.J. 1988. Structural comparison between oxidized and reduced Escherichia coli thioredoxin. Proton NMR and CD studies. Biochemistry 27: 5000–5008. [DOI] [PubMed] [Google Scholar]
- Holmgren, A. 1972. Tryptophan fluorescence study of conformational transitions of the oxidized and reduced form of thioredoxin. J. Biol. Chem. 247: 1992–1998. [PubMed] [Google Scholar]
- Holmgren, A. 1976. Hydrogen donor system for Escherichia coli ribonucleoside-diphosphate reductase dependent upon glutathione. Proc. Natl. Acad. Sci. 73: 2275–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren, A. 1989. Thioredoxin and glutaredoxin systems. J. Biol. Chem. 264: 13963–13966. [PubMed] [Google Scholar]
- Holmgren, A., Söderberg, B.O., Eklund, H., and Brändén, C.I. 1975. Three-dimensional structure of Escherichia coli thioredoxin-S2 to 2.8 Å resolution. Proc. Natl. Acad. Sci. 72: 2305–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, M.L., Roseman, N.A., Slabaugh, M.B., and Mathews, C.K. 1993. Vaccinia virus ribonucleotide reductase. Correlation between deoxyribonucleotide supply and demand. J. Biol. Chem. 268: 7155–7162. [PubMed] [Google Scholar]
- Huber, D., Cha, M.-I., Debarbieux, L., Planson, A.-G., Cruz, N., López, G., Tasayco, M.L., Chaffotte, A., and Beckwith, J. 2005. A selection for mutants that interfere with folding of Escherichia coli thioredoxin-1 in vivo. Proc. Natl. Acad. Sci. 102: 18872–18877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng, M.F. and Dyson, H.J. 1996. Direct measurement of the aspartic acid 26 pK a for reduced Escherichia coli thioredoxin by 13C NMR. Biochemistry 35: 1–6. [DOI] [PubMed] [Google Scholar]
- Jeng, M.F., Campbell, A.P., Begley, T., Holmgren, A., Case, D.A., Wright, P.E., and Dyson, H.J. 1994. High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin. Structure 2: 853–868. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47: 110–119. [DOI] [PubMed] [Google Scholar]
- Kallis, G.B. and Holmgren, A. 1980. Differential reactivity of the functional sulfhydryl groups of cysteine-32 and cysteine-35 present in the reduced form of thioredoxin from Escherichia coli . J. Biol. Chem. 255: 10261–10265. [PubMed] [Google Scholar]
- Katti, S.K., LeMaster, D.M., and Eklund, H. 1990. Crystal structure of thioredoxin from Escherichia coli at 1.68 Å resolution. J. Mol. Biol. 212: 167–184. [DOI] [PubMed] [Google Scholar]
- Kelley, R.F., Wilson, J., Bryant, C., and Stellwagen, E. 1986. Effects of guanidine hydrochloride on the refolding kinetics of denatured thioredoxin. Biochemistry 25: 728–732. [DOI] [PubMed] [Google Scholar]
- Kleywegt, G.J. and Jones, T.A. 1996. Making the most of your search model. CCP4/ESF-EACBM Newslett . Protein Crystallogr. 32: 32–36. [Google Scholar]
- Kumar, J.K., Tabor, S., and Richardson, C.C. 2004. Proteomic analysis of thioredoxin-targeted proteins in Escherichia coli . Proc. Natl. Acad. Sci. 101: 3759–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladbury, J.E., Wynn, R., Hellinga, H.W., and Sturtevant, J.M. 1993. Stability of oxidized Escherichia coli thioredoxin and its dependence on protonation of the aspartic acid residue in the 26 position. Biochemistry 32: 7526–7530. [DOI] [PubMed] [Google Scholar]
- Laurent, T.C., Moore, E.C., and Reichard, P. 1964. Enzymatic synthesis of deoxyribonucleotides. IV. Isolation and characterization of thioredoxin, the hydrogen donor from Escherichia coli B. J. Biol. Chem. 239: 3436–3444. [PubMed] [Google Scholar]
- LeMaster, D.M. 1996. Structural determinants of the catalytic reactivity of the buried cysteine of Escherichia coli thioredoxin. Biochemistry 35: 14876–14881. [DOI] [PubMed] [Google Scholar]
- Maier, C.S., Schimerlik, M.I., and Deinzer, M.L. 1999. Thermal denaturation of Escherichia coli thioredoxin studied by hydrogen/deuterium exchange and electrospray ionization mass spectrometry: Monitoring a two-state protein unfolding transition. Biochemistry 38: 1136–1143. [DOI] [PubMed] [Google Scholar]
- Martin, J.L. 1995. Thioredoxin–a fold for all reasons. Structure 3: 245–250. [DOI] [PubMed] [Google Scholar]
- Matsushita, K., Inoue, T., Adachi, O., and Toyama, H. 2005. Acetobacter aceti possesses a proton motive force-dependent efflux system for acetic acid. J. Bacteriol. 187: 4346–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel, U. and Gottschalk, G. 1985. The internal pH of Acetobacterium wieringae and Acetobacter aceti during growth and production of acetic acid. Arch. Microbiol. 143: 47–51. [Google Scholar]
- Nakano, S., Fukaya, M., and Horinouchi, S. 2006. Putative ABC transporter responsible for acetic acid resistance in Acetobacter aceti . Appl. Environ. Microbiol. 72: 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasteur, L. 1868. Etudes sur le vinaigre, sa fabrication, ses maladies, moyens de les prévenir. Nouvelles observations sur la conservation des vins par la chaleur. Gauthier-Villars, Paris, France.
- Perrakis, A., Sixma, T.K., Wilson, K.S., and Lamzin, V.S. 1997. wARP: Improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr. D Biol. Crystallogr. 53: 448–455. [DOI] [PubMed] [Google Scholar]
- Pflugrath, J.W. 1999. The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55: 1718–1725. [DOI] [PubMed] [Google Scholar]
- Pleasants, J.C., Guo, W., and Rabenstein, D.L. 1989. A comparative study of the kinetics of selenol/diselenide and thiol/disulfide exchange reactions. J. Am. Chem. Soc. 111: 6553–6558. [Google Scholar]
- Potterton, E., Briggs, P., Turkenburg, M., and Dodson, E. 2003. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59: 1131–1137. [DOI] [PubMed] [Google Scholar]
- Ritz, D. and Beckwith, J. 2001. Roles of thiol-redox pathways in bacteria. Annu. Rev. Microbiol. 55: 21–48. [DOI] [PubMed] [Google Scholar]
- Sandalova, T., Zhong, L., Lindqvist, Y., Holmgren, A., and Schneider, G. 2001. Three-dimensional structure of a mammalian thioredoxin reductase: Implications for mechanism and evolution of a selenocysteine-dependent enzyme. Proc. Natl. Acad. Sci. 98: 9533–9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setterdahl, A.T., Chivers, P.T., Hirasawa, M., Lemaire, S.D., Keryer, E., Miginiac-Maslow, M., Kim, S.K., Mason, J., Jacquot, J.P., and Longbine, C.C., et al. 2003. Effect of pH on the oxidation-reduction properties of thioredoxins. Biochemistry 42: 14877–14884. [DOI] [PubMed] [Google Scholar]
- Singh, R. and Whitesides, G.M. 1990. Comparisons of rate constants for thiolate-disulfide interchange in water and in polar aprotic solvents using dynamic proton NMR line shape analysis. J. Am. Chem. Soc. 112: 1190–1197. [Google Scholar]
- Steiner, P. and Sauer, U. 2001. Proteins induced during adaptation of Acetobacter aceti to high acetate concentrations. Appl. Environ. Microbiol. 67: 5474–5481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner, P. and Sauer, U. 2003. Overexpression of the ATP-dependent helicase RecG improves resistance to weak organic acids in Escherichia coli . Appl. Microbiol. Biotechnol. 63: 293–299. [DOI] [PubMed] [Google Scholar]
- Thelander, L. and Reichard, P. 1979. Reduction of ribonucleotides. Annu. Rev. Biochem. 48: 133–158. [DOI] [PubMed] [Google Scholar]
- Vagin, A. and Teplyakov, A. 1997. MOLREP: An automated program for molecular replacement. J. Appl. Crystallogr. 30: 1022–1025. [Google Scholar]
- Vagin, A.A., Steiner, R.A., Lebedev, A.A., Potterton, L., McNicholas, S., Long, F., and Murshudov, G.N. 2004. REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 60: 2184–2195. [DOI] [PubMed] [Google Scholar]
- Whitesides, G.M., Lilburn, J.E., and Szajewski, R.P. 1977. Rates of thiol-disulfide interchange reactions between mono- and dithiols and Ellman's reagent. J. Org. Chem. 42: 332–338. [Google Scholar]
- Wilson, N.A., Barbar, E., Fuchs, J.A., and Woodward, C. 1995. Aspartic acid 26 in reduced Escherichia coli thioredoxin has a pK a > 9. Biochemistry 34: 8931–8939. [DOI] [PubMed] [Google Scholar]
- Wynn, R., Cocco, M.J., and Richards, F.M. 1995. Mixed disulfide intermediates during the reduction of disulfides by Escherichia coli thioredoxin. Biochemistry 34: 11807–11813. [DOI] [PubMed] [Google Scholar]