Abstract
Women diagnosed with breast cancer before the age of 40 years who have a strong family history of breast and/or ovarian cancer were selected from an Australian population-based case-control-family study for large deletion screening within the BRCA1 promoter. Deletions within the BRCA1 promoter region are usually not detected by the methods applied in routine clinical mutation detection strategies. Fifty-one of the 66 women (77%) who met our inclusion criteria were tested for promoter deletions using linkage disequilibrium analysis of two BRCA1 polymorphic sites (C/G1802 and Pro871Leu) and Multiplex Ligation-dependent Probe Amplification. Two cases of BRCA1 promoter deletion involving exons 1A-2, and exons 1A-23, were detected. The morphology of the breast cancers arising in these women with BRCA1 promoter deletions was consistent with the morphology associated with other germline BRCA1 mutations. Large genomic deletions that involve the promoter regions of BRCA1 make up 20% (2/10) of all known BRCA1 mutations in this group of young women with a strong family history of breast and ovarian cancer. Our data supports the inclusion of testing for large genomic alterations in the BRCA1 promoter region in routine clinical mutation detection within BRCA1.
Keywords: BRCAI promoter, large deletions, early-onset breast cancer
Introduction
Large genomic alterations within BRCA1 are now known to represent a non-trivial proportion of deleterious, clinically relevant, BRCA1 mutations. Early studies of genomic alterations reported several large deletions, many with evidence of founder effects (for example, 1), but the molecular methods were laborious, technically challenging and usually required a large amount of genomic DNA. More recently, Hogervorst et al., (2003) described a multiplex ligation-dependent probe amplification (MLPA) test to identify large genomic alterations in BRCA1(2). This methodology is easily applied to genomic DNA and larger scale screening for genomic alterations in BRCA1.
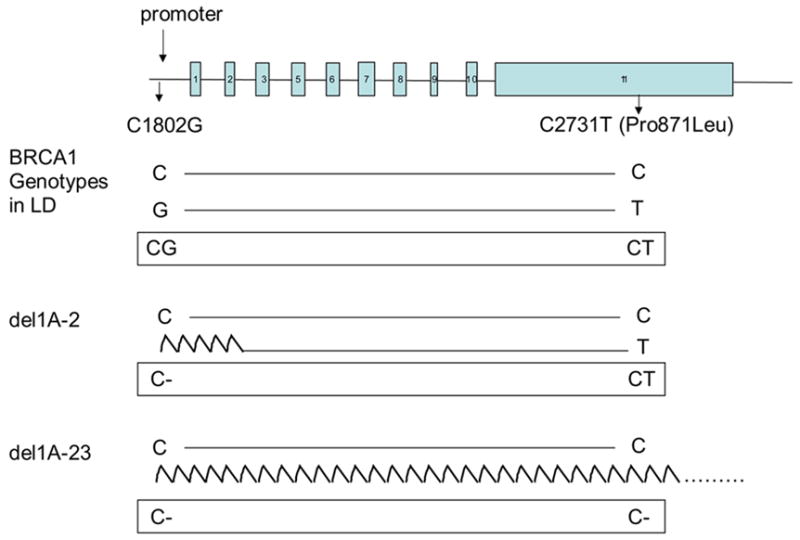
As it is the site of transcription initiation, a genetic mutation within the promoter could potentially disrupt normal expression of the BRCA1 protein. The 5′ end of BRCA1 lies head to head with the NBR2 gene (Next to BRCA1 gene 2) and the two genes share a bi-directional promoter that is capable of transcription in both directions. Two promoter sites, α and & β, exist for BRCA1 resulting in the transcription of alternate mRNA transcripts with varying first exons (1A or 1B) (3, 4). The relevance of mutation screening within the BRCA1 promoter has also been demonstrated. In a family known to be BRCA1-linked and to lack the BRCA1 transcript from the allele associated with disease susceptibility, Swensen et al., (1997) identified a 14kb deletion in BRCA1 that removed exon 1A, 1B and exon 2 (5). Brown et al., (2002) using a method described by Catteau et al. (1999) that utilized the strong linkage disequilibrium identified between the two BRCA1 variants, C1802G and C2731T (Pro871Leu), reported a large genomic deletion in the 5′ region of BRCA1 that included NBR2, ΨBRCA1 and NBR1 in an Australian multiple-case breast cancer family (6, 7).
The proportion of women with breast cancer, especially young women, whose cancers are attributable to deletions within the promoter region of BRCA1 is currently unknown. Although methods vary between laboratories and services, deletions within the BRCA1 promoter region are usually not detected by the methods applied in the majority of routine clinical mutation detection strategies. To quantitate this proportion we identified 66 women from our Australian population-based case-control-family study of breast cancer who were diagnosed with breast cancer before the age of 40 years and had a strong family history of breast and/or ovarian cancer (two or more first- or second- degree relatives affected with breast and/or ovarian cancer). Previous mutation screening performed on the germline DNA of these 66 cases had included screening within BRCA1, BRCA2 and ATM using a variety of mutation detection techniques (8, 9). This identified that 15 (23%) of these 66 cases to have detectable BRCA1 (n=8), BRCA2 (n=6) or specific ATM (n=1) (9) mutations. The study was approved by the ethics committees of The University of Melbourne and The Cancer Council Victoria.
Materials
The Australian Breast Cancer Family Registry
The Australian Breast Cancer Family Registry (ABCFR) is a population-based,case-control-family study of breast cancer (in which cases, control subjects and their relatives were administered the same questionnaire) and an emphasis on early-onset disease, that was carried out in Melbourne and Sydney, Australia (10,11,12). Sixty six women were identified within the ABCFR to be diagnosed with breast cancer before the age of 40 years and to have a strong family history of breast and/or ovarian cancer (two or more first- or second- degree relatives affected with breast and/or ovarian cancer). Previous mutation screening performed on the germline DNA of these 66 cases had included screening within BRCA1, BRCA2 and ATM using a variety of mutation detection techniques (8, 9). This identified that 15 (23%) of these 66 cases to have detectable BRCA1 (n=8), BRCA2 (n=6) or specific ATM (n=1) (9) mutations, Table 1. The study was approved by the ethics committees of The University of Melbourne and The Cancer Council Victoria.
Table 1.
Proband Cancer Family History.
| Families | affectedsa | cancersb | breast cancers | ovarian cancers | mutation carriersc |
|---|---|---|---|---|---|
| 6 | 3 | 3 | 3 (1 male) | 0 | |
| 1 | 3 | 3 | 2 | 1 | |
| 2 | 3 | 3 | 2 | 1 | |
| 1 | 3 | 3 | 2 | 1 | |
| 3 BRCA1, 1 BRCA2, | |||||
| 29 | 3 | 3 | 3 | 1 | 1 ATM |
| 1 | 3 | 4 | 3 | 1 | 1 BRCA1 |
| 1 | 3 | 4 | 4 | 0 | 1 BRCA2 |
| 15 | 4 | 4 | 4 | 0 | 3 BRCA1, 2 BRCA2 |
| 2 | 4 | 4 | 3 | 1 | 1 BRCA2, 1 CHEK2 |
| 1 | 4 | 4 | 2 | 2 | |
| 2 | 4 | 5 | 4 | 1 | |
| 2 | 4 | 5 | 5 | 0 | 1 BRCA1, 1 BRCA2 |
| 1 | 5 | 5 | 5 | 0 | |
| 1 | 6 | 7 | 7 | 0 | |
| 1 | 6 | 6 | 6 | 0 |
total number of family members affected with breast or ovarian cancer (including proband and their first- and second-degree relatives)
total number of cancers in proband and first- and second-degree relatives.
Methods
Promoter deletion screening
Genotyping for BRCA1 variants, C1802G (β-promoter, Genbank Accession number U37574) and C2731T (Pro871Leu, exon 11, U14680) was performed for 51/66 women who met our criteria (Table 1) using the method described by Catteau et al., (1999) (7).
Multiplex Ligation-dependent Probe Amplification (MLPA)
MLPA was carried out on 125ng of DNA using the SALSA P002-BRCA1 Exon Copy Number Test Kit (MRC-Holland) and analyzed on an ABI 3730 analyzer using Genotyper software. For data analysis, peak areas of each probe were normalized by dividing each peak area by the total peak area of all probes for a particular sample. This normalized value was divided by the average normalized value of that probe (obtained from all samples). MLPA was performed in duplicate.
Tumour morphology assessment
Histology slides were reviewed as described in Armes et al., (1998) (13).
Results
Genotyping for BRCA1 variants, C1802G and C2731T (Pro871Leu, exon 11) was performed for 51/66 women who met our criteria (Table 1) using the method described by Catteau et al., (1999) (7). Of these tested cases, 46 (90%) were found to be in accordance with the expected linkage disequilibrium, and 28 (55%) of genotypes were uninformative for assessment of BRCA1 promoter deletions as they were homozygous at both polymorphic sites (indicated by # in Table 2). Five cases (10%) were not in linkage disequilibrium, and of these three were apparent homozygous (CC) at C1802G and heterozygous (CT) at C2731T (Pro871Leu). This genotype was suggestive of a BRCA1 promoter deletion (indicated by* in Table 2). One individual was apparent homozygous (CC) at C1802G and apparent homozygous (TT) at C2731T (Pro871Leu), while another was heterozygous (CG) at C1802G and apparent homozygous (TT) at C2731T (Pro871Leu).
Table 2.
Genotyping and MLPA results
| C1802G (promoter) | C2731T Pro871Leu (exon 11) | Number of cases | MLPA for promoter deletions | Other known mutation | reference |
|---|---|---|---|---|---|
| CC | CC | 22# | negative | 6 BRCA1, 4 BRCA2 | (14,15) |
| C- | C- | 1# | deletion exons 1A-23 | none | |
| CG | CT | 18 | negative | 2 BRCA1, 1 BRCA2 | (14,15) |
| GG | TT | 5# | negative | none | |
| CC | CT | 2* | negative | none | |
| C- | CT | 1* | deletion exons 1A, 1B, 2 | none | |
| CC | TT | 1 | negative | ATM, c.4258C>T (p.Leu1420Phe), | (9) |
| CG | TT | 1 | negative | 1 BRCA2 | (14) |
| Total 51 | |||||
Key to table;, genotype supportive of promoter deletion; C-, hemizygocity;
genotypes uninformative for detection of BRCA1 promoter deletions.
Only one of the three cases that were apparent CC at C1802G and CT at C2731T (Pro871Leu) and therefore suspected of BRCA1 promoter deletions showed a reduction in signal strength in the MLPA analysis. This reduction was observed for exons 1A, 1B and 2 (del exon1A-2). A second individual (with an uninformative apparent CC C1802G and apparent CC C2731T (Pro871Leu) genotype) showed a loss of exons 1A-23 via MLPA analysis. Profiles of the cases containing deletions and a normal DNA sample are shown in Figure 2.
Figure 2.
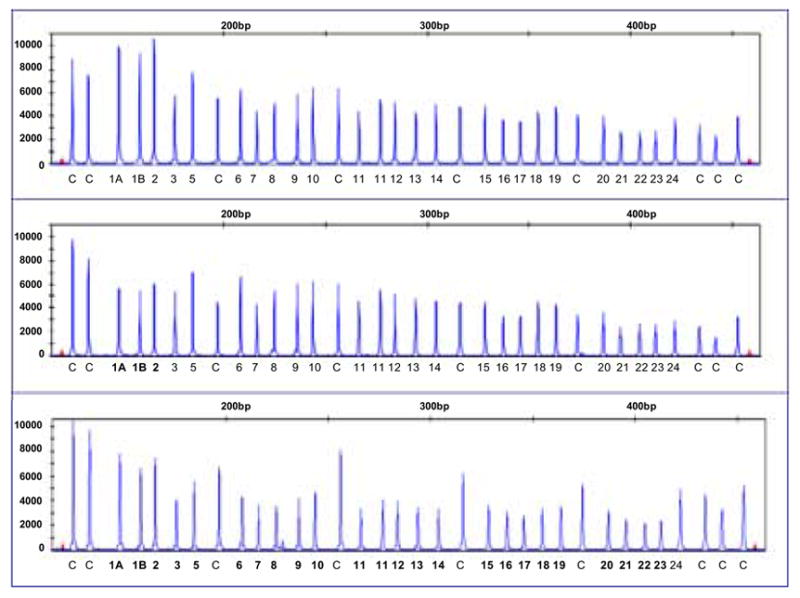
MLPA profiles showing no large genomic alterations in BRCA1 (top panel); loss within exons 1A, 1B and 2 (centre panel); and loss of exons 1A to 23 (bottom panel). Exons of loss are indicated in bold. Control fragments are distributed throughout the profile and indicated with C.
BRCA1, del exon 1A-2
The cancer family history of the case carrying the BRCA1, del exon 1A-2 mutation included personal diagnosis at age 29 years of a grade 3, infiltrating ductal carcinoma of the breast. Histopathology review of the breast cancer revealed several features that were consistent with the possibility of a BRCA1 mutation, including a high mitotic index (125 mitotic cells per 10 hpf), minimal tubule formation, a circumscribed growth pattern, an intense lymphocytic infiltrate, necrosis, trabecular growth pattern and lack of ER and PR protein expression (via immunohistochemistry). Two maternal and paternal aunts of the case had been diagnosed with breast cancer at the age of 30 and 51 years respectively, and her paternal uncle had been diagnosed with kidney cancer at the age of 55 years. DNA samples were available for both parents of the case and were genotyped at C1802G and C2731T and screened for large genomic alterations in BRCA1 via MLPA. The mother was found to be apparent homozygous for both variants (GG at C1802G and TT at C2731T) and the father was found to be apparent homozygous at both positions (CC at CG C1802G and CC at C2731T). MLPA results showed a loss of signal strength of BRCA1 exons 1A, 1B and 2 in the DNA sample from the mother and hence the apparent homozygous GG at C1802G was actually hemizygous G- at C1802G.
BRCA1, del exon 1A-23
The cancer family history of the case identified with the BRCA1, del exon 1A-23 mutation included personal diagnosis at age 34 years of a grade 3, infiltrating ductal carcinoma of the breast. Histopathology review of the breast cancer revealed several features that were consistent with the possibility of a BRCA1 mutation, including a high mitotic index (56 mitotic cells per 10 hpf), minimal tubule formation, a circumscribed growth pattern, a moderate lymphocytic infiltrate, necrosis, a trabecular growth pattern and lack of ER and PR protein expression (via immunohistochemistry). The mother and maternal grandmother of the case had been diagnosed with breast cancer at the age of 28 and 45 years respectively. DNA was not available for testing from any other family members.
Discussion
We have identified two women diagnosed with breast cancer before the age of 40 years, and a strong family history of breast cancer, who carry large genomic deletions in BRCA1 that involved the promoter region. MLPA detected both of these deletions. The BRCA1 1A-23 deletion could not be detected using the linkage disequilibrium genotypying method as both variants, in the promoter and exon 11, were hemizygous due to the deletion. The other mutation detected by MLPA was a BRCA1 1A-2 deletion. Deletions of this type have been described as an artifact of MLPA due to incomplete denaturation of DNA in CpG regions. We do not think that this is of concern to our data as a deletion in the promoter in this case is supported by the genotyping data, this mutations was present in the germline of the mother of this case and genotyping within the family was consistent with the presence of this mutation. In addition, there is a previously described case carrying a known mutation of this type detected by another method (6).
Two additional cases had genotypes suggestive of BRCA1 promoter deletions (CC at C1802G and CT at C2731T (Pro871Leu) that were not supported by MLPA analysis. One of the 146 normal (unaffected) controls in the report of Catteau et al. (1999) also had this combination of genotypes and two individuals had CG at C1802G and CC at C2731T (Pro871Leu) showing that although very high there is not complete linkage disequilibrium between these two variants (7). One of the cases we detected with 1802CC and 2731CT genotype was diagnosed with an atypical medullary type cancer and thus had many histological features associated with carrying a BRCA1 germline mutation. The second case had some features associated with BRCA1 mutations as it was a grade 3 infiltrating ductal carcinoma that had a high mitotic count (22/10hpf) and a circumscribed growth pattern but it lacked a pushing margin, syncytial growth pattern, a lymphocytic infiltrate and necrosis. The morphological features of the cancers arising in these cases suggests that these cases may carry germline BRCA1 mutations as yet unidentified by our current and extensive BRCA1 mutation screening (8).
We found two other cases with genotypes inconsistent with complete linkage disequilibrium between C1802G and C2731T (Pro871Leu). One case had genotype CC C1802G and TT at C2731T (Pro871Leu) and was known to carry the ATM missense mutation c.4258C>T (p.Leu1420Phe) (9). The second carried the genotype CG C1802G and TT at C2731T (Pro871Leu) and was known to carry the BRCA2 germline protein truncating mutation 6503delTT. It is intriguing that 3/5 cases whose genotypes were inconsistent with complete linkage disequilibrium carried germline mutations in key molecules in DNA repair pathways.
We identified BRCA1 promoter deletions in two (4%) of the young women with breast cancer with a strong family history of breast and ovarian cancer selected from our Australian population-based study. BRCA1 promoter region deletions, therefore, represent 2/10 (20%) of all known BRCA1 germline mutations identified to date in these young women. Our data suggests that it is beneficial to include a screen for deletions within the BRCA1 promoter region in routine clinical BRCA1 mutation detection.
Figure 1. The genotyped BRCA1 polymorphisms and outcomes.
Acknowledgments
The Australian Breast Cancer Family Study was supported by the National Health and Medical Research Council of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation (Australia), and the U.S. National Cancer Institute, National Institutes of Health, under Request for Application CA-95-003 as part of the Breast Cancer Family Registry, and through cooperative agreements with the Fox Chase Cancer Center; Huntsman Cancer Institute, Columbia University; Northern California Cancer Center; Cancer Care Ontario; and The University of Melbourne. Fragment analysis was done at the Australian Genome Research Facility (AGRF). The AGRF receives support from the Australian Commonwealth Government.
Footnotes
Conflict of interest statement
None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Petrij-Bosch A, Peelen T, van Vliet M, et al. BRCA1 genomic deletions are major founder mutations in Dutch breast cancer patients. Nat Genet. 1997;17:341–5. doi: 10.1038/ng1197-341. Erratum in: Nat Genet. 1997, 17:503. [DOI] [PubMed] [Google Scholar]
- 2.Hogervorst FB, Nederlof PM, Gille JJ, et al. Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res. 2003;63:1449–53. [PubMed] [Google Scholar]
- 3.Brown MA, Nicolai H, Xu C-F, et al. Regulation of BRCA1. Nature. 1994;372:733. doi: 10.1038/372733a0. 1994. [DOI] [PubMed] [Google Scholar]
- 4.Xu C-F, Chambers JA, Solomon E. Complex regulation of the BRCA1 gene. J Biol Chem. 1997;272:20994–20997. doi: 10.1074/jbc.272.34.20994. [DOI] [PubMed] [Google Scholar]
- 5.Swensen J, Hoffman M, Skolnick MH, Neuhausen SL. Identification of a 14kb deletion involving the promoter region of BRCA1 in a breast cancer family. Hum Mol Genet. 1997;6:1513–1517. doi: 10.1093/hmg/6.9.1513. [DOI] [PubMed] [Google Scholar]
- 6.Brown MA, Lo L-J, Catteau A, et al. Germline BRCA1 promoter deletions in UK and Australian familial breast cancer patients: identification of a novel deletion consistent with BRCA1:ΨBRCA1 recombination. Hum Mutat. 2002;19:435–442. doi: 10.1002/humu.10055. [DOI] [PubMed] [Google Scholar]
- 7.Catteau A, Xu C-F, Brown MA, et al. Identification of a C/G polymorphism in the promoter region of the BRCA1 gene and its use as a marker for rapid detection of promoter deletions . Br J Cancer. 1999;79:759–763. doi: 10.1038/sj.bjc.6690122. 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dite GS, Jenkins MA, Southey MC, et al. Familial risks, early-onset breast cancer, and BRCA1 and BRCA2 germline mutations. J Natl Cancer Inst. 2003;95:448–57. doi: 10.1093/jnci/95.6.448. [DOI] [PubMed] [Google Scholar]
- 9.Chenevix-Trench G, Spurdle AB, Gatei M, et al. Dominant negative ATM mutations in breast cancer families. J Natl Cancer Inst. 2002;94:205–15. doi: 10.1093/jnci/94.3.205. Erratum in: J Natl Cancer Inst 2002, 94, 952. [DOI] [PubMed] [Google Scholar]
- 10.Hopper JL, Giles GG, McCredie MR, Boyle P. Background rational and protocol for a case-control-family study of breast cancer. Breast. 1994;3:79–86. [Google Scholar]
- 11.McCredie MR, Diet GS, Giles GG, Hopper JL. Breast cancer in Australian women under the age of 40. Cancer Causes and Control. 1998;9:189–98. doi: 10.1023/a:1008886328352. [DOI] [PubMed] [Google Scholar]
- 12.Hopper JL, Chenevix-Trench G, Jolley D, et al. Design and analysis issues in a population-based, case-control-family study of the genetic epidemiology of breast cancer and the Cooperative Family Registry fro Breast Cancer Studies (CFRBCS) Monogr Natl Cancer Inst. 1999;26:95–100. doi: 10.1093/oxfordjournals.jncimonographs.a024232. [DOI] [PubMed] [Google Scholar]
- 13.Armes JE, Egan AJM, Southey MC, et al. The histologic phenotypes of breast carcinoma occurring before age 40 years in women with and without BRCA1 or BRCA2 germline mutations: a population-based study. Cancer. 1998;83:2335–2345. [PubMed] [Google Scholar]
- 14.Hopper JL, Southey MC, Dite GS, et al. Population-based estimate of the average age-specific cumulative risk of breast cancer for a defined set of protein-truncating mutations in BRCA1 and BRCA2. Australian Breast Cancer Family Study. Cancer Epidemiol Biomarkers Prev. 1999;8:741–7. [PubMed] [Google Scholar]
- 15.Southey MC, Tesoriero AA, Andersen CR, et al. BRCA1 mutations and other sequence variants in a population-based sample of Australian women with breast cancer. Br J Cancer. 1999;79:34–9. doi: 10.1038/sj.bjc.6690008. [DOI] [PMC free article] [PubMed] [Google Scholar]


