Abstract
Using colony blot hybridization with stx2 and eae probes and agglutination in anti-O157 lipopolysaccharide serum, we isolated stx2-positive and eae-positive sorbitol-fermenting (SF) enterohemorrhagic Escherichia coli (EHEC) O157:NM (nonmotile) strains from initial stool specimens and stx-negative and eae-positive SF E. coli O157:NM strains from follow-up specimens (collected 3 to 8 days later) from three children. The stx-negative isolates from each patient shared with the corresponding stx2-positive isolates fliCH7, non-stx virulence traits, and multilocus sequence types, which indicates that they arose from the stx2-positive strains by loss of stx2 during infection. Analysis of the integrity of the yecE gene, a possible stx phage integration site in EHEC O157, in the consecutive stx2-positive and stx-negative isolates demonstrated that yecE was occupied in stx2-positive but intact in stx-negative strains. It was possible to infect and lysogenize the stx-negative E. coli O157 strains in vitro using an stx2-harboring bacteriophage from one of the SF EHEC O157:NM isolates. The acquisition of the stx2-containing phage resulted in the occupation of yecE and production of biologically active Shiga toxin 2. We conclude that the yecE gene in SF E. coli O157:NM is a hot spot for excision and integration of Shiga toxin 2-encoding bacteriophages. SF EHEC O157:NM strains and their stx-negative derivatives thus represent a highly dynamic system that can convert in both directions by the loss and gain of stx2-harboring phages. The ability to recycle stx2, a critical virulence trait, makes SF E. coli O157:NM strains ephemeral EHEC that can exist as stx-negative variants during certain phases of their life cycle.
Sorbitol-fermenting (SF) enterohemorrhagic Escherichia coli (EHEC) O157:NM strains (nonmotile) were first identified in 1988 in Germany (28) and have now emerged as a cause of human diseases, including life-threatening hemolytic-uremic syndrome (HUS), in additional countries (4, 7, 8, 17, 20, 22, 37). In Germany, SF EHEC O157:NM strains account for >10% of sporadic cases of HUS (20, 22), and after EHEC O157:H7, they are the second most common cause of HUS (20, 22). They have also caused several outbreaks in Germany (5, 41, 42) and the United Kingdom (3, 18).
The genes encoding Shiga toxins (Stx) (44), the major virulence factors of EHEC, are located in temperate lambdoid bacteriophages which are integrated in the host genome during lysogenic growth (2, 25, 29, 35, 48, 53). A lytic phage cycle can be induced in EHEC strains using sublethal doses of UV light (35) or subtherapeutic concentrations of various antibiotics (23, 33, 35, 45, 47, 58). The existence of stx genes in a multitude of E. coli serotypes (2, 55) is attributable to transduction with stx-converting phages (2, 25, 45). Transduction in vivo has been demonstrated in several animal models including mice (1), sheep (14), and insects (39).
Both loss and transfer of the stx gene appear to occur during human infection and can lead to a change in the pathotype of the infecting strain (10, 21, 34). We have demonstrated the excretion of stx2-positive EHEC O157:NM in the initial stool specimen (collected 8 days after the onset of prodromal diarrhea) of an HUS patient, followed by the excretion of stx-negative E. coli O157:NM in the follow-up stool specimen collected 3 days later (34). Comparison of stx gene losses in SF EHEC O157:NM and non-SF EHEC O157:H7 isolates showed a significantly higher proportion of stx-negative strains (12.7% versus 0.8%) among SF E. coli O157:NM (21). This loss of stx genes has important diagnostic implications if laboratories rely on toxin detection to find these organisms late in illness (34). Furthermore, the loss of stx complicates epidemiological investigations (11) and might even influence the outcome of the disease (21).
One of integration sites for stx2-harboring bacteriophages in the chromosome of SF EHEC O157:NM is the yecE gene (11). In this study of the dynamic processes in the evolution of the genome in SF EHEC O157:NM, we compared the integrity of the phage integration site in stx2-positive SF EHEC O157:NM strains and their stx-negative derivatives and investigated the ability of stx2 phages from SF EHEC O157 patients’ isolates to lysogenize stx-negative variants of SF EHEC O157:NM.
MATERIALS AND METHODS
Bacterial strains.
Three SF EHEC O157:NM strains and their corresponding stx-negative derivatives were isolated from stool specimens collected 8 to 11 days after the onset of diarrhea and in subsequent stool specimens from the same patients collected 3 to 8 days later, respectively. All patients had HUS. Pair A1-A2 has been partially described previously (34). Pairs B1-B2 and C1-C2 are from a 15-month-old boy and a 20-month-old girl, respectively. In each pair, isolate 1 is stx2 positive and isolate 2 is stx negative. The stx2-positive SF EHEC O157:NM strain from the initial stool specimen from patient B (B1; 258/98) has been described previously (11). Strains were isolated as described previously (20, 21, 34). Briefly, stool specimens were inoculated into Hajna broth and grown, and the broth was then enriched for E. coli O157 using immunomagnetic separation with Dynabeads coated with anti-E. coli O157 antibody (Dynal, Oslo, Norway). Beads with attached E. coli O157 were cultured on sorbitol-MacConkey (SMAC) agar (Oxoid, Basingstoke, United Kingdom). Overnight cultures harvested into saline were screened for stx1, stx2, rfbO157, and eae by PCR (19, 34). SF E. coli O157 was isolated from PCR-positive cultures using colony blot hybridization with stx2 and eae probes and slide agglutination with antiserum specific for the E. coli O157 lipopolysaccharide (21, 34).
PCR.
PCR analyses were performed as described previously (11, 48). stx2, rfbO157, and sfpA (a specific marker for SF E. coli O157:NM) (19) were detected using primer pairs LP43 and LP44, O157-F and O157-R, and sfpA-U and sfpA-L, respectively (19, 20); stx2 genes were subtyped using PCR with primers GK3 and GK4 and restriction of the amplicon with HaeIII (20). Genes encoding other toxins (cdt-V and EHEC-hlyA) and adhesins (eae, efa1, lpfO157-OI141, and lpfO157-OI154), other plasmid-carried genes (katP, espP, and etpD), and terE (used as a marker for the gene cluster encoding tellurite resistance) (24, 38) were detected using established protocols (9, 12, 26, 27, 49, 51). The eae genes were subtyped (56), and the genotype of the fliC gene, encoding the structural subunit of the H antigen, was determined as described previously (49, 57). The intact or occupied status of yecE was tested using primers EC10 and EC11 (16). The integration of an stx2-carrying bacteriophage homologous to phage Φ258320 (originating from SF EHEC O157:NM strain 258/98) (11) in yecE in wild-type EHEC strains and lysogens was identified by PCR with primers Int-258320 and EC11, which connect the integrase gene of phage Φ258320 with yecE (11).
Multilocus sequence typing.
Internal fragments of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) were analyzed as described previously (54), except for use of a newly designed forward primer for icd (5′-CGATTATCCCTTACATTGAAG-3′), which yielded more reliable sequencing results. Alleles and sequence types were assigned in accordance with the E. coli multilocus sequence typing website (http://web.mpiib-berlin.mpg.de/mlst/dbs/Ecoli).
Induction of stx2-converting phages from SF EHEC O157:NM and transduction experiments.
The stx2-converting bacteriophage Φ258320 was induced from SF EHEC O157:NM strain B1 (258/98) (11) using 0.5 μg/ml of mitomycin C (Sigma-Aldrich, Deisenhofen, Germany) (45). One hundred microliters of the phage lysate was mixed with 100 μl of the log-phase culture (l06 to 107 CFU) of an stx-negative SF E. coli O157:NM strain (A2, B2, or C2) and 125 μl of 0.1 M CaCl2 solution and incubated for 2 h at 37°C without shaking. After that, the mixture was transferred into 4 ml of Luria-Bertani (LB) broth and incubated for 48 h at 37°C and 180 rpm. Tenfold dilutions of the culture were inoculated on LB agar, and overnight growths harvested into 1 ml of saline were screened for stx2 by PCR with primers LP43 and LP44. The PCR-positive cultures were restreaked on LB agar, and plates containing ∼200 well-separated colonies were used to isolate stx2-containing colonies (putative lysogens) by colony blot hybridization with the stx2 probe (20, 34). The stx2-positive colonies were subcultured three times on LB agar and tested by PCR for stx2 after the third passage to identify stable lysogens (45); the lysogens were checked for the maintenance of stx2 at 3-month intervals for 1 year. E. coli strain C600, which can be lysogenized with phage Φ258320 (11), was used as a positive control in the transduction experiments.
Phenotypes.
Sorbitol fermentation was detected on SMAC after overnight incubation. β-d-Glucuronidase activity was determined on Difco nutrient agar with 4-methylumbelliferyl β-d-glucuronide (Becton Dickinson, Heidelberg, Germany). Production of EHEC hemolysin was detected on enterohemolysin agar (Sifin, Berlin, Germany) and resistance to tellurite on cefixime-tellurite-SMAC (9). Stx production was investigated using a latex agglutination assay (Verotox-F; Denka Seiken Co., Ltd., Tokyo, Japan). Stx cytotoxicity titers in culture supernatants were assessed using the Vero cell assay (30). Production of cytolethal distending toxin V (CDT-V) was tested using the Chinese hamster ovary cell assay (26).
RESULTS
Characteristics of stx2-positive and stx-negative SF E. coli O157:NM strains isolated from sequential stool specimens.
All three initial EHEC O157:NM strains isolated from stool specimens from the three patients and all three subsequent stx-negative isolates shared putative virulence genes encoding non-Stx toxins (EHEC hemolysin and CDT-V) and various adhesins (Table 1). Moreover, all six strains displayed an identical combination of plasmid-carried genes (presence of EHEC-hlyA, etpD, and sfpA and absence of katP and espP). Each of the strains contained rfbO157 and fliCH7 but lacked the terE gene and, accordingly, failed to grow on cefixime-tellurite-SMAC. All strains fermented sorbitol, produced β-d-glucuronidase, and secreted CDT-V in similar amounts, but all failed to express the enterohemolytic phenotype (Table 1). Moreover, they shared identical sequence types (ST11), indicating identical clonal backgrounds (Table 1). The finding that the stx2-positive and stx-negative intrapatient isolates share stx-independent characteristics suggested that the latter organisms descended from the former ones by the loss of stx2 during infection.
TABLE 1.
Molecular and phenotypic traits of consecutive stx2-positive and stx-negative SF E. coli O157 isolates from patients A, B, and C
| Strain | Molecular traita
|
Phenotypeb
|
STc | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rfbO157 | fliCH7 | stx2 | EHEC hlyA | cdt-V | eae | efa1d | sfpA | lpfAO157-OI141 | lpfAO157-OI154 | terE | katP | espP | etpD | SF/ GUD | EHEC-Hly | CDT | Ter | ||
| A1 | + | + | + | + | + | γ | + | + | + | + | − | − | − | + | +/+ | − | 1:16 | − | 11 |
| A2 | + | + | − | + | + | γ | + | + | + | + | − | − | − | + | +/+ | − | 1:8 | − | 11 |
| B1 | + | + | + | + | + | γ | + | + | + | + | − | − | − | + | +/+ | − | 1:8 | − | 11 |
| B2 | + | + | − | + | + | γ | + | + | + | + | − | − | − | + | +/+ | − | 1:8 | − | 11 |
| C1 | + | + | + | + | + | γ | + | + | + | + | − | − | − | + | +/+ | − | 1:8 | − | 11 |
| C2 | + | + | − | + | + | γ | + | + | + | + | − | − | − | + | +/+ | − | 1:4 | − | 11 |
The presence of the genes was determined by PCR as described in Materials and Methods; +, presence of the gene; −, absence of the gene.
Phenotypes were determined as described in Materials and Methods. SF, sorbitol fermentation; GUD, production of β-d-glucuronidase; EHEC-Hly, production of EHEC hemolysin; CDT, production of CDT-V (titers determined using Chinese hamster ovary cells); Ter, tellurite resistance. +, presence of the phenotype; −, absence of the phenotype.
ST, sequence type.
A complete efa1 gene (ca. 10 kb) was identified in all strains using PCRs targeting the 5′, middle, and 3′ regions of the gene (27).
Integrity of yecE in stx2-positive and stx-negative SF E. coli O157:NM strains.
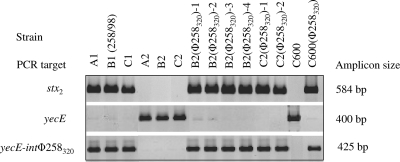
yecE was occupied in each of the stx2-positive strains of the original infection (A1, B1, and C1) (Table 2; Fig. 1, lanes 1 to 3) and was intact in all three stx-negative strains (A2, B2, and C2) (Table 2; Fig. 1, lanes 4 to 6).
TABLE 2.
Presence of stx2 genes and occupation of yecE with an stx2-harboring phage in SF E. coli O157:NM parental stx2-positive strains, their stx-negative derivatives, and lysogens
| Patient's isolate or lysogen | Result of PCR targeting:
|
Stx2 titer (latex agglutination assay)c | Stx cytotoxicity titer (Vero cell assay) | ||
|---|---|---|---|---|---|
| stx2 | yecEa | yecE-intΦ258320b | |||
| A1 | + | − | + | 1:64 | 1:128 |
| A2 | − | + | − | <1:2 | <1:2 |
| B1 | + | − | + | 1:256 | 1:1,024 |
| B2 | − | + | − | <1:2 | <1:2 |
| C1 | + | − | + | 1:128 | 1:256 |
| C2 | − | + | − | <1:2 | <1:2 |
| B2(Φ258320)-1 | + | − | + | 1:64 | 1:64 |
| B2(Φ258320)-2 | + | − | + | 1:64 | 1:128 |
| B2(Φ258320)-3 | + | − | + | 1:128 | 1:256 |
| B2(Φ258320)-4 | + | − | + | 1:128 | 1:128 |
| C2(Φ258320)-1 | + | − | + | 1:128 | 1:256 |
| C2(Φ258320)-2 | + | − | + | 1:64 | 1:128 |
| C600 | − | + | − | <1:2 | <1:2 |
| C600(Φ258320) | + | − | + | 1:16 | 1:32 |
+, the locus is intact; −, the locus is occupied by foreign DNA.
+, a phage with the int gene homologous to that of phage Φ258320 is integrated in yecE; −, the phage is absent.
Verotox-F (Denka Seikesn, Tokyo, Japan).
FIG. 1.
PCR analysis of occupation of yecE in SF EHEC O157:NM clinical isolates, their stx-negative derivatives, and lysogens. The strains tested, PCR targets, and lengths of PCR amplicons are listed across the top and to the left and right of the rows of amplicons, respectively. Strain B2 (258/98), harboring phage Φ258320 integrated in yecE (11), and E. coli K-12 C600, which has yecE intact (13), were used as controls. In PCR targeting yecE, the presence of an amplicon indicates that the locus is intact, whereas the absence of an amplicon (or a very weak amplicon) indicates that the locus is occupied by foreign DNA. In PCR connecting yecE with the integrase gene (int) of phage Φ258320, the presence of an amplicon indicates that a phage with a homologous int gene is integrated in yecE; the absence of an amplicon indicates the absence of such phage.
Transduction of stx-negative SF E. coli O157:NM with an stx2-harboring phage from SF EHEC O157:NM.
A stable (for at least 12 months) infection with phage Φ258320 could be achieved in two of the three stx-negative SF E. coli O157:NM strains. The remaining strain (A2) could be transiently infected but lost the phage after the second passage on LB agar. Four stable lysogens (among 246 transduced colonies) originated from the cognate stx-negative strain B2, an stx-negative derivative of strain B1 (258/98) (transduction frequency of 1.6%) and were designated B2(Φ258320)-1 to B2(Φ258320)-4. Two stable lysogens (among 221 transduced colonies) originated from a noncognate stx-negative strain (C2) derived from EHEC strain C1 (transduction frequency of 0.9%) and were designated C2(Φ258320)-1 and C2(Φ258320)-2. The lysogenization of the stx-negative strains with stx2-harboring phage Φ258320 resulted in the acquisition of stx2 by each of the lysogens, as demonstrated by PCR with primers LP43-LP44 (Table 2; Fig. 1, lanes 7 to 12). Moreover, phage Φ258320 also transduced the stx2 gene into the control strain E. coli C600 (Table 2; Fig. 1, lane 14).
Stx production.
The latex agglutination assay confirmed that all three parental stx2-containing SF EHEC O157:NM strains produced Stx2 (titers of 1:64 to 1:256) and that their stx-negative derivatives did not (titers of <1:2) (Table 2). Each of the six lysogens produced Stx2 in titers (1:64 to 1:128) comparable to that of the donor strain B1 (1:256) (Table 2). Stx2 produced by the lysogens was cytotoxic toward Vero cells (Table 2), indicating that the biological activity is similar to that of Stx2 produced by the wild-type SF EHEC O157:NM strains.
Phage integration sites.
In each lysogen, the acquisition of the stx2-harboring phage Φ258320 resulted in the occupation of yecE (Table 2; Fig. 1, lanes 7 to 12), which had been intact in the stx-negative recipient strains B2 and C2 (Table 2; Fig. 1, lanes 5 and 6, respectively). We confirmed that yecE in the lysogens was occupied by phage Φ258320, using PCR to link yecE with the integrase gene of phage Φ258320. The PCR produced an amplicon of 425 bp in strain B1 (258/98) (and also A1 and C1) (Fig. 1, lanes 1 to 3) and each of the lysogens (Fig. 1, lanes 7 to 12), demonstrating that all these strains contained a phage homologous to phage Φ258320 integrated in yecE. As expected, phage Φ258320 also integrated into yecE in the control E. coli strain C600 (Fig. 1, lane 14). Taken together, these data demonstrate that stx-negative SF E. coli O157:NM strains can be converted to EHEC O157:NM via transduction with an stx2-harboring phage originating from an EHEC O157:NM strain and that yecE plays an essential role in this process.
DISCUSSION
One noteworthy aspect of EHEC infections is that the infecting organism can undergo major genotype and phenotype changes during the course of infection in humans (10, 21, 34). We have demonstrated interconversion between various pathotypes of SF E. coli O157:NM, in which EHEC O157:NM strains harboring stx2 lose stx2 and stx-negative E. coli O157:NM can be transduced by an stx2-harboring phage from an EHEC O157 patient's isolate. In these events, yecE is a hot spot for integration and excision of stx2 phages. The ability to undergo bidirectional conversion from one pathotype to another enables recycling of an important pathogenic trait in SF E. coli O157:NM and indicates that these organisms can respond to environmental influences with a high degree of flexibility and that this property increases their capacity to survive under a variety of conditions in the human host and reservoir(s).
Although yecE is only rarely occupied by phages in EHEC O157:H7 (16, 46), our analysis of consecutive stx2-positive and stx-negative SF E. coli O157:NM strains confirms our previous observation that yecE is an important integration site for Stx2-encoding phages in SF EHEC O157:NM strains (11). It is of interest that up to now no other function is known for the yecE gene or its product. yecE in all stx2-positive SF EHEC O157:NM strains was occupied by an stx2-converting phage, whereas in all stx-negative SF E. coli O157:NM strains the yecE genes were intact (Fig. 1). Lysogenization of stx-negative strains with stx2-containing phage Φ258320 originating from a SF EHEC O157:NM clinical isolate resulted in the occupation of yecE in the lysogens (Fig. 1). The presence of phage Φ258320 in yecE in the genomes of the lysogens could be confirmed by the signal elicited from all lysogens in the PCR connecting yecE with the integrase gene of phage Φ258320 (Fig. 1). Together with acquiring the stx2 gene, all lysogens also acquired the ability to produce Stx2 cytotoxic to Vero cells. This demonstrates that via transduction with the stx2 phage, the stx-negative, eae-positive SF E. coli O157:NM strains were converted to biologically potent EHEC. In contrast to the case for SF EHEC O157:NM, stx2 phages in EHEC O157:H7, including both sequenced strains EDL933 and Sakai (24, 32, 38, 40) and other clinical isolates (6, 47), use wrbA as a common genomic integration site, whereas yehV is frequently used for integration of stx2 phages in EHEC O157:H7 isolated from cattle (46). In addition, other, mostly yet-unidentified chromosomal loci have been proposed to serve as integration sites for stx2-harboring phages in the genome of EHEC O157:H7 (6, 36, 47). Although it is presently unknown why yecE, which is intact in E. coli O157:H7 strains EDL933 and Sakai (24, 38) and other isolates (unpublished data), does not regularly serve as an stx2 phage integration site in these organisms (46), this fact is in accordance with a recent observation that the selection of an stx phage integration site preferentially depends on the host strain rather than on the phage (46). The greater stability of stx2 in EHEC O157:H7 than in SF EHEC O157:NM (21, 34) might be attributable to the difference in the integration sites for stx2-harboring phages in the two organisms.
The observation that stx-negative SF E. coli O157:NM strains occur more frequently than stx-positive SF E. coli O157:NM strains in the gastrointestinal tracts of animals (31, 43, 59) suggests that an stx-negative phase might be a normal occurrence in the life cycle of SF EHEC O157:NM in the environment. The stx-negative condition might enable the pathogens to avoid lysis triggered by various phage-inducing stimuli, such as H2O2 (52), thereby conferring a selective advantage to survive under different conditions. If analogous to E. coli O157:H7 (15), the absence of an stx phage would not influence the ability of stx-negative SF E. coli O157 organisms to colonize the intestines of animals and thus to be spread in the environment via fecal contamination. We propose that such stx-negative, eae-positive SF E. coli O157:NM strains, which can be converted to EHEC by transduction with stx2 phages from EHEC O157 in vitro, represent “EHEC wannabes.” The acquisition of an stx phage by such organisms not only makes them highly pathogenic for the human host but also could play a role in their ecology. It has been recently shown that the carriage of an Stx-encoding prophage increases the rate of survival of E. coli O157:H7 within grazing protozoa (Tetrahymena pyriformis) (50) and thus in the environment. Apparently, both gain and loss of stx phages can contribute to the evolution of SF E. coli O157:NM. Because of their ability to recycle stx2, which is the major virulence trait, SF E. coli O157:NM can be considered ephemeral EHEC strains that can exist as stx-negative organisms during certain phases of their life cycle.
A change in the pathotype of the infecting strain during infection has consequences regarding the laboratory diagnosis and epidemiological investigations. Loss of the stx gene makes it impossible to correctly identify stx-negative variants using methods based on the detection of only stx and/or Stx. To overcome this limitation, we recommend the parallel use of procedures independent of the presence of stx, in particular the detection of eae using PCR (34). In the case of SF E. coli O157:NM, detection of O157 lipopolysaccharide antigen by slide agglutination and/or its encoding gene using PCR and, especially, detection of the sfpA gene, which is a unique marker for SF E. coli O157:NM (19), allow identification of both stx-positive and stx-negative strains (19). Furthermore, loss of stx genes in strains that are related epidemiologically can change the pulsed-field gel electrophoresis pattern (11), thereby complicating epidemiological investigations of SF E. coli O157:NM disease (11). Therefore, the possibility of an altered pulsed-field gel electrophoresis pattern arising from the loss of stx should be always considered when interpreting the DNA fingerprints of potentially epidemiologically related SF E. coli O157:NM strains.
Ongoing studies in our laboratories aim to investigate the conditions favoring the loss of stx and the possibility of promoting the excision of stx and conversion to less pathogenic stx-negative EHEC. In the absence of specific anti-EHEC treatment, such an intervention would offer a new and powerful preventive and therapeutic approach.
Acknowledgments
This study was supported by a grant from the EU Network ERA-NET PathoGenoMics (project number 0313937C), by a grant from the EU Network of Excellence EuroPathoGenomics (number LSHB-CT-2005-512061), by a grant from the Interdisciplinary Center of Clinical Research (IZKF) Münster (project no. Ka2/061/04), and by 973 Program grant 2005CB522904 from the Ministry of Science and Technology, People's Republic of China.
We thank Phillip I. Tarr (Washington University School of Medicine, St. Louis, MO) for helpful discussion during preparation of the manuscript.
Footnotes
Published ahead of print on 2 November 2007.
REFERENCES
- 1.Acheson, D. W., J. Reidl, X. Zhang, G. T. Keusch, J. J. Mekalanos, and M. K. Waldor. 1998. In vivo transduction with Shiga toxin 1-encoding phage. Infect. Immun. 66:4496-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, H. E. 2007. Stx-phages: drivers and mediators of the evolution of STEC and STEC-like pathogens. Future Microbiol. 2:165-174. [DOI] [PubMed] [Google Scholar]
- 3.Allison, L., S. Harding, M. Locking, K. Pollock, J. Evans, H. Knight, G. Foster, J. Cowden, and M. Hanson. 2006. Sorbitol-fermenting E. coli O157 in Scotland—laboratory investigation of an outbreak of haemolytic ureamic syndrome, abstr. P01.1.01, p. 4. Abstr. 6th Int. Symp. Shiga Toxin (Verocytotoxin)-Producing Escherichia coli Infect. (VTEC 2006), Melbourne, Australia.
- 4.Allison, L. 2002. HUS due to a sorbitol-fermenting verotoxigenic E. coli O157 in Scotland. Eurosurveill. Wkly. 44:021031. [Google Scholar]
- 5.Ammon, A., L. R. Petersen, and H. Karch. 1999. A large outbreak of hemolytic uremic syndrome caused by an unusual sorbitol-fermenting strain of Escherichia coli O157:H−. J. Infect. Dis. 179:1274-1277. [DOI] [PubMed] [Google Scholar]
- 6.Besser, T. E., N. Shaikh, N. J. Holt, P. I. Tarr, M. E. Konkel, P. Malik-Kale, C. W. Walsh, T. S. Whittam, and J. L. Bono. 2007. Greater diversity of Shiga toxin-encoding bacteriophage insertion sites among Escherichia coli O157:H7 isolates from cattle than in those from humans. Appl. Environ. Microbiol. 73:671-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bettelheim, K. A., M. Whipp, S. P. Djordjevic, and V. Ramachandran. 2002. First isolation outside Europe of sorbitol-fermenting verocytotoxigenic Escherichia coli (VTEC) belonging to O group O157. J. Med. Microbiol. 51:713-714. [DOI] [PubMed] [Google Scholar]
- 8.Bielaszewska, M., H. Schmidt, M. A. Karmali, R. Khakhria, J. Janda, K. Blahova, and H. Karch. 1998. Isolation and characterization of sorbitol-fermenting Shiga toxin (verocytotoxin)-producing Escherichia coli O157:H− strains in the Czech Republic. J. Clin. Microbiol. 36:2135-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bielaszewska, M., P. I. Tarr, H. Karch, W. Zhang, and W. Mathys. 2005. Phenotypic and molecular analysis of tellurite resistance among enterohemorrhagic Escherichia coli O157:H7 and sorbitol-fermenting O157:NM clinical isolates. J. Clin. Microbiol. 43:452-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bielaszewska, M., R. Prager, R. Köck, A. Mellmann, W. Zhang, H. Tschäpe, P. I. Tarr, and H. Karch. 2007. Shiga toxin gene loss and transfer in vitro and in vivo during enterohemorrhagic Escherichia coli O26 infection in humans. Appl. Environ. Microbiol. 73:3144-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bielaszewska, M., R. Prager, W. Zhang, A. W. Friedrich, A. Mellmann, H. Tschäpe, and H. Karch. 2006. Chromosomal dynamism in progeny of outbreak-related sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM. Appl. Environ. Microbiol. 72:1900-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bielaszewska, M., W. Zhang, P. I. Tarr, A. Sonntag, and H. Karch. 2005. Molecular profiling and phenotype analysis of Escherichia coli O26:H11 and O26:NM: secular and geographic consistency of enterohemorrhagic and enteropathogenic isolates. J. Clin. Microbiol. 43:4225-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 14.Cornick, N. A., A. F. Helgerson, V. Mai, J. M. Ritchie, and D. W. K. Acheson. 2006. In vivo transduction of an Stx-encoding phage in ruminants. Appl. Environ. Microbiol. 72:5086-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornick, N. A., A. F. Helgerson, and V. Sharma. 2007. Shiga toxin and Shiga toxin-encoding phage do not facilitate Escherichia coli O157:H7 colonization in sheep. Appl. Environ. Microbiol. 73:344-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Greve, H., C. Qizhi, F. Deboeck, and J. P. Hernalsteens. 2002. The Shiga toxin VT2-encoding bacteriophage varphi297 integrates at a distinct position in the Escherichia coli genome. Biochim. Biophys. Acta 1579:196-202. [DOI] [PubMed] [Google Scholar]
- 17.Eklund, M., M. Bielaszewska, U. Nakari, H. Karch, and A. Siitonen. 2006. Molecular and phenotypic profiling of sorbitol-fermenting Escherichia coli O157:H− human isolates from Finland. Clin. Microbiol. Infect. 12:634-641. [DOI] [PubMed] [Google Scholar]
- 18.Eurosurveillance Editorial Team. 2006. E. coli O157 infections in the UK. Euro. Surveill. 11:E060601.2. [PubMed] [Google Scholar]
- 19.Friedrich, A. W., K. V. Nierhoff, M. Bielaszewska, A. Mellmann, and H. Karch. 2004. Phylogeny, clinical associations, and diagnostic utility of the pilin subunit gene (sfpA) of sorbitol-fermenting, enterohemorrhagic Escherichia coli O157:H− J. Clin. Microbiol. 42:4697-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedrich, A. W., M. Bielaszewska, W. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 21.Friedrich, A. W., W. Zhang, M. Bielaszewska, A. Mellmann, R. Köck, A. Fruth, H. Tschäpe, and H. Karch. 2007. Prevalence, virulence profiles, and clinical significance of Shiga toxin-negative variants of enterohemorrhagic Escherichia coli O157 infection in humans. Clin. Infect. Dis. 45:39-45. [DOI] [PubMed] [Google Scholar]
- 22.Gerber, A., H. Karch, F. Allerberger, H. M. Verweyen, and L. B. Zimmerhackl. 2002. Clinical course and the role of Shiga toxin-producing Escherichia coli infection in the hemolytic-uremic syndrome in pediatric patients, 1997-2000, in Germany and Austria: a prospective study. J. Infect. Dis. 186:493-500. [DOI] [PubMed] [Google Scholar]
- 23.Grif, K., M. P. Dierich, H. Karch, and F. Allerberger. 1998. Strain-specific differences in the amount of Shiga toxin released from enterohemorrhagic Escherichia coli O157 following exposure to subinhibitory concentrations of antimicrobial agents. Eur. J. Clin. Microbiol. Infect. Dis. 17:761-766. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 25.Herold, S., H. Karch, and H. Schmidt. 2004. Shiga toxin-encoding bacteriophages—genomes in motion. Int. J. Med. Microbiol. 294:115-121. [DOI] [PubMed] [Google Scholar]
- 26.Janka, A., M. Bielaszewska, U. Dobrindt, L. Greune, M. A. Schmidt, and H. Karch. 2003. Cytolethal distending toxin gene cluster in enterohemorrhagic Escherichia coli O157:H− and O157:H7: characterization and evolutionary considerations. Infect. Immun. 71:3634-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janka, A., M. Bielaszewska, U. Dobrindt, and H. Karch. 2002. Identification and distribution of the enterohemorrhagic Escherichia coli factor for adherence (efa1) gene in sorbitol-fermenting Escherichia coli O157:H−. Int. J. Med. Microbiol. 292:207-214. [DOI] [PubMed] [Google Scholar]
- 28.Karch, H., H. Böhm, H. Schmidt, F. Gunzer, S. Aleksic, and J. Heesemann. 1993. Clonal structure and pathogenicity of Shiga-like toxin-producing, sorbitol-fermenting Escherichia coli O157:H−. J. Clin. Microbiol. 31:1200-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karch, H., H. Schmidt, C. Janetzki-Mittmann, J. Scheef, and M. Kroger. 1999. Shiga toxins even when different are encoded at identical positions in the genomes of related temperate bacteriophages. Mol. Gen. Genet. 262:600-607. [DOI] [PubMed] [Google Scholar]
- 30.Karmali, M. A., M. Petric, C. Lim, P. C. Fleming, G. S. Arbus, and H. Lior. 1985. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J. Infect. Dis. 151:775-782. [DOI] [PubMed] [Google Scholar]
- 31.Lee, J. H., and S. Choi. 2006. Isolation and characteristics of sorbitol-fermenting Escherichia coli O157 strains from cattle. Microbes Infect. 8:2021-2026. [DOI] [PubMed] [Google Scholar]
- 32.Makino, K., K. Yokoyama, Y. Kubota, C. H. Yutsudo, S. Kimura, K. Kurokawa, K. Ishii, M. Hattori, I. Tatsuno, H. Abe, T. Iida, K. Yamamoto, M. Onishi, T. Hayashi, T. Yasunaga, T. Honda, C. Sasakawa, and H. Shinagawa. 1999. Complete nucleotide sequence of the prophage VT2-Sakai carrying the verotoxin 2 genes of the enterohemorrhagic Escherichia coli O157:H7 derived from the Sakai outbreak. Genes Genet. Syst. 74:227-239. [DOI] [PubMed] [Google Scholar]
- 33.Matsushiro, A., K. Sato, H. Miyamoto, T. Yamamura, and T. Honda. 1999. Induction of prophages of enterohemorrhagic Escherichia coli O157:H7 with norfloxacin. J. Bacteriol. 181:2257-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mellmann, A., M. Bielaszewska, L. B. Zimmerhackl, R. Prager, D. Harmsen, H. Tschäpe, and H. Karch. 2005. Enterohemorrhagic Escherichia coli in human infection: in vivo evolution of a bacterial pathogen. Clin. Infect. Dis. 41:785-792. [DOI] [PubMed] [Google Scholar]
- 35.O'Brien, A. D., J. W. Newland, S. F. Miller, R. K. Holmes, H. W. Smith, and S. B. Formal. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694-696. [DOI] [PubMed] [Google Scholar]
- 36.Ohnishi, M., J. Terajima, K. Kurokawa, K. Nakayama, T. Murata, K. Tamura, Y. Ogura, H. Watanabe, and T. Hayashi. 2002. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl. Acad. Sci. USA 99:17043-17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orth, D., K. Grif, M. P. Dierich, and R. Würzner. 2006. Sorbitol-fermenting Shiga toxin-producing Escherichia coli O157: indications for an animal reservoir. Epidemiol. Infect. 134:719-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 39.Petridis, M., M. Bagdasarian, M. K. Waldor, and E. Walker. 2006. Horizontal transfer of Shiga toxin and antibiotic resistance genes among Escherichia coli strains in house fly (Diptera: Muscidae) gut. J. Med. Entomol. 43:288-295. [DOI] [PubMed] [Google Scholar]
- 40.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robert Koch-Institut. 2002. Häufung Sorbitol-fermentierender E. coli O157:H− in mehreren Bundesländern. Epidemiol. Bull. 15:123. [Google Scholar]
- 42.Robert Koch-Institut. 2003. Ein HUS-Ausbruch durch sorbitol-fermentierende EHEC des Serovars O157:H−: Untersuchungsergebnisse und Lehren für die Surveillance. Epidemiol. Bull. 22:171-175. [Google Scholar]
- 43.Rogerie, F., A. Marecat, S. Gambade, F. Dupond, P. Beaubois, and M. Lange. 2001. Characterization of Shiga toxin-producing E. coli and O157 serotype E. coli isolated in France from healthy domestic cattle. Int. J. Food Microbiol. 63:217-223. [DOI] [PubMed] [Google Scholar]
- 44.Sandvig, K. 2001. Shiga toxins. Toxicon 39:1629-1635. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt, H., M. Bielaszewska, and H. Karch. 1999. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage φ3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:3855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serra-Moreno, R., J. Jofre, and M. Muniesa. 2007. Insertion site occupancy by stx2 bacteriophages depends on the locus availability of the host strain chromosome. J. Bacteriol. 189:6645-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaikh, N., and P. I. Tarr. 2003. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: integrations, excisions, truncations, and evolutionary implications. J. Bacteriol. 185:3596-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, H. W., P. Green, and Z. Parsell. 1983. Vero cell toxins in Escherichia coli and related bacteria: transfer by phage and conjugation and toxic action in laboratory animals, chickens and pigs. J. Gen. Microbiol. 129:3121-3137. [DOI] [PubMed] [Google Scholar]
- 49.Sonntag, A., R. Prager, M. Bielaszewska, W. Zhang, A. Fruth, H. Tschäpe, and H. Karch. 2004. Phenotypic and genotypic analyses of enterohemorrhagic Escherichia coli O145 strains from patients in Germany. J. Clin. Microbiol. 42:954-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steinberg, K. M., and B. R. Levin. 2007. Grazing protozoa and the evolution of the Escherichia coli O157:H7 Shiga toxin-encoding prophage. Proc. Biol. Sci. 274:1921-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toma, C., E. Martinez Espinosa, T. Song, E. Miliwebsky, I. Chinen, S. Iyoda, M. Iwanaga, and M. Rivas. 2004. Distribution of putative adhesins in different seropathotypes of Shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 42:4937-4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner, P. L., D. W. Acheson, and M. K. Waldor. 2001. Human neutrophils and their products induce Shiga toxin production by enterohemorrhagic Escherichia coli. Infect. Immun. 69:1934-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wagner, P. L., and M. K. Waldor. 2002. Bacteriophage control of bacterial virulence. Infect. Immun. 70:3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wirth, T., D. Falush, R. Lan, F. Colles, P. Mensa, L. H. Wieler, H. Karch, P. R. Reeves, M. C. J. Maiden, H. Ochman, and M. Achtman. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.World Health Organization. 1999. Zoonotic non-O157 Shiga toxin-producing Escherichia coli (STEC), p. 1-30. In Report of a WHO Scientific Working Group meeting, Berlin, Germany, 22-23 June 1998. World Health Organization, Geneva, Switzerland.
- 56.Zhang, W. L., B. Kohler, E. Oswald, L. Beutin, H. Karch, S. Morabito, A. Caprioli, S. Suerbaum, and H. Schmidt. 2002. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J. Clin. Microbiol. 40:4486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, W., A. Mellmann, A. Sonntag, L. Wieler, M. Bielaszewska, H. Tschäpe, H. Karch, and A. W. Friedrich. 2007. Structural and functional differences between disease-associated genes of enterohaemorrhagic Escherichia coli O111. Int. J. Med. Microbiol. 297:17-26. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, X., A. D. McDaniel, L. E. Wolf, G. T. Keusch, M. K. Waldor, and D. W. Acheson. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664-670. [DOI] [PubMed] [Google Scholar]
- 59.Zweifel, C., M. Kaufmann, J. Blanco, and R. Stephan. 2006. Significance of Escherichia coli O157 in sheep at slaughter in Switzerland. Schweiz. Arch. Tierheilkd. 148:289-295. [DOI] [PubMed] [Google Scholar]