Abstract
PDZ-domain–containing proteins such as PSD-95 have been implicated in the targeting and clustering of membrane proteins. Biochemical and immunohistochemical studies indicate that PSD-95 recognizes COOH-terminal S/TXV sequences present in Kv1 K+ channels. However, the effect of binding a PDZ domain on a target protein has not been studied in live cells. In the present study, a green fluorescent protein–Kv1.4 fusion protein is used to study the effect of PSD-95 on channel movement. Fluorescence recovery after photobleaching showed that PSD-95 can immobilize K+ channels in the plasma membrane in an all-or-none manner. Furthermore, time lapse imaging showed that channel clusters formed in the presence of PSD-95 are stable in size, shape, and position. As expected from previous reports, two green fluorescent protein–tagged COOH-terminal variants of Kv1.4, Δ15 and V655A, are not clustered by PSD-95. However, coexpression of PSD-95 with V655A, but not Δ15, leads to the appearance of PSD-95 immunoreactivity in the plasma membrane. Furthermore, fluorescence recovery after photobleaching studies show that V655A channels are immobilized by PSD-95. Thus, V655A channels can interact with PSD-95 in a manner that leads to channel immobilization, but not clustering. These experiments document for the first time that PSD-95 immobilizes target proteins. Additionally, the data presented here demonstrate that the structural requirements for protein clustering and immobilization by PSD-95 are distinct.
Keywords: green fluorescent protein, Kv1.4, fluorescence recovery after photobleaching, channel immobilization
introduction
The targeting of membrane proteins, such as ion channels and receptors, to discrete plasma membrane domains is critical to cellular function. This is particularly true in neurons due to their dependence on electrochemical signaling for cell to cell communication. Examples of protein targeting in neurons include the aggregation of sodium channels at the node of Ranvier, post-synaptic accumulation of N-methyl-d-arginine receptors, and localization of voltage-gated K+ channels to axons and terminals (Salzer, 1997; Sheng et al., 1992, 1993; Rhodes et al., 1995; Maletic-Savatic et al., 1995; Muller et al., 1996). It is also frequently important for membrane proteins to be complexed with down-stream effector molecules or anchored to structural elements. Recently, molecules containing PDZ domains have been suggested to play a role in the targeting of proteins to discrete membrane regions and the formation of membrane protein complexes (Brenman and Bredt, 1997; Tsunoda et al., 1997; Irie et al., 1997; Tejedor et al., 1997; Thomas et al., 1997; for reviews, see Gomperts, 1996; Sheng 1996; Kornau et al., 1997).
PDZ domains, which act as protein–protein interaction modules similar to Src homology 2 (SH2) and SH3 domains, bind consensus sequences typically in the COOH terminus of their target proteins (Songyang et al., 1997). One group of PDZ-containing proteins found in neuronal tissue is the PSD-95/SAP90 family that includes SAP97/hdlg, chapsyn-110/PSD-93, SAP102, and PSD-95/SAP90. These proteins localize to sites of cell–cell contact. For example, PSD-95 is associated with synapses in both the pre- and postsynaptic regions. PDZ domains of some members of the PSD-95 family recognize the consensus sequence tS/TxV (Kim et al., 1995; Kornau et al., 1995; Kim and Sheng, 1996; Muller et al., 1996). Both the NR2 N-methyl-d-arginine receptor subunit and the Kv1.4 K+ channel subunit end in the consensus sequence ES/TDV. Biochemical and immunohistochemical studies have shown that PSD-95 and SAP97 bind to and cluster these proteins (Kim et al., 1995; Kornau et al., 1995; Kim and Sheng, 1996). Furthermore, these studies have indicated that the terminal valine of the Kv1.4 is essential for interaction with PSD-95 (Kim et al., 1995). Due to technical limitations, the interactions of PDZ-containing-proteins with their target proteins have not been studied in live cells. Therefore, the dynamic nature of this interaction and its effect on target mobility has not been examined.
Fluorescence recovery after photobleaching (FRAP)1 can be used in live cells to quantitate lateral diffusion of a labeled protein in a cellular membrane. FRAP can also be used to determine the fraction of labeled protein that exists in an immobile, or nearly immobile, state within the bilayer (Edidin, 1994). For example, previous studies using a technique similar to FRAP have found that 75% of plasma membrane voltage-gated K+ channels in skeletal muscle are immobile. Furthermore, channels that were mobile diffused at a very slow rate (D = 5 × 10−11 cm2/s) (Weis et al., 1986), where D is the diffusion coefficient. These findings indicate that voltage-gated K+ channels are among the least mobile class of proteins examined (Edidin, 1994). However, the molecular interactions limiting K+ channel movement were unknown.
In the present study, an enhanced green fluorescent protein (EGFP)-Kv1.4 fusion protein was used to investigate this subunit's interaction with PSD-95 for the first time in live cells. FRAP and time lapse imaging were used to determine if interaction with PSD-95 can contribute to the high level of K+ channel immobilization. The interactions of PSD-95 with EGFP-Kv1.4 COOH-terminal variants were also examined. The data from these studies indicate that there are different structural requirements for the clustering and immobilization of Kv1.4 channels by PSD-95. Additionally, these data suggest a potential for a greater multiplicity of functions and binding partners for PDZ-containing proteins than previously believed.
materials and methods
Construction of EGFP-Kv1.4, EGFP-Kv1.4Δ15, and EGFP-Kv1.4V655A
EGFP-Kv1.4 was constructed by subcloning of SacI-Bg1II fragment of rat Kv1.4 cDNA (RK3 [Roberds et al., 1991], nucleotides 555–2693 [Stühmer et al., 1989]) into SacI-BamHI site of EGFP-C1 (Clontech), followed by shifting the frame at BglII site of the vector. The resulting fusion construct lacks the first nine amino acids of Kv1.4. Instead, the seven amino acids SGLRISIS were introduced between EGFP and Kv1.4.
EGFP-Kv1.4V655A and EGFP-Kv1.5 Δ15 were generated with EGFP-Kv1.4 by a PCR-based method. The obtained fusion protein constructs were sequenced to verify introduced changes.
Cell Culture and Transfections
HEK 293 cells were obtained from American Type Culture Collection and cultured in DMEM supplemented with 10% fetal bovine serum. Cells were passed with trypsin-EDTA solution at 1:10 dilution on a polylysine-coated glass coverslip or plastic dishes. 1 d after passage, cells were transfected with plasmid DNA (1 μg/ 35-mm dish or 5 μg/100-mm dish) by the calcium phosphate precipitation method (Transfinity; Gibco-BRL). Cotransfection of PSD-95 with channel DNA was typically carried out at a 1:1 ratio. However, in the case of observing clusters in HEK293 cells, transfection was carried out at a 1:5 ratio (channel:PSD-95).
Electrophysiological Recordings
Standard whole-cell voltage clamp recording was performed with an EPC-9 patch clamp amplifier (HEKA Electronik) using the PULSE program (HEKA Electronik) on a power Macintosh computer. Patch pipettes were filled with a solution containing (mM) 140 KCl, 1 MgCl2, 10 Na-HEPES, 10 EGTA, 3 MgATP, pH 7.3. Bath solution contained (mM) 140 NaCl, 5.4 KCl, 0.8 MgCl2, 10 Na-HEPES, 10 glucose, pH 7.3.
Western Blot Analysis
Post-nuclear membrane fraction was prepared from HEK 293 cells mock-transfected or transfected with one of the EGFP-Kv1.4 plasmids by differential centrifugation. In brief, cells grown on plastic dishes were collected with phosphate-buffered saline supplemented with 1 mM EDTA and homogenized in a 10-ml solution consisting of 10 mM Tris-HCl, pH 7.5, 0.25 M sucrose, 1 mM EDTA, and 0.2 mM phenylmethyl sulfonate using a dounce pestle. The homogenate was centrifuged at 1,000 g for 10 min to remove nuclei and undisrupted cells. Post-nuclei membrane fraction was then obtained by centrifugation of the supernatant at 100,000 g for 60 min. EGFP-channel fusion proteins were extracted in 1% Triton X-100 solution containing 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 0.2 mM phenylmethyl sulfonate by sonication, followed by removing unsolubilized materials by centrifugation.
Protein concentration was determined with a protein assay solution (Bio-Rad Laboratories) using human IgG as a standard. Triton extracts (25 μg protein) were separated on a 7.5% SDS gel and transferred to nitrocellulose membrane. The membrane was coated with 5% nonfat dry milk, probed with polyclonal anti– GFP antibody (Clontech) at 1/2,000 dilution, and then with anti–rabbit IgG conjugated with peroxidase. Immunoreactive proteins were detected with enhanced chemiluminescence solutions (NEN Research Products).
Fluorescence Recovery After Photobleaching
HEK 293 cells transfected with either EGFP-Kv1.4, Δ15, or V655A were imaged with a 2001CSLM (Molecular Dynamics Inc.) using an excitation wavelength of 488 and standard FITC optics. Bleaching was achieved by rapid scan of a defined line with the laser at 100% intensity. A 512 × 512 pixel area (0.17 μm/pixel) was then imaged at 3% power every 20–30 s for a total of 10 min. To plot recovery, the integral of the fluorescence intensity within the bleach spot was determined for each time point. Nonlinear regression analysis was used to derive a t 1/2. A diffusion coefficient was then calculated as D = (3r 2/4t 1/2)γ (r = 1/2 width of bleach area, γ is a correction factor that takes into account the effect of percent bleach). Three measurements were used to calculate the immobile fraction, fluorescence in the bleach area before bleach (F pre), fluorescence immediately after bleach (F post), and fluorescence in the bleached area after steady state had been reached (F recov). Immobile fraction was then calculated as F im = (F pre) − (F recov)/(F pre) − (Fpost). Error bars represent SEM.
A potential concern with the FRAP technique is the potential for erroneous results caused by photodamage to cells. To examine the extent to which photodamage during bleaching might have contributed to our results, a double bleach paradigm was used. Diffusion coefficients and immobile fractions calculated for first and second bleaches were not significantly different (data not shown). Furthermore, a comparison of diffusion coefficients for GFP-tagged secretory granules derived by FRAP and independently by particle tracking yielded similar results (Burke et al., 1997). Finally, as all experiments were carried out using the same bleaching protocol, differences in mobile fraction observed for the wild-type channel and its variants could not be due to photodamage. Thus, photodamage is not a problem with our GFP-based FRAP experiments
Time Lapse Imaging
COS1 cells cotransfected with EGFP-Kv1.4 and PSD-95 were imaged on a conventional epifluorescence microscope every 30 s over a 10-min period with a cooled CCD camera (Photometrics) driven by Ratio Tool software from Inovision. Time-lapse movies were then created and examined for cluster movement.
PSD-95 Immunofluorescence
HEK 293 cells were either mock transfected, transfected with PSD-95 alone, or transfected with PSD-95 at a 1:5 ratio (PSD-95: channel) with either wild type (wt), Δ15, or V655A DNA. 2 d after transfection, cells were fixed with 4% paraformaldehyde for 30 min at room temperature. Fixed cells were then rinsed in PBS and incubated for 2 min in PBS containing 0.2% triton. Cells were again rinsed, and then incubated for 30 min in PBS containing 1.0% BSA. An anti–PSD-95 monoclonal antibody was applied at a 1:5 dilution in PBS-BSA and incubated for 1 h at 37°C. A rhodamine conjugated anti–mouse secondary antibody (Boehringer Mannheim) was then used to image PSD-95 localization via fluorescent confocal microscopy as described above.
results
EGFP-Kv1.4 Localizes to the Plasma Membrane to Form Functional Channels
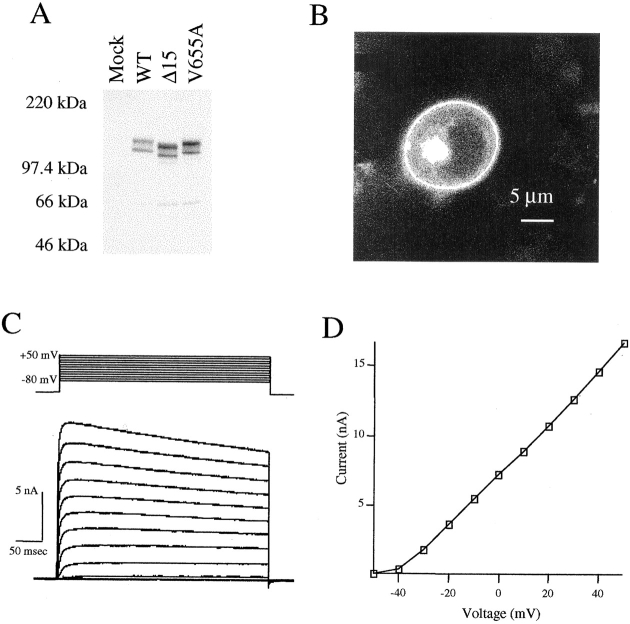
Our initial studies were directed at demonstrating that EGFP-tagged Kv1.4 expresses efficiently, targets to the plasma membrane, and forms functional K+ channels when expressed in HEK 293 cells. To verify protein expression, qualitative Western blot analysis with an anti– EGFP antibody was performed on postnuclear membranes isolated from transfected cells (Fig. 1 A, WT). Western blot analysis of Kv1.4 will typically yield two immunoreactive bands, one at 97 and another at 87 kD (e.g., Takimoto et al., 1995). The higher molecular weight band represents a heavily glycosylated form of the protein (our unpublished data). Analysis of the EGFP-tagged channel subunit also yielded two immunoreactive bands, with molecular weights appropriate for the fusion protein. Western blot analysis was also carried out on cells expressing two EGFP-tagged COOH-terminal variants of Kv1.4. In the first variant, EGFP-Kv1.4CΔ15 (Δ15), the final 15 amino acids of the fusion construct were deleted. The slight downward shift of the immunoreactive bands reflects this deletion (Fig. 1 A, Δ15). The other variant examined was EGFP-Kv1.4V655A (V655A) (Fig. 1 A). In this construct, the final valine (V655 in wild-type Kv1.4) was mutated to an alanine. Again, two immunoreactive bands were seen with molecular weights of 120 and 110 kD. Therefore, all three constructs were expressed efficiently.
Figure 1.
Cell surface expression of EGFP-Kv1.4. (A) A polyclonal antibody against EGFP was used for Western blot analysis of a P2 membrane fraction isolated from 293 cells transfected with either EGFP-Kv1.4, Δ15, or V655A. EGFP-Kv1.4 immunoreactive bands were seen at 120 and 110 kD, the expected molecular weights for the two glycosylation states of the fusion protein. (B) Confocal microscopy showing the cell surface localization of EGFP-Kv1.4 in HEK 293 cells. (C) Whole cell patch clamp recording from HEK 293 cells transfected with EGFP-Kv1.4. Membrane potential was stepped from −50 to +50 mV in 10-mV, 500-ms steps. (D) Peak current vs. voltage curve.
We next used confocal microscopy to examine the localization of the EGFP-Kv1.4 fusion protein in transfected HEK 293 cells. In Fig. 1 B, the single plane imaging capability of the confocal microscope shows a clear “ring” of fluorescence. This is indicative of a plasma membrane localization for the EGFP-tagged protein. Similar localization and fluorescence levels were also observed for Δ15 and V655A (data not shown).
Finally, whole cell patch clamp recordings were carried out to demonstrate the presence of functional channels in HEK 293 cells transfected with the EGFP-Kv1.4 construct (Fig. 1, C and D). Large voltage-gated currents were seen and activation was consistent with the known properties of Kv1.4 channels. However, no fast inactivation was evident. This reflects loss of the “ball and chain” inactivation caused by the substitution of the first nine amino acids of Kv1.4 with the EGFP moiety. Taken together, these data indicate that the EGFP-Kv1.4 fusion protein was efficiently produced and targeted to the cell surface when expressed in HEK 293 cells. Additionally, tagged subunits were capable of forming functional voltage-gated K+ channels.
EGFP-tagged Kv1.4 Forms Clusters in the Presence of PSD-95
Coexpression of PSD-95 with wt Kv1.4 has been shown previously to lead to the formation of channel clusters at the cell surface in COS7 cells (Kim et al., 1995; Kim and Sheng, 1996). Therefore, channel clustering in the presence of PSD-95 was used as a determinant of the EGFP-tagged subunit's ability to undergo normal molecular interactions. Fig. 2 A (wt) shows that PSD-95 induces clustering of EGFP-Kv1.4 fluorescence in COS1 cells. Clusters were also observed in HEK 293 cells cotransfected with EGFP-Kv1.4 and PSD-95 (Fig. 2 B).
Figure 2.
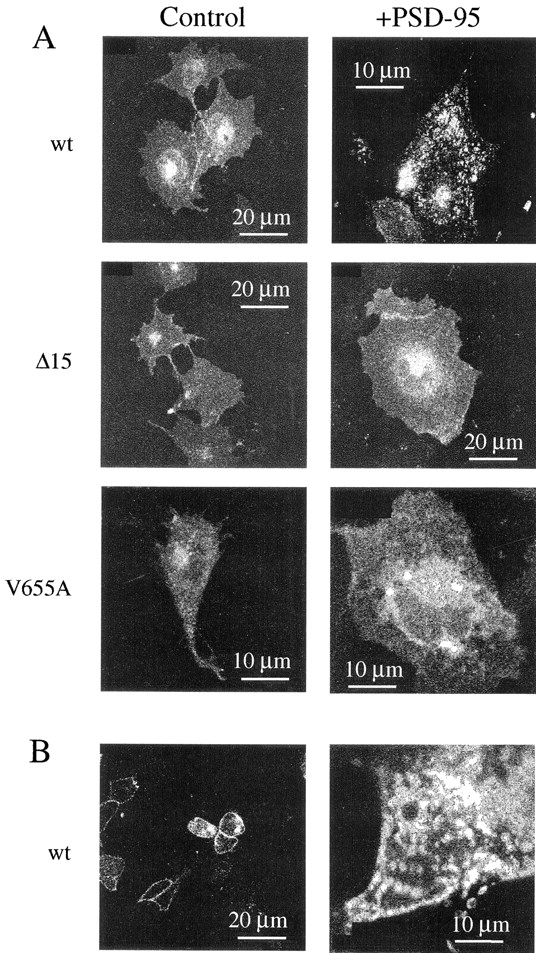
Clustering of EGFP-Kv1.4 in the presence of PSD-95. (A) Confocal images of COS1 cells transfected with EGFP-Kv1.4 (wt), Δ15, or V655A in the absence (Control) or presence (+PSD-95) of PSD-95. (B) Confocal image of HEK 293 cells transfected with EGFP-Kv1.4 in the absence or presence of PSD-95.
The interaction of Kv1.4 with PSD-95 is believed to occur through a COOH-terminal domain that includes the subunit's final four amino acids, ETDV. It has been reported that disruption of this region in wt Kv1.4 can inhibit the interaction of the channel with PSD-95 (Kim et al., 1995; Kim and Sheng, 1996). Therefore, to further examine the appropriateness of the molecular interactions of the EGFP-tagged subunit, two COOH-terminal variants, Δ15 and V655A, were tested for their ability to form clusters in the presence of PSD-95. As expected from previous reports (Kim et al., 1995; Kim and Sheng, 1996), the EGFP-tagged COOH-terminal variants did not form clusters when coexpressed with PSD-95 in COS1 cells (Fig. 2 A) or in HEK 293 cells (data not shown). These data indicate that the EGFP-tagged Kv1.4 subunit is capable of normal molecular interaction with PSD-95.
PSD-95-induced Channel Clusters Are Stable Structures
Previously, the clustering of K+ channels by PSD-95 has been studied by immunohistochemical approaches in fixed cells (Kim and Sheng, 1996; Horio et al., 1997). Thus, the dynamic properties of clusters have not been studied. Hence, it is unclear whether clusters are stationary structures or whether they move within the plasma membrane. Similarly, the reversibility of the interaction of PSD-95 with channels has not been measured in live cells. Use of the EGFP-tagged Kv1.4 construct allowed us to address these issues with time lapse imaging and FRAP. In COS1 cells, time lapse imaging of PSD-95-induced Kv1.4 channel clusters showed that these structures are immobile over a 10-min duration (Fig. 3, A and B). Additionally, analysis of these images revealed almost no change in the shape of individual clusters during the observation period. Therefore, PSD-95-induced channel clusters are stable structures. Furthermore, the immobility of the clusters supports a role for PSD-95 in anchoring protein clusters to plasma membrane microdomains such as the synapse.
Figure 3.
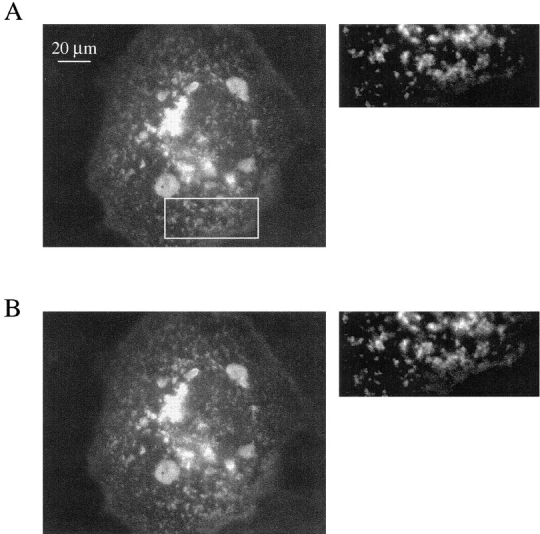
PSD-95 induced EGFP-Kv1.4 channel clusters are immobile, stable structures. (A) Epifluorescence image of a COS1 cell transfected with EGFP-Kv1.4 and PSD-95. This is the first image in a series of 20 images taken every 30 s. The box indicates the area shown to the right that has been contrast enhanced and expanded 180% from the original image. (B) The final image in the series taken 10 min after the image in A. The same region was expanded from this image to demonstrate the lack of change in cluster location, size, and shape over the observation period.
COOH-Terminal Alterations Can Decrease the Fraction of Immobile Kv1.4 Channels as Determined by FRAP
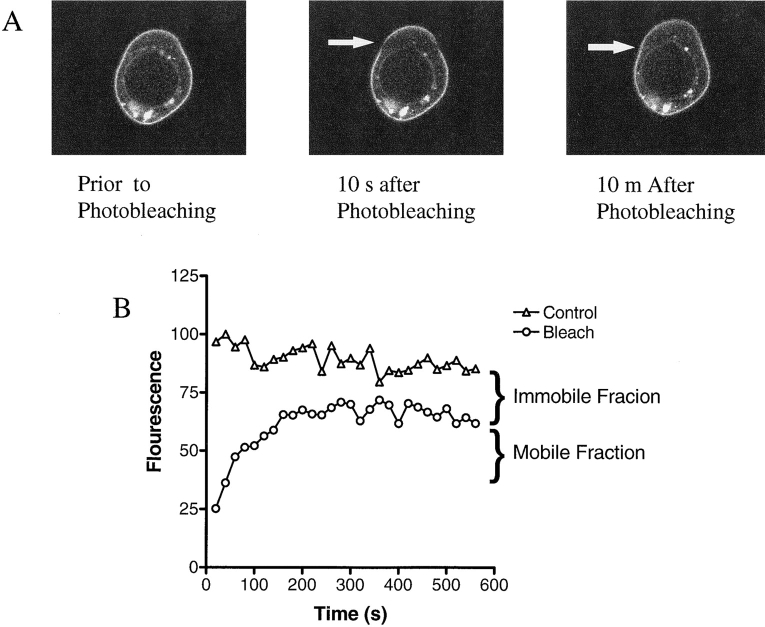
In initial FRAP experiments, we determined the mobility of the EGFP-Kv1.4 molecule expressed alone in HEK 293 cells. These cells were used because their round shape makes them advantageous for confocal FRAP experiments (Levitan, 1998). A brief, high intensity line-scan resulted in the photobleaching of EGFP-Kv1.4 molecules within a defined region (Fig. 4 A). The rate of recovery of fluorescence within this region was then monitored until steady state had been reached. Recovery data was plotted as shown in Fig. 4 B to derive a t 1/2. From the derived t 1/2, D was calculated for each individual experiment using the equation D = (3r 2/ 4t 1/2)γ (Feldman et al., 1981). The average diffusion coefficient for EGFP-Kv1.4 expressed alone was 5.6 × 10−11 cm2/s (n = 16) (Fig. 5 A). This agrees well with the previously published value of 5 × 10−11 cm2/s for voltage-gated K+ channel in frog muscle (Weis et al., 1986). Thus, mechanisms for limiting diffusion of channels appear to be conserved in a variety of cell types. Diffusion coefficients for Δ15 and V655A were also determined and were not found to be statistically different from that derived for EGFP-Kv1.4 (Fig. 5 A).
Figure 4.
Fluorescence recovery after photobleaching. (A) A typical FRAP experiment for cells transfected with EGFP-Kv1.4 is shown. The arrow in the center and final images points to the bleach region. Note the fluorescence recovery in the final image (10 min after photobleaching). Recovery was monitored every 30 s for a total of 10 min. (B) Graph showing a typical recovery curve for a bleached region and data from a control region. Note that steady state is achieved during the recovery period. A diffusion coefficient and immobile fraction for EGFP-Kv1.4 was obtained from this data as described in materials and methods.
Figure 5.
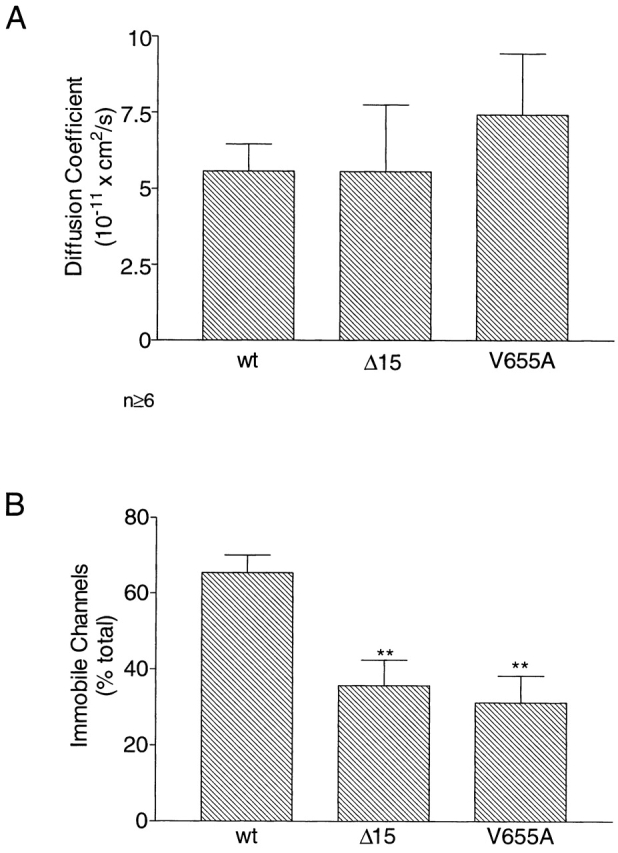
The COOH-terminal region of EGFP-Kv1.4 is important to channel mobility in the absence of PSD-95. (A) Comparison of diffusion coefficient for EGFP-Kv1.4 (wt), Δ15, and V655A when transfected alone in HEK 293 cells. (B) Comparison of immobile fractions for EGFP-Kv1.4 (wt), Δ15, and V655A when transfected alone in HEK 293 cells. P values were determined by Bonferroni test between data for wt and data for each of the COOH-terminally altered fusion proteins. n ≥ 7; **P < 0.005.
The fraction of immobile channels was also calculated from FRAP data. Surprisingly, when EGFP-Kv1.4 was expressed in HEK 293 cells an average of 65.4 ± 4.6% (n = 12) (Fig. 5 B, wt) of the channels were found to be immobile. This value is comparable with results obtained in skeletal muscle (Weis et al., 1986). We suggest that this high percentage is due to the existence of an endogenous HEK 293 cell protein that is capable of interacting with and immobilizing the channel. Immunocytochemistry carried out with an anti– PSD-95 antibody gave no indication of endogenous PSD-95. Therefore, this protein is likely to be distinct from PSD-95. Interestingly, the fraction of immobile channels for both the deleted and mutated forms of the protein were decreased by an average of 48% compared with EGFP-Kv1.4 (Fig. 5 B). These data indicate that an endogenous HEK 293 protein, distinct from PSD-95, requires the COOH-terminal valine of the Kv1.4 subunit to immobilize the channel for prolonged periods.
Translocation of PSD-95 to the Plasma Membrane in the Presence of V655A
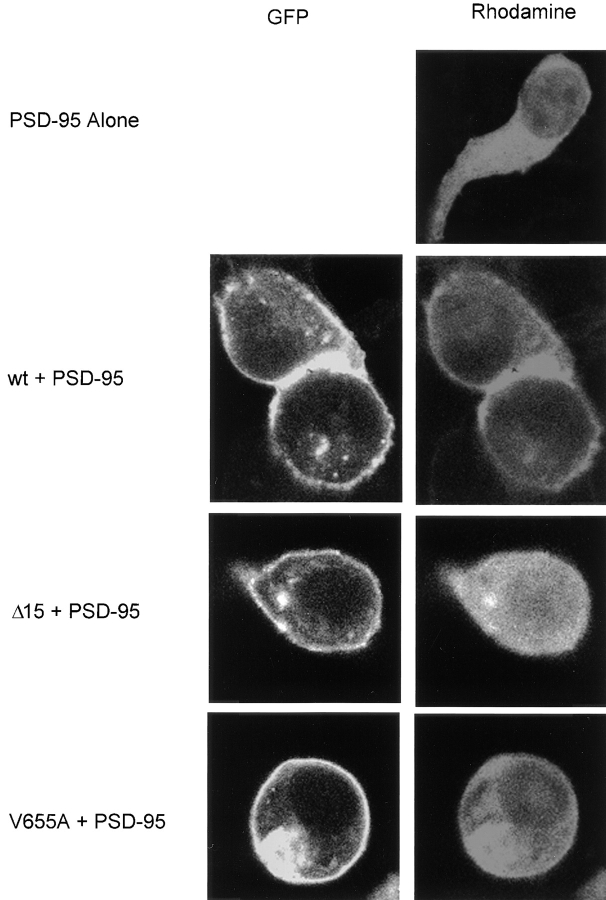
To further study the interactions of GFP-tagged Kv1.4 with PSD-95, immunohistochemical localization of PSD- 95 was carried out in the presence and absence of channel. These studies were carried out in HEK 293 cells transfected with a fivefold excess of channel DNA to enhance our ability to image changes in PSD-95 localization. These conditions did not lead to the formation of channel clusters with the wild-type channel. Expressed alone, PSD-95 displayed a diffuse cytoplasmic pattern of localization (Fig. 6, PSD-95 Alone). However, when coexpressed with the wild-type channel, a ring of cell-surface PSD-95 immunofluorescence can be observed. This localization corresponds to that observed for EGFP-Kv1.4 (Fig. 6, wt + PSD-95). Localization of PSD-95 was also examined in the presence of channel variants. In the presence of Δ15, PSD-95 maintains a diffuse cytoplasmic distribution (Fig 6, Δ15 + PSD-95). However, coexpression of PSD-95 with V655A led to colocalization of the two proteins at both the cell membrane and intracellular sites (Fig. 6, V655A + PSD-95). This finding suggests that while PSD-95 cannot cluster the V665A mutant, it may still maintain the ability to interact with this protein.
Figure 6.
Translocation of PSD-95 to the plasma membrane in the presence of wt Kv1.4 and V655A. Confocal images of HEK 293 cells transfected with PSD-95 alone or PSD-95 and EGFP-tagged Kv1.4 channels (wt) or a tagged variant (Δ15, V655A). Images showing the localization of the fluorescently tagged channels are shown in the first column (GFP). Immunocytochemical localization of PSD-95 with a rhodamine-cojugated secondary antibody is shown in the second column (Rhodamine). Images for each cell were collected at a single focal plane using standard optics for FITC and rhodamine.
Both EGFP-Kv1.4 and V655A, but Not Δ15, Are Immobilized by PSD-95
To investigate whether a nonclustering interaction with PSD-95 can alter channel mobility, FRAP experiments were carried out in HEK 293 cells. The diffusion coefficient for the wild-type channel was unaltered in the presence of PSD-95 (compare Figs. 5 A and 7 A). However, an ∼20% increase over control conditions in the number of immobile channels was detected when EGFP-Kv1.4 was coexpressed with PSD-95 (Fig. 7 B). Because an endogenous mechanism limits the availability of mobile channels to ∼1/3 of the total, this result is significant (i.e., most available channels are immobilized). Furthermore, this result implies that binding of the wild-type channel by PSD-95 must be stable within the 10-min duration of the FRAP experiment. Additionally, binding and immobilization appears to be an all-or-none phenomenon so that only those channels that had not interacted with PSD-95 participate in fluorescence recovery and the calculation of the diffusion coefficient.
Figure 7.
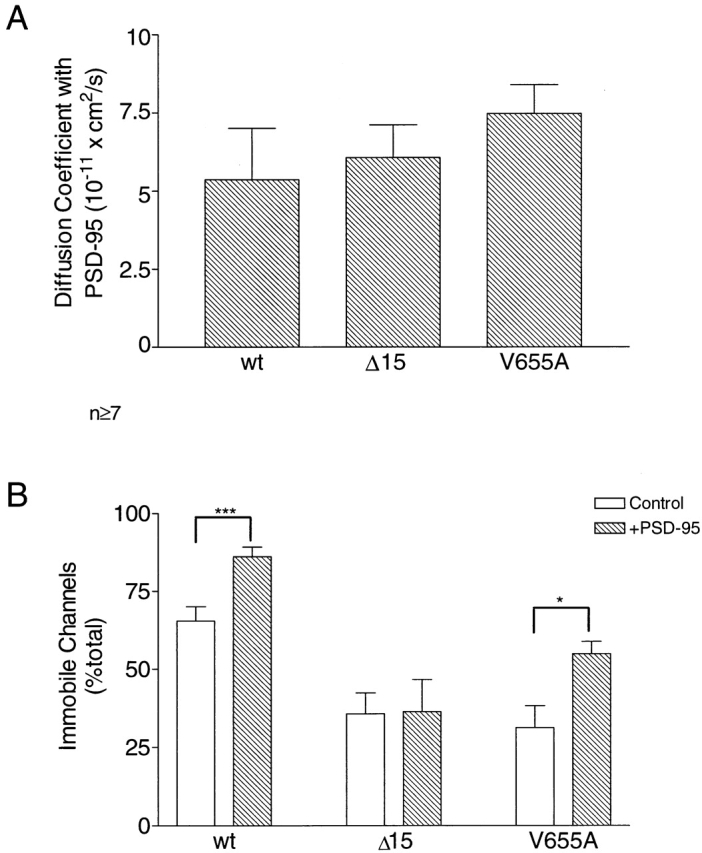
V655A is immobilized when coexpressed with PSD-95. (A) Comparison of diffusion coefficients for EGFP-Kv1.4 (wt), Δ15, and V655A when coexpressed with PSD-95 in HEK 293 cells. (B) Comparison of control data from Fig. 5 B with immobile fractions observed for EGFP-Kv1.4 (wt), Δ15, and V655A in the presence of PSD-95. Note that wt and V655A are immobilized by PSD-95. In contrast, Δ15 is not affected. Data based on the means from 7–15 experiments are shown. P values were derived by Student's t test comparing immobile fractions for each channel type in the presence and absence of PSD-95. Error bars represent the SEM. n ≥ 7; *P < 0.05; ***P < 0.001.
The mobilities of Δ15 and V655A in the presence of PSD-95 were also measured. Cotransfection of PSD-95 had no effect on the diffusion coefficients for these variants. Furthermore, no statistically significant effect was evident on the percent of immobile Δ15 channels (Fig. 7 B), as expected from previous biochemical and clustering studies (Kim et al., 1995; Kim and Sheng, 1996). This indicates that the COOH-terminal 15 amino acid deletion did indeed inhibit the interaction of PSD-95 and the channel.
Biochemical and clustering assays would predict that coexpression of PSD-95 with V655A should have no effect on the mobility of the mutated channel. In contrast, the translocation studies discussed above (Fig. 6) suggest an interaction between the two proteins. FRAP experiments provide further support for this interaction: coexpression of the two proteins resulted in a 22% increase in V655A channel immobilization, as compared with control conditions (Fig. 7 B). This is similar to the increase in immobilization observed for the wild-type channel in the presence of PSD-95. Therefore, despite mutation of the terminal valine to an alanine, which was thought to prevent PSD-95 binding to the Kv1.4 subunit, PSD-95 is still capable of immobilizing V655A channels. Because this channel is not clustered, these data indicate that there are different structural requirements for the immobilization and clustering of K+ channels by PSD-95.
discussion
Immobilization and Clustering of Channels by PSD-95
In this report, we provide the first live cell evidence that the interaction of PSD-95 with the Kv1.4 subunit leads to channel immobilization. The incomplete recovery of fluorescence observed in FRAP experiments indicates that few bound channels were freed from PSD-95 and became diffusible during the 10-min recovery period. Time lapse imaging of PSD-95 induced channel clusters also supports this interpretation: channel clusters were found to be immobile structures that are stable in shape and size over a period of minutes (Fig. 4). These findings argue for a long residence time, or slow dissociation rate, for the PSD-95/Kv1.4 complex. Furthermore, these data support the hypothesis that PSD-95 immobilizes target proteins at specific plasma membrane sites in vivo.
Experiments with truncated (Δ15) and mutated (V655A) forms of the EGFP-tagged Kv1.4 protein showed that PSD-95 did not form cell surface clusters with these COOH-terminal variants (Fig. 2 A). However, in contrast to previous reports concluding that the V655A mutant is incapable of interacting with PSD-95 (Kim et al., 1995; Kim and Sheng, 1996), we found that these channels were immobilized in the presence of PSD-95. In fact, FRAP experiments indicated that the interaction of PSD-95 with V655A is sustained for many minutes. Most surprisingly, immobilization of V655A, a variant that is not clustered, occurred at a level comparable with that observed for wild-type channels.
This finding has two important implications for our understanding of the function of PDZ-containing proteins. First, the ability of PSD-95 to stably interact with the V655A mutant in a cellular environment suggests a need to consider a broader range of potential binding partners for PDZ domains. By definition, the PSD-95/ V655A interaction occurs via a mechanism independent of the protein's mutated terminal valine. The crystal structure for the third PDZ domain in PSD-95 has shown that there are a number of other sites of interaction with target proteins (Doyle et al., 1996). The importance of these sites may have been underestimated in studies using biochemical techniques (such as yeast-two-hybrid, immunoprecipitation, and filter overlay) due to the loss of the native conformation of both the PDZ domain and the target proteins. Therefore, a population of proteins that has not yet been identified may exist that binds to PSD-95 PDZ domains. Additionally, it is possible that other PDZ-containing proteins will also have multiple populations of binding partners.
Second, the ability of PSD-95 to immobilize, but not to cluster, the V655A mutant suggests that the structural characteristics of PDZ-binding sequences determine the consequences of binding. This may indicate versatility in PDZ domain function that has not been previously suggested. For example, one set of target proteins with a binding sequence containing COOH-terminal valines would be clustered by a PDZ domain interaction. In contrast, a second group of proteins with a structurally distinct binding site (e.g., with COOH-terminal alanines) would only be immobilized upon interaction with the same PDZ domain. In principle, the latter type of substrate would compete for PDZ domain binding and could inhibit clustering of bona fide tS/TXV substrates. Thus, the ability to either cluster membrane proteins or to immobilize them at a low density suggests a broader and more complex role of PDZ domain proteins on control of cellular functions such as the ligand sensitivity or localized ion fluxes.
Endogenous Mechanisms for Channel Immobilization Exist in Many Cell Types
When expressed alone in HEK 293 cells, 65% of EGFP-Kv1.4 channels were immobile. Channels that were mobile diffused slowly, D = 5.6 × 10−11 cm2/s. This is nearly 100× slower than the expected D of 1–4 × 10−9 cm2/s for an integral membrane protein able to diffuse freely in a fluid lipid bilayer (Saffman and Delbruck, 1975). Therefore, there are mechanisms endogenous to HEK 293 cells for the immobilization of K+ channels. The immobile fraction and diffusion coefficient derived in our FRAP studies were similar to those previously reported for voltage-gated K+ channels in frog muscle (Weis et al., 1986). Thus, these mechanisms for immobilization seem to be conserved in different cell types.
Expressed alone, the Δ15 and V655A variants had immobile pools approximately half that observed for EGFP-Kv1.4. This was not due to the presence of endogenous PSD-95, however, because none was detected by immunofluorescence in mock transfected cells. Furthermore, V655A channels are immobilized by PSD-95. Therefore, at least one endogenous mechanism in HEK 293 cells involves interaction with the COOH terminus of Kv1.4. Despite truncation of the COOH terminus, 34% of Δ15 channels remained immobile. Additionally, the diffusion coefficient for mobile channels was not altered under any conditions. Thus, there are also mechanisms for immobilizing channels and slowing their diffusion that do not involve the COOH terminus. In the case of Shaker-type K+ channels, one potential deterrent to free diffusion is the existence of an extracellular site for N-glycosylation between the S1 and S2 domains. Extracellular glycosylation has been shown to affect membrane protein diffusion. For example, deletion of extracellular N-linked glycosylation sites in class I MHC molecules led to an increase in the diffusion coefficient (Edidin and Wei, 1982; Weir and Edidin, 1988). Thus, multiple sites of interaction may determine the mobility of plasma membrane proteins.
Conclusion
In the current study, we showed that GFP-tagged Kv1.4 can be used to study channel movement and distribution in live cells. Our results indicate that binding of PSD-95 leads to all or none immobilization of channels. Furthermore, we found that channel clusters formed in the presence of PSD-95 are stable structures. In addition, our finding that V655A maintains the ability to interact with PSD-95 in vivo suggests the potential for a wider array of proteins capable of interacting with PDZ domains than deduced from earlier in vitro studies. Finally, it was demonstrated that PSD-95 binding may have varied consequences for membrane proteins: it can either induce formation of macromolecular clusters or simply immobilize binding partners. Thus, the cellular role of proteins containing PDZ domains is likely to be even more complex and varied than previously believed.
Acknowledgments
We thank Dr. Morgan Sheng (Howard Hughes Medical Institute, Massachusetts General Hospital, Boston, MA) for the PSD-95 cDNA and Dr. James Trimmer (State University New York, Stony Brook, NY) for anti–PSD-95 antibodies.
This work was supported by grants from the National Institutes of Health to E.S. Levitan (HL55312 and a minority supplement to N.A. Burke [NS32385]). E.S. Levitan is an American Heart Association Established Investigator.
Abbreviations used in this paper
- EGFP
enhanced green fluorescent protein
- FRAP
fluorescence recovery after photobleaching
- wt
wild type
references
- Brenman JE, Bredt DS. Synaptic signaling by nitric oxide. Curr Opin Neurobiol. 1997;7:374–378. doi: 10.1016/s0959-4388(97)80065-7. [DOI] [PubMed] [Google Scholar]
- Burke NV, Han W, Takimoto K, Li D, Watkins SP, Levitan ES. Neuronal peptide release is limited by secretory granule release. Neuron. 1997;19:1095–1102. doi: 10.1016/s0896-6273(00)80400-6. [DOI] [PubMed] [Google Scholar]
- Doyle DA, Lee A, Lewis J, Eujoon K, Sheng M, MacKinnon R. Crystal structure of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- Edidin, M. 1994. Mobility and Proximity in Biological Membranes. S. Damjanovich, M. Edidin, J. Szollosi, and L. Tron, editors. CRC Press, Boca Raton, FL. 109–135.
- Edidin M, Wei T. Lateral diffusion of H-2 antigens on mouse fibroblasts. J Cell Biol. 1982;95:453–462. doi: 10.1083/jcb.95.2.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman EL, Axelrod D, Schwartz M, Heathcock AM, Agaranoff BW. Studies on the localization of newly added membrane in growing neurites. J Neurobiol. 1981;12:591–598. doi: 10.1002/neu.480120607. [DOI] [PubMed] [Google Scholar]
- Gomperts SN. Clustering membrane proteins: it's all coming together with the PSD-95/SAP90 protein family. Cell. 1996;84:659–662. doi: 10.1016/s0092-8674(00)81043-0. [DOI] [PubMed] [Google Scholar]
- Horio Y, Hibino H, Inanobe A, Yamada M, Ishii M, Tada Y, Kurachi Y. Clustering and enhanced activity of an inwardly rectifying potassium channel, Kir4.1, by an anchoring protein, PSD-95/SAP90. J Biol Chem. 1997;272:12885–12888. doi: 10.1074/jbc.272.20.12885. [DOI] [PubMed] [Google Scholar]
- Irie M, Hata Y, Takeuchi M, Ichtchenko K, Touoda A, Hirao K, Takai T, Rosahi TW, Sudhof TC. Binding of neuroligins to PSD-95. Science. 1997;277:1511–1514. doi: 10.1126/science.277.5331.1511. [DOI] [PubMed] [Google Scholar]
- Kim E, Niethammer M, Rothchild A, Jan Y, Sheng M. Clustering of Shaker-type K+channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- Kim E, Sheng M. Differential K+channel clustering activity of PSD-95 and SAP97, two related membrane-associated putative guanylate kinases. Neuropharmacology. 1996;35:993–1000. doi: 10.1016/0028-3908(96)00093-7. [DOI] [PubMed] [Google Scholar]
- Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- Kornau H, Seeburg PH, Kennedy MB. Interaction of ion channels and receptors with PDZ domain proteins. Curr Opin Neurobiol. 1997;7:368–373. doi: 10.1016/s0959-4388(97)80064-5. [DOI] [PubMed] [Google Scholar]
- Levitan ES. Tagging of K+channels with GFP to study mobility and interactions with other proteins. Methods Enzymol. 1998;294:47–58. doi: 10.1016/s0076-6879(99)94006-5. [DOI] [PubMed] [Google Scholar]
- Maletic-Savatic M, Lenn NJ, Trimmer JS. Differential spatiotemporal expression of K+channel polypeptides in rat hippocampal neurons developing in situ and in vitro. J Neurosci. 1995;15:3840–3851. doi: 10.1523/JNEUROSCI.15-05-03840.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller BM, Kistner U, Kindler S, Chung WJ, Kuhlendahl S, Fenster SD, Lau L, Veh RW, Huganir RL, Gundelfinger ED, Garner CC. SAP 102, a novel postsynaptic protein that interacts with NMDA receptor complexes in vivo. Neuron. 1996;17:255–265. doi: 10.1016/s0896-6273(00)80157-9. [DOI] [PubMed] [Google Scholar]
- Rhodes KJ, Keilbaugh SA, Barrezueta NX, Lopez KL, Trimmer JS. Association and colocalization of K+channel alpha- and beta-subunit polypeptides in rat brain. J Neurosci. 1995;15:5360–5371. doi: 10.1523/JNEUROSCI.15-07-05360.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberds SL, Tamkun MM. Cloning and tissue-specific expression of five voltage-gated potassium channel cDNAs expressed in rat heart. Proc Natl Acad Sci USA. 1991;88:1798–1802. doi: 10.1073/pnas.88.5.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffman PG, Delbruck M. Brownian motion in biological membranes. Proc Natl Acad Sci USA. 1975;72:3111–3113. doi: 10.1073/pnas.72.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer JL. Clustering sodium channels at the node of Ranvier: close encounters of the axon-glia kind. Neuron. 1997;18:843–846. doi: 10.1016/s0896-6273(00)80323-2. [DOI] [PubMed] [Google Scholar]
- Sheng M, Tsaur ML, Jan YN, Jan LY. Subcellular segregation of two A-type K+channel proteins in rat central neurons. Neuron. 1992;9:271–284. doi: 10.1016/0896-6273(92)90166-b. [DOI] [PubMed] [Google Scholar]
- Sheng M, Liao YJ, Jan YN, Jan LY. Presynaptic A-current based on heteromultimeric K+channels detected in vivo. Nature. 1993;365:72–75. doi: 10.1038/365072a0. [DOI] [PubMed] [Google Scholar]
- Sheng M. PDZs and receptor/channel clustering: rounding up the latest suspects. Neuron. 1996;17:575–578. doi: 10.1016/s0896-6273(00)80190-7. [DOI] [PubMed] [Google Scholar]
- Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton AC, Chan JM, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- Stühmer W, Ruppersburg JP, Schroter KH, Stocker M, Giese KP, Perschke A, Baumann A, Pongs O. Molecular basis of functional diversity of voltage-gated potassium channels in mammalian brain. EMBO (Eur Mol Biol Organ) J. 1989;8:3235–3244. doi: 10.1002/j.1460-2075.1989.tb08483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto K, Gealy R, Fomina AF, Trimmer JS, Levitan ES. Inhibition of voltage-gated K+channel gene expression by the neuropeptide thyrotropin-releasing hormone. J Neurosci. 1995;15:449–457. doi: 10.1523/JNEUROSCI.15-01-00449.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejedor FJ, Bokhari A, Rogero O, Gorczyca M, Zhang J, Kim E, Sheng M, Budnik V. Essential role for dlg in synaptic clustering of Shaker K+channels in vivo. J Neurosci. 1997;17:152–159. doi: 10.1523/JNEUROSCI.17-01-00152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas U, Kim E, Kuhlendahl S, Koh YH, Gundelfinger ED, Sheng M, Garner CC, Budnik V. Synaptic clustering of the cell adhesion molecule fasciclin II by discs-large and its role in the regulation of presynaptic structure. Neuron. 1997;19:787–799. doi: 10.1016/s0896-6273(00)80961-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda S, Sierralta J, Yumel S, Bodner R, Suzuki E, Becker A, Socolich M, Zucker CS. A multivalent PDZ-domain protein assembles signalling complexes in a G-protein-coupled cascade. Nature. 1997;388:243–249. doi: 10.1038/40805. [DOI] [PubMed] [Google Scholar]
- Weis RE, Roberts WW, Stühmer W, Almers W. Mobility of voltage-dependent channels and lectin receptors in the sarcolemma of frog skeletal muscle. J Gen Physiol. 1986;87:955–983. doi: 10.1085/jgp.87.6.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wier M, Edidin M. Constraint of the translational diffusion of a membrane glycoprotein by its external domains. Science. 1988;242:412–414. doi: 10.1126/science.3175663. [DOI] [PubMed] [Google Scholar]