Abstract
The alternative sigma factor σB contributes to transcription of stress response and virulence genes in diverse gram-positive bacterial species. The composition and functions of the Listeria monocytogenes and Listeria innocua σB regulons were hypothesized to differ due to virulence differences between these closely related species. Transcript levels in stationary-phase cells and in cells exposed to salt stress were characterized by microarray analyses for both species. In L. monocytogenes, 168 genes were positively regulated by σB; 145 of these genes were preceded by a putative σB consensus promoter. In L. innocua, 64 genes were positively regulated by σB. σB contributed to acid stress survival in log-phase cells for both species but to survival in stationary-phase cells only for L. monocytogenes. In summary, (i) the L. monocytogenes σB regulon includes >140 genes that are both directly and positively regulated by σB, including genes encoding proteins with importance in stress response, virulence, transcriptional regulation, carbohydrate metabolism, and transport; (ii) a number of L. monocytogenes genes encoding flagellar proteins show higher transcript levels in the ΔsigB mutant, and both L. monocytogenes and L. innocua ΔsigB null mutants have increased motility compared to the respective isogenic parent strains, suggesting that σB affects motility and chemotaxis; and (iii) although L. monocytogenes and L. innocua differ in σB-dependent acid stress resistance and have species-specific σB-dependent genes, the L. monocytogenes and L. innocua σB regulons show considerable conservation, with a common set of at least 49 genes that are σB dependent in both species.
A sigma factor is a dissociable protein subunit that directs bacterial RNA polymerase holoenzyme to recognize a promoter sequence upstream of a gene prior to transcription initiation. New associations between alternative sigma factors and core RNA polymerase essentially reprogram promoter recognition specificities of the enzyme in response to changing environmental conditions, thus allowing expression of new sets of target genes appropriate for the conditions. The alternative sigma factor σB, encoded by sigB, has been identified as contributing to the general stress response in several gram-positive bacteria, including Bacillus subtilis (31), Bacillus licheniformis (7), Bacillus anthracis (24), Bacillus cereus (68), Listeria monocytogenes (2, 70), Staphylococcus aureus (75), and Corynebacterium glutamicum (47).
L. monocytogenes is a non-spore-forming facultative intracellular pathogen that causes listeriosis, a serious invasive disease in both animals and humans. To establish a food-borne bacterial infection, L. monocytogenes must have the ability to survive under a variety of stress conditions, including those encountered in a wide range of nonhost environments and food matrices, as well as under rapidly changing conditions encountered during gastrointestinal passage (exposure to organic acids, bile salts, and osmotic gradients) and subsequent stages of infection (e.g., in the intracellular environment). L. monocytogenes σB is activated following exposure to a number of environmental stress conditions (2) and contributes to bacterial survival under acid and oxidative stresses and during carbon starvation (13, 21, 22, 70). In addition, transcription of several virulence genes, including prfA, bsh, inlA, and inlB, is at least partially σB dependent (41-43, 53, 58, 59, 65, 66). Further, a ΔsigB null mutant has reduced invasiveness in human intestinal epithelial cells (42) and reduced virulence in intragastrically inoculated guinea pigs (27). A previous study using a subgenomic microarray comprised of 208 L. monocytogenes-specific probes (41) identified 55 σB-dependent L. monocytogenes genes with ≥1.5-fold-higher transcript levels in the parent strain than in an isogenic sigB null mutant. However, as both the B. subtilis (33) and the S. aureus (4) σB regulons appear to be comprised of at least 150 genes, we hypothesized that the initial efforts to characterize the L. monocytogenes σB regulon (41) likely revealed less than half of the genes comprising the regulon in this species. A more complete identification of the L. monocytogenes σB regulon will enhance our understanding of the contributions of this key regulator to stress response and virulence in this important food-borne pathogen.
The nonpathogenic Listeria innocua is the Listeria species most closely related to L. monocytogenes (56). Previous comparative studies of these two species have already furthered our understanding of L. monocytogenes virulence. For example, while comparisons of these two species revealed a high level of genome synteny (29), identification and characterization of genes found in L. monocytogenes, but absent from L. innocua, have led to the discovery of virulence genes not previously recognized as such in L. monocytogenes (e.g., aut [9]). We thus reasoned that comparative transcriptome analyses of the σB regulons in L. monocytogenes and L. innocua would provide further insight into the role of σB in virulence and stress response. For example, we hypothesized that the organisms may have evolved different σB regulons and/or σB-dependent gene expression patterns to enable appropriate responses to the different sets of stress conditions likely to be encountered by an orally transmitted pathogen (L. monocytogenes) or a nonpathogenic saprophyte (L. innocua). We thus used an L. monocytogenes whole-genome microarray, which was modified to include additional probes targeting putative σB-dependent L. innocua genes, to identify additional members of the L. monocytogenes σB regulon and to initially characterize σB-dependent L. innocua genes, using a previously reported L. monocytogenes ΔsigB strain (70) as well as a newly constructed L. innocua ΔsigB strain. The isogenic parent and ΔsigB strain pairs for both species were also assayed for stress survival to allow comparison of the phenotypic consequences of sigB deletions in L. monocytogenes and L. innocua.
MATERIALS AND METHODS
Media, bacterial strains, and mutant construction.
L. monocytogenes serotype 1/2a strain 10403S (5) and its isogenic sigB null mutant (FSL A1-254 [70]) as well as L. innocua strain FSL C2-008 (73), which was isolated in the year 2000 from smoked fish plant A, as described by Hoffman et al. (35), and its isogenic sigB null mutant (FSL R4-009) were used throughout this study. The L. innocua ΔsigB strain was created by using essentially the same method and primers previously used to create the L. monocytogenes ΔsigB strain (70). Briefly, a sigB fragment with a 297-bp in-frame deletion was created from L. innocua FSL C2-008 using splicing by overlap extension PCR and cloned into the shuttle vector pKSV7. The resulting plasmid (pSR2) was electroporated into L. innocua, and transformants were serially passaged at 40°C in brain heart infusion broth (BHI; Difco, Detroit, MI) containing 7.5 μg of chloramphenicol/ml to select for chromosomal integration of the plasmid. Chromosomal integrants were serially passaged in BHI at 30°C to allow excision of the plasmid to yield a single copy of sigB. Five passages were required for chromosomal integration of the plasmid and for subsequent excision. Retention of the desired ΔsigB allele was confirmed by PCR using primers LmsigB-15 and LmsigB-16 (as previously described [70]) and sequencing of the PCR products using the Big Dye terminator system and an Applied Biosystems 3700 sequencer at the Cornell University BioResource Center (Ithaca, NY).
Stock cultures were stored at −80°C in BHI containing 15% glycerol. Cultures were streaked onto BHI agar and incubated at 37°C for 24 h to obtain isolated colonies for inoculation of overnight cultures. Cells were grown in BHI broth at 37°C with shaking (250 rpm; series 25 incubator shaker; New Brunswick Scientific, Edison, NJ) unless otherwise indicated. Preliminary growth experiments in BHI at 37°C showed no growth pattern differences between parent and ΔsigB strains for either species.
HMM profiling.
Hidden Markov models (HMMs) were built using HMMER version 2.3.2 (http://hmmer.janelia.org) with the default HMMER null model settings. The −10 and −35 sequences for 34 σB-dependent promoters from L. monocytogenes (see Fig. S1 and Table S1 in the supplemental material) were aligned manually and used to train the HMM; separate alignments were created using spacers of 13 to 16 bases between the −35 and −10 regions. Separate models were built for the forward and reverse orientations. The calibrated HMM was used to perform an HMM σB consensus promoter search against the genomic sequences for L. monocytogenes EGD-e and L. innocua CLIP 11262 (29) using the default null model. While the genome sequence for L. monocytogenes 10403S, the parent strain used here, has recently become available (Listeria monocytogenes Sequencing Project [http://www.broad.mit.edu]), the sequence has not been completely closed or annotated and was thus not used for the initial HMM search. PERL scripts were employed to identify coding sequences located between 20 and 350 bases downstream of a predicted σB-dependent promoter. High-scoring putative σB-dependent promoters, as predicted by HMM (e-value, ≤0.01), were identified 20 to 350 nucleotides (nt) upstream of 225 L. monocytogenes open reading frames (ORFs) (see Table S2 in the supplemental material).
A second HMM was built with the HMMER null model settings modified to reflect the GC content of the L. monocytogenes genome (A/T = 0.31, G/C = 0.19). When the p1 parameter was lowered from 0.999 to 0.966, 10 times more putative σB motifs (domains) were returned than had been generated by the default HMMER settings, which enabled an additional comparison with the σB promoter sequences previously identified and with the whole-genome microarray data. All other steps of the HMM search were performed as described above.
The results of these two HMM searches, in conjunction with visual evaluation of sequences upstream of genes identified as σB dependent by the microarray, were used to identify genes or operons preceded by putative σB promoters. Transcriptional terminators as defined by de Hoon et al. (17) were used to delimit operons. Promoters identified by different approaches were classified into one of seven different categories depending on the approach that had been used to identify a given σB-dependent promoter (see Table S3 in the supplemental material for details).
Cell collection and growth for RNA isolation.
L. monocytogenes and L. innocua parent and ΔsigB strains were exposed to different stress conditions before cells were collected for RNA isolation for subsequent microarray experiments. Stress conditions included (i) growth to early stationary phase or (ii) exposure of log-phase (optical density at 600 nm [OD600] of 0.4) cells to salt stress (BHI with 0.3 M NaCl; an osmolarity level similar to that encountered by bacteria in the small intestine [66]). Optical density measurements were performed using a model DU640 spectrophotometer (Beckman, Fullerton, CA). For each strain, an isolated colony was inoculated into 5 ml of BHI broth and grown overnight; diluted 1:100 in 5 ml of fresh, prewarmed BHI; grown to an OD600 of 0.4; diluted 1:100 in 50 ml of fresh, prewarmed BHI (in a 300-ml sidearm flask); and then grown as follows. For early stationary phase, cells were grown to an OD600 of 1.0, followed by incubation for an additional 3 h. A total of 10 ml of stationary-phase culture was collected for RNA isolation. For exposure of log-phase cells to salt stress, cells were grown to an OD600 of 0.4, and then a 10-ml aliquot of these cells was added to an equal volume of prewarmed BHI containing 0.6 M NaCl to yield a final concentration of 0.3 M NaCl. After incubation at 37°C for 10 min with shaking, cells were harvested and used for RNA isolation. Three independent cell collections and RNA isolations were performed for each strain under each experimental condition; experiments were designated as LMO-NaCl and LIN-NaCl (i.e., exposure of L. monocytogenes or L. innocua to 0.3 M NaCl, respectively) or LMO-STAT and LIN-STAT (i.e., growth of L. monocytogenes or L. innocua to early stationary phase, respectively).
RNA isolation.
RNA isolation was performed using the RNAprotect Bacteria reagent and the RNeasy Midi kit (Qiagen, Valencia, CA). Briefly, a 2× volume of RNAprotect Bacteria reagent was added to bacterial cells exposed to a given stress condition, followed by vortexing and incubation at room temperature for 5 min. Cells were then pelleted by centrifugation and stored at −80°C until RNA isolation. For each experimental stress condition, total RNA isolations, DNase treatments, and phenol-chloroform extractions were performed essentially as described in the work of Kazmierczak et al. (41) with the following modifications. Cells were treated with 20 mg of lysozyme/ml (Fisher, Hampton, NH) followed by sonication (Sonicator 3000; Misonix, Farmingdale, NY) on ice at 21 W (three 30-s bursts on ice). DNase treatment of RNA was performed with 40 units RQ1 DNase enzyme (Promega, Madison, WI) in the presence of 400 units RNasin Plus RNase inhibitor (Promega) for 1 h at 37°C. Following the final 100% chloroform extraction, RNA was ethanol precipitated and stored at −80°C overnight. Precipitated RNA was centrifuged, washed in 70% and 100% ethanol, and then resuspended in RNase-free water. RNA was checked for quality by agarose gel electrophoresis and UV spectrophotometer readings at 260 and 280 nm with the NanoDrop-1000 (NanoDrop Technologies, Wilmington, DE).
DNA isolation.
Chromosomal DNA for use as positive-control spots on the microarray was isolated from overnight cultures of L. monocytogenes 10403S and L. innocua FSL C2-008 essentially as described by Flamm et al. (23). DNA was resuspended in printing buffer containing 1× SSC (0.15 M sodium chloride, 0.015 M sodium citrate) with 0.0025% Sarkosyl.
Microarray design and construction.
DNA microarrays were designed to include all ORFs identified in the sequenced genome of L. monocytogenes EGD-e (29). Seventy-mer oligonucleotides representing 2,857 L. monocytogenes EGD-e ORFs were obtained from Qiagen Operon Array-Ready Oligo Sets. EGD-e and L. monocytogenes 10403S both represent the same L. monocytogenes lineage (II), serotype (1/2a), and ribotype (DUP-1039C); therefore, probes designed using the EGD-e genome were expected to hybridize well with 10403S genes (11). Using the unfinished genome sequence for strain 10403S (Listeria monocytogenes Sequencing Project [http://www.broad.mit.edu]), we verified cross-hybridization identities (CHIs) between the EGD-e probes and the target genes in strain 10403S; CHI is the percent identity of a given microarray probe to the corresponding homologous gene in another strain or species. Overall, 2,107, 2,578, and 2,695 of the EGD-e probes showed CHI values of 100, ≥95, and ≥90, respectively; only 45 probes showed CHI values of <90. A total of 117 of the EGD-e-based microarray probes were not detected in the 10403S genome (11). We thus concluded that the array used here would allow comprehensive identification of differentially expressed genes in strain 10403S, with the possibility of some false negatives (i.e., inability to identify genes targeted by a probe with a low CHI or genes present in the 10403S genome and absent in the EGD-e genome).
For each 70-mer, the CHI of the probe to the corresponding ORF in L. innocua CLIP 11262 was provided by Qiagen. CHI values ranged from 32 (low identity to an L. innocua ORF) to 100 (complete identity to an L. innocua ORF). High-scoring putative σB-dependent promoters, as predicted by HMM (e-value, ≤0.01), were identified 20 to 350 nt upstream of 232 L. innocua ORFs (see Table S4 in the supplemental material). With the goal of achieving appropriate hybridization between microarray probes and the transcripts from these putative σB-dependent L. innocua genes, L. innocua-specific microarray probes (with a CHI of 100) were designed for 57 L. innocua ORFs that either had a CHI of <90 (28 genes) or were absent entirely from the L. monocytogenes microarray (29 genes). Probes were designed for an additional four genes with e-values slightly above the cutoff of 0.01. Probe design was performed using the ArrayOligoSelector program (http://arrayoligosel.sourceforge.net/) (6) and the L. innocua CLIP 11262 genome (29) (see Table S5 in the supplemental material). L. innocua probes were synthesized by Operon Biotechnologies (Huntsville, AL).
DNA microarrays were constructed to include 70-mer oligonucleotides representing (i) 2,857 ORFS from L. monocytogenes EGD-e, (ii) the 61 L. innocua-specific probes, and (iii) five yeast ORFS (act1, mfa1, mfa2, ras1, and ste3) as negative controls (74). Oligonucleotides were resuspended in printing buffer (1× SSC plus 0.0025% Sarkosyl) to a final concentration of 25 μM and spotted in duplicate onto UltraGAPs slides (Corning, Corning, NY) using a custom-built XYZ microarrayer at the Cornell University Microarray Core Facility (Ithaca, NY). Serial (1:2) dilutions in printing buffer of L. monocytogenes and L. innocua chromosomal DNA (representing 133.3 to 1.0 ng/μl) were spotted in duplicate on the microarray as positive controls. Salmon sperm DNA (0.1 μg/μl in 1× SSC plus 0.005% Sarkosyl) was also spotted as printing quality controls. Slides were UV cross-linked (Spectrolinker XL-1000 UV cross-linker; Spectronics Corporation, Westbury, NY) with 300 mJ energy and then stored under desiccation.
cDNA labeling and microarray hybridization.
The SuperScript Plus Indirect cDNA Labeling System for DNA Microarrays (Invitrogen, Carlsbad, CA) was used according to the manufacturer's protocol to synthesize and differentially label cDNA from 10 μg total RNA. The QIAquick PCR purification kit (Qiagen) was used to purify cDNA after the overnight, 42°C reverse transcription reaction and the dye coupling reaction (an additional 35% guanidine-HCl wash step was included during purification). For each purification, RNase-free water was used for the final elution step. Purified, labeled cDNA was quantified with UV spectrophotometer readings (Nanodrop Technologies) at wavelengths appropriate to determine the frequency of incorporation, which is defined as the number of labeled nucleotides incorporated per 1,000 nt of cDNA. cDNA samples with frequencies of incorporation between 20 and 50 were used for microarray hybridization. Purified, labeled cDNAs for the parent strain and the corresponding ΔsigB strain (differentially labeled with fluorescent dyes) were combined and dried. The combined target was resuspended in 1× hybridization buffer containing 0.5× formamide, 5× SSC, 0.1% sodium dodecyl sulfate, 0.1 mM dithiothreitol, and 600 μg/ml salmon sperm DNA in diethyl pyrocarbonate-treated water (J. Gilbert, H. Hasseman, R. Cline, and J. Hnath, The Institute for Genomic Research Standard Operating Procedure #M008, “Hybridization of Labeled DNA Probes”). cDNA target samples were vortexed for 1 min and incubated at 95°C for 5 min (repeated twice), before a brief centrifugation to cool the sample.
Slide blocking, hybridization, and posthybridization washing were performed as previously described (49). Slides were centrifuged to dry and scanned with a GenePix 4000B Scanner (Molecular Devices, Sunnyvale, CA) at the Microarray Core Facility using the following general scanning parameters: pixel size, 10; focus position, 20; lines to average, 1; laser power, 100%. The auto-photomultiplier tube gain function was used to scan the slides with an acceptable photomultiplier tube gain range of 400 to 900.
Microarray replicates and statistical analysis.
Raw TIFF images were automatically gridded and analyzed using the GenePix Pro 6.0 software. Spots flagged from the data and removed from the subsequent analysis included empty spots, spots saturated in both channels, and spots of poor morphology. Data were preprocessed (background corrected and normalized) using the LIMMA package (61) available from the BioConductor software project for the R programming environment (R version 2.2.1) (28).
To achieve robust ratios for low-intensity spots, “normexp” background correction was employed using the LIMMA package (with offset of 100). Print-tip loess normalization was performed to account for within-array spatial bias and intensity bias. To assist with the within-array normalization process, control spots known a priori to be nondifferentially expressed were given increased weight during normalization. Control spots were chosen to span the entire range of intensities (A values in a minus-versus-add plot of the data) and included genomic DNA spots (with dilutions spanning from middle to high intensities) and printing and negative controls (to account for low intensities). To enable comparison of results across arrays, scale normalization was performed.
The LIMMA package was also used for differential expression analysis (62). Briefly, a linear model was fitted to the normalized data, followed by empirical Bayes smoothing to calculate moderated t-statistics and B-statistics. To account for duplicate spots, the “duplicateCorrelation” function was used (63). P values were adjusted for multiple comparisons by controlling for the false discovery rate. Genes with an adjusted P value of <0.05 were considered statistically significant, and a change of ≥2.0-fold was used as a cutoff for identification of differentially expressed genes. Genes were considered to be positively regulated by σB if transcript levels were significantly and at least 2.0-fold higher in the parent strain than in the ΔsigB strain (represented as positive changes in Table S6 in the supplemental material). Genes were considered to be negatively regulated in a σB-dependent manner if transcript levels were significantly and at least 2.0-fold lower in the parent strain than in the ΔsigB strain (represented as negative changes in Table S7 in the supplemental material).
TaqMan quantitative reverse transcriptase-PCR (qRT-PCR).
TaqMan probe and primer sets for L. monocytogenes rpoB, gap, opuCA, gadA, and fri have been reported previously (10, 40, 59, 66); TaqMan MGB (minor groove binder) probe and primer sets for L. innocua lin0285 (rpoB), lin0289, lin0942 (fri), lin1467 (opuCA), lin1992 (cspD), lin2528 (gadA), lin2553 (gap), and lin2891 and for L. monocytogenes lmo0265, lmo1879 (cspD), and lmo2748 (see Table S8 in the supplemental material) were designed using Primer Express 1.0 software (Applied Biosystems, Foster City, CA).
qRT-PCR was performed using TaqMan One-Step RT-PCR Master Mix Reagent, Multiscribe RT, and the ABI Prism 7000 Sequence Detection System (all from Applied Biosystems) as described by Sue et al. (66). Each qRT-PCR was run in duplicate on each reaction plate, and RT-negative reactions (excluding the Multiscribe RT) were run in parallel to account for any possible genomic DNA contamination in each qRT-PCR. Standard curves for each target gene were included for each assay to allow for absolute quantification of cDNA levels, and data were analyzed using the ABI Prism 7000 Sequence Detection System software as previously described (66). Normalization and log transformation were performed as described by Kazmierczak et al. (40). As microarray analyses indicated that transcript levels for L. innocua gap were lower in the ΔsigB strain, providing evidence of σB-dependent regulation for gap in the tested strain, statistical analyses of transcript levels for L. innocua genes were also repeated using transcript levels normalized to rpoB transcript levels only. With the exception of transcript levels for L. innocua cspD (see Results and Discussion for details), normalization of L. innocua gene transcript levels to either rpoB alone or both rpoB and gap identified the same differences as significant.
The same RNA that had been isolated for the microarray experiments was also used in the qRT-PCR assays, with the exception of one LIN-NaCl and one LIN-STAT replicate. RNA was isolated from an additional set of cells grown under each of these conditions to provide sufficient RNA for three replicate qRT-PCR assays.
Acid stress and ethanol stress experiments.
L. monocytogenes and L. innocua parent and ΔsigB cultures were characterized in a series of stress survival assays, including (i) exposure to HCl-acidified BHI broth (pH 2.5) (21), (ii) exposure to synthetic gastric fluid (SGF; pH 2.5) (15), and (iii) exposure to BHI plus 16.5% (vol/vol) ethanol (21, 48). Exposure to BHI-HCl and SGF was performed using both log- and early-stationary-phase cells, while ethanol exposure was performed only with early-stationary-phase cells. Log-phase cells (i.e., OD600 of 0.4) were grown as detailed above for RNA isolation experiments. For early-stationary-phase cells, overnight cultures were subcultured 1:100 into 10 ml fresh BHI and grown to an OD600 of 0.8 for 1 h (a condition previously shown to activate σB [65]); this growth protocol was used to allow direct comparisons of results with previously reported survival experiments (13, 21). For acid exposure, 1 ml of culture was pelleted and resuspended in 1 ml of either BHI-HCl (pH 2.5) or SGF (pH 2.5), followed by incubation at 37°C with shaking for 10 min or 60 min for log- or early-stationary-phase cells, respectively. For ethanol survival, early-stationary-phase cells were pelleted and resuspended in 1 ml of BHI plus 16.5% (vol/vol) ethanol, followed by incubation at 37°C with shaking for 120 min. For all experiments, Listeria numbers were enumerated by plating appropriate serial dilutions in phosphate-buffered saline on BHI agar (in duplicate); enumeration was performed immediately prior to stress exposure and after stress exposure for the incubation times specified. Plates were incubated at 37°C for 48 h.
Swarming assay.
Swarming abilities of L. monocytogenes and L. innocua strains were evaluated on semisoft agar; the L. monocytogenes ΔflaA strain (55) was included as a negative control. Strains were initially grown for 24 h at 37°C on BHI agar and BHI agar containing 0.3 M NaCl. Colonies from BHI and BHI plus 0.3 M NaCl were used to stab-inoculate BHI semisoft agar (0.4%) and BHI plus 0.3 M NaCl semisoft agar (0.4%), respectively. Swarming ability was assessed by measuring colony area using SigmaScan Pro 5.0 (SPSS Inc., Chicago, IL) for each strain after incubation at 30°C and 37°C for 72 h. Growth area for each mutant strain was normalized to the growth area of the parent strain, which was set at 100%.
Statistical analyses.
Statistical analyses were performed using SAS Version 9.1 (SAS Institute Inc., Cary, NC). Two-sided two-sample t tests were used for analysis of the qRT-PCR data, while a two-sided one-sample t test was used for analysis of the swarming data. Statistical analysis of the stress survival data was performed using a general linear model with Tukey's multiple comparison analysis.
Microarray data accession number.
Raw and normalized microarray data in MIAME format are available at the NCBI Gene Expression Omnibus (GEO) data repository (20) under accession number GSE7492.
RESULTS AND DISCUSSION
Microarray, qRT-PCR, HMM, and phenotypic characterization of L. monocytogenes and L. innocua ΔsigB strains provided data indicating that (i) the L. monocytogenes σB regulon includes >140 genes that are both directly and positively regulated by σB, including genes encoding proteins with importance in stress response, virulence, transcriptional regulation, metabolism, and transport; (ii) a number of L. monocytogenes genes encoding flagellar proteins show higher transcript levels in a ΔsigB mutant, suggesting indirect effects of σB on motility and chemotaxis; and (iii) even though the L. monocytogenes and L. innocua σB regulons show considerable conservation with a core of at least 49 genes that appear to be σB dependent in both Listeria spp., L. monocytogenes and L. innocua both have species-specific σB-dependent genes and show differences in σB-dependent stress resistance phenotypes (i.e., acid stress resistance).
Microarray analyses identify >140 L. monocytogenes genes directly and positively regulated by σB.
Whole-genome microarrays were used in competitive hybridization experiments with RNA isolated from L. monocytogenes 10403S and an isogenic ΔsigB strain to define the σB regulon under two conditions that result in high σB activity, including (i) exposure of log-phase cells to salt stress (65) and (ii) entry into stationary phase (2, 65). Statistical analysis was performed separately for the two data sets. Genes were identified as putatively σB regulated under at least one of these conditions if they met both of the following criteria: (i) an adjusted P value of <0.05 and (ii) an at least 2.0-fold difference in mRNA transcript levels between the ΔsigB mutant and parent strain. Using these criteria, 168 genes showed ≥2.0-fold-higher transcript levels in the L. monocytogenes parent than in the ΔsigB mutant (Table 1; see also Table S6 in the supplemental material); these genes are considered to be positively regulated by σB. A total of 128 genes showed ≥2.0-fold-higher transcript levels in the L. monocytogenes ΔsigB mutant than in the parent strain (see Table S7 in the supplemental material); these genes are considered to be negatively regulated by σB.
TABLE 1.
Functional categories of genes identified by microarray analyses as positively regulated by σB
| Category | No. of σB-dependent genes with or without putative σB-dependent promotera,b
|
|||
|---|---|---|---|---|
|
L. monocytogenes (n = 168)
|
L. innocua (n = 64)
|
|||
| With promoter | Without promoter | With promoter | Without promoter | |
| Stress | 11 (clpC, ctc, sepA) | 1 (lmo2064) | 7 (clpC, ctc, sepA) | 1 (cspDc) |
| Virulence and virulence associated | 5 (bsh, hfq, inlAB) | 1 (hfq) | ||
| Transcriptional regulation | 7 (rsbV, hrcA) | 3 (lmo0402, lmo0445) | 5 (rsbV, rpoD) | 1 (lin0615) |
| Transport and transport systems | 21 (gbuA, opuCA) | 5 (lmo0607, lmo0653) | 6 (opuCA) | |
| Metabolism | 37 (ldh, lmo2571) | 8 (lmo0043, lmo0913) | 14 (ldh, lin2716) | 1 (lin0913) |
| DNA metabolism and transport | 2 (lmo0996, lmo1606) | 1 (lin1647) | ||
| Protein synthesis and modification | 3 (lmo1698, lmo2511) | 1 (lin2655) | ||
| Cell envelope and cellular processes | 6 (lmo0610, lmo0880) | 1 (lmo1291) | 3 (lin0372, lin0879) | |
| Unknown and hypothetical | 52 (lmo0953) | 6 (lmo2671) | 19 (lin0952) | 4 (lin0483) |
Numbers of genes in a given category are indicated; examples of genes are listed in parentheses; boldface indicates genes identified as σB dependent in both L. monocytogenes and L. innocua.
Putative σB-dependent promoters were identified by HMM or visual analyses as detailed in Table S3 in the supplemental material.
Not confirmed as σB dependent by qRT-PCR.
Among the 168 genes identified as positively regulated by σB through microarray analysis, 83 genes (49%) had an HMM-predicted putative σB-dependent promoter sequence either directly upstream of a specified ORF or upstream of the first gene of an operon, suggesting direct σB regulation of these genes. As the initial HMM-based approach used stringent criteria for promoter identification (i.e., e-value of ≤0.01 with distance from start codon of between 20 and 350 nt [see Table S2 in the supplemental material]), the upstream regions of the remaining 85 σB-dependent genes were reevaluated for evidence of a σB-dependent promoter sequence using both visual inspection and HMM-identified putative promoters with higher e-values and less-stringent distance criteria. These reevaluations identified a putative σB-dependent promoter upstream of an additional 62 genes to yield a total of 145 genes that appear to have a σB-dependent promoter and thus are predicted to be directly regulated by σB (see Tables S3 and S6 in the supplemental material).
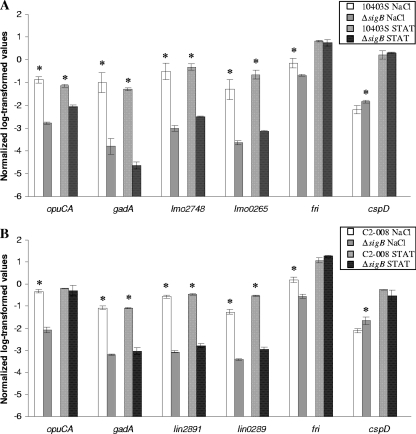
A total of 72 genes (including 49 preceded by a σB-dependent promoter directly upstream either of a specified ORF or of the first gene of an operon) were positively regulated by σB in both salt-stressed and stationary-phase cells (see Fig. S2 in the supplemental material). A total of 58 and 38 genes showed σB-dependent transcript levels only in salt-stressed cells (e.g., the inlAB operon) or only in stationary-phase cells (e.g., the lmo0398-0400 operon, encoding a fructose-specific phosphotransferase system [PTS]), respectively (see Table S6 in the supplemental material). Interestingly, among the 26 σB-dependent L. monocytogenes genes related to transport, 11 genes were induced in a σB-dependent manner only in salt-stressed cells, demonstrating the importance of σB in facilitating transport, e.g., of compatible solutes, during salt and/or osmotic stress. qRT-PCR of fri (lmo0943/lin0942), which encodes Fri, a nonheme iron-binding ferritin, confirmed that fri is positively regulated by σB in salt-stressed, but not stationary-phase, L. monocytogenes and L. innocua (Fig. 1). This is consistent with the microarray data, which showed higher transcript levels in both the L. monocytogenes and the L. innocua parent strain exposed to salt (compared to the ΔsigB strain; adjusted P < 0.05 for L. innocua and adjusted P < 0.1 for L. monocytogenes), while showing no differences in stationary-phase cells between the two species (see Table S6 in the supplemental material). qRT-PCR thus confirmed that some genes show σB-dependent transcription only under some stress conditions.
FIG. 1.
qRT-PCR confirmation of selected genes identified by microarray data as σB dependent in L. monocytogenes 10403S (A) and in L. innocua FSL C2-008 (B). qRT-PCRs were designed to be specific for the six selected L. monocytogenes and L. innocua genes (see Table S8 in the supplemental material); correlation plots for microarray and qRT-PCR results are provided as Fig. S8 in the supplemental material. The six genes were selected to represent genes with homologues in both species; homologous genes are presented in the same order in each figure, e.g., lmo2748 and lin2891 are homologous. Transcript levels for the test genes were normalized to the geometric mean of the transcript levels for housekeeping genes rpoB and gap; an asterisk indicates that normalized transcript levels for the parent and ΔsigB strain differed significantly for cells exposed to a given stress condition (salt stress or stationary phase). As microarray analyses indicated that transcript levels for L. innocua gap were lower in the ΔsigB strain, statistical analyses of transcript levels for L. innocua genes were also repeated using transcript levels normalized to rpoB only; except for cspD (see text), these statistical analyses identified the same differences as significant as the analyses based on transcript levels normalized to the geometric mean of rpoB and gap transcript levels.
The genes positively regulated by σB were classified into nine functional categories (Table 1; see also Table S6 in the supplemental material). As expected, a number of genes related to stress response displayed σB-dependent transcription including the general stress response genes ctc and lmo1601, as well as ltrC, which is involved in cold stress response (76, 77), and gadA, which encodes a glutamate decarboxylase important for acid resistance (14). Several virulence and virulence-associated genes also showed σB-dependent expression including bsh (encoding the bile salt hydrolase), hfq (encoding an RNA-binding protein), and various internalin genes (e.g., the inlAB operon [see Fig. S3 in the supplemental material] and inlD). While the contributions of σB to the gastrointestinal stages of infection have been well documented (27), σB-dependent genes also appear to contribute to intracellular infection by L. monocytogenes. For example, of the 26 σB-dependent genes previously identified to be upregulated in either intravacuolar or intracytosolical L. monocytogenes (12), 23 were confirmed as σB dependent in our microarray analyses (see Table S9 in the supplemental material). Three members of the universal stress protein (Usp) family, found to be upregulated intracellularly by Chatterjee et al. (12), were also found to be σB dependent, including lmo2748, which was also confirmed as σB dependent by qRT-PCR (Fig. 1; see also Table S6 in the supplemental material). Further analyses on the overlaps between the σB regulon identified here and L. monocytogenes genes that are upregulated during intracellular replication and survival (e.g., references 12 and 38) will likely shed additional insight on the contributions of σB to transcriptional regulation during infection.
The 168 genes positively regulated by σB included 26 operons with more than one gene identified as σB-dependent (these operons contained from two to eight genes identified as σB dependent; see Fig. S3 and S4 in the supplemental material), including a number of operons involved with carbohydrate metabolism. L. monocytogenes genes and operons positively regulated by σB and involved in carbohydrate metabolism include a mannose-specific PTS operon (lmo0784 to lmo0781; lmo0784 was previously identified as σB dependent), a fructose-specific PTS (lmo0398 to lmo0400, not previously reported as σB dependent), and an operon (lmo0348 to lmo0341) that includes genes encoding enzymes in the pentose phosphate pathway, as well as a gene involved in galactose metabolism (lmo0539). In addition, two two-gene operons encoding dihydroxyacteone kinases were also identified as positively regulated by σB (see Fig. S4 and Table S6 in the supplemental material). Except for the fructose-specific PTS operon, all other genes mentioned above were preceded by a putative σB-dependent promoter, suggesting direct regulation by σB. σB-dependent transcription of genes involved in carbohydrate metabolism suggests that σB may be important for energy metabolism, most likely during exposure to energy stress conditions, consistent with the observation that B. subtilis σB is activated during energy stress (8, 69).
Interestingly, seven genes encoding transcriptional regulators were found to be positively regulated by σB, including lmo2460, which encodes CggR, a transcriptional regulator highly similar to the B. subtilis central glycolytic gene regulator (18); lmo2460 is preceded by a σB consensus promoter. hrcA, which encodes a negative regulator of chaperone proteins and is involved in resistance to heat stress (32) also showed lower transcript levels in the ΔsigB strain and is preceded by a σB-dependent promoter. In addition, the last two genes in the four-gene clpC operon (ctsR-mcsA-mcsB-clpC), which has a σB-dependent promoter preceding mcsA, also showed evidence of σB-dependent transcription in salt-stressed cells. HrcA and CtsR both regulate transcription of heat shock genes in L. monocytogenes (25, 32, 39, 54). Regulatory interactions between L. monocytogenes σB and PrfA, a transcriptional regulator of virulence gene transcription, have been reported previously (45), including coregulation of genes by both proteins (50, 52) and σB-dependent transcription of prfA (53, 58, 59). Our microarray data suggest additional important contributions of σB to multiple L. monocytogenes regulatory networks based on evidence of interactions between L. monocytogenes σB and other regulators.
We also confirmed that the L. monocytogenes σB regulon includes a number of stress response genes (e.g., gadA, ctc, and clpC) and virulence genes (e.g., inlAB, inlD, bsh, and hfq), as well as genes encoding transporters and cell surface proteins. The size of the L. monocytogenes σB regulon identified by our microarray analyses is consistent with the σB regulons identified in other bacteria including B. subtilis (127 genes) (57) and S. aureus (198 genes) (4). Previously only ∼55 L. monocytogenes genes had been classified as σB dependent, predominantly through use of a subgenomic microarray targeting 208 L. monocytogenes genes (41). Our microarray results confirmed 45 of the 55 L. monocytogenes genes identified as σB dependent by Kazmierczak et al. (41). Six of the 10 genes not confirmed by the present study (lmo0524, lmo1539, lmo2389, lmo2399, pdhA, and qoxA) had statistically significant differences (adjusted P < 0.05) in transcript levels between the parent and ΔsigB strain in at least one data set, but the differences did not meet our minimum 2.0-fold criterion. While we conclude that we have identified the majority of the L. monocytogenes σB regulon, further studies using cells exposed to different stress conditions, followed by qRT-PCR and promoter mapping strategies, will likely reveal some additional members of the L. monocytogenes σB regulon. Genes identified here as showing significant evidence for σB dependence with changes below the 2.0-fold cutoff represent likely candidates for additional members of the σB regulon.
A number of L. monocytogenes genes encoding flagellar proteins show higher transcript levels in a ΔsigB mutant, suggesting indirect effects of σB on motility and chemotaxis.
While σB clearly is important as a positive regulator of transcription in Listeria and in a number of other gram-positive bacteria, we also identified 128 genes that showed higher transcript levels in the L. monocytogenes ΔsigB strain (see Table S7 in the supplemental material), suggesting that σB also contributes to negative regulation of gene transcription, likely through indirect means, such as through positive regulation of a repressor. Interestingly, hly (lmo0202) showed significantly higher transcript levels in the L. monocytogenes ΔsigB strain, suggesting indirect negative regulation of this virulence gene by σB; these findings are consistent with previously reported results that showed increased hemolysis activity in the L. monocytogenes ΔsigB strain compared to that in the L. monocytogenes 10403S parent strain (13, 53). Similar to our findings, others have also reported genes that appear to be indirectly repressed by σB, including 53 genes in S. aureus (4).
A total of 91 genes showed lower transcript levels in the parent strain only in salt-stressed cells (LMO-NaCl), including 21 genes encoding different ribosomal subunit proteins; 11 of these genes are located in a 19-gene locus (lmo2614 to lmo2632). These data suggest that σB indirectly controls transcription of L. monocytogenes genes involved in translation, consistent with findings that rpsJ, which encodes the ribosomal protein S10, was found to be negatively regulated by σB in B. subtilis (57).
Interestingly, 16 genes located in a 28-gene locus (lmo0691 to lmo0718) that encodes flagellar structural components also showed higher transcript levels in the ΔsigB strain in cells exposed to NaCl; this locus is one of two large operons that encode the flagellar apparatus (72) (see Fig. S5 in the supplemental material). Specific comparisons between the previously reported L. monocytogenes DegU regulon (72) and the σB-repressed L. monocytogenes genes reported here identified a total of 23 genes in the DegU regulon that are repressed by σB. When the cutoff for genes differentially regulated by σB was relaxed from a difference of ≥2.0-fold to a difference of >1.3-fold (still with an adjusted P < 0.05), 41 genes determined to be positively regulated by the two-component response regulator DegU at 24°C (including 18 flagellum-specific genes and three chemotaxis-specific genes) (72), were also found to be negatively regulated by σB in salt-stressed cells. lmo1699 (encoding a protein similar to methyl-accepting chemotaxis proteins) and lmo1700 (encoding an unknown protein), which represent an operon within the DegU regulon, also were found to be negatively regulated by σB in salt-stressed cells; these two genes displayed the greatest differences between the ΔsigB strain and the parent strain (−4.7 and −5.3 for lmo1699 and lmo1700, respectively). Phenotypic characterization of swarming ability of the L. monocytogenes and L. innocua ΔsigB strains supported the relevance of the transcriptome analyses and determined that in bacteria grown at 30°C in BHI with and without 0.3 M NaCl, the ΔsigB strain consistently showed small, but statistically significant, increases in swarming compared to the parent strain (see Fig. S6 in the supplemental material). When bacteria were grown at 37°C (a temperature that permits limited swarming in 10403S but no swarming in EGD-e [30]), the L. innocua ΔsigB strain still showed small, but statistically significant, increases in swarming compared to the isogenic parent strain (see Fig. S6 in the supplemental material), despite limited microarray-based evidence for significantly higher transcript levels for the flagellar genes in the L. innocua ΔsigB. This apparent contradiction is likely explained by the observation that these genes, which showed <2.0-fold difference, were targeted by probes with relatively low CHI values (see Fig. S5 in the supplemental material).
Overall, our data indicate that σB negatively regulates genes encoding proteins related to motility and chemotaxis in response to salt stress and that DegU and σB coregulate a number of genes, in particular genes related to motility and chemotaxis. Specifically, our data provide support for the hypothesis that σB controls transcription of a gene encoding a protein that suppresses transcription of selected motility and chemotaxis genes, although a specific gene(s) responsible for this effect remains to be identified. For example, microarray data did not show effects of the sigB deletion on transcript levels for mogR, which encodes a transcriptional repressor of flagellar and motility genes (30). While hrcA, which encodes a negative regulator, is positively regulated by σB, microarray experiments showed no evidence that HrcA regulates flagellar genes (37). Previous studies have shown that an L. monocytogenes ΔdegU strain as well as a ΔmogR strain has attenuated virulence in mouse models of infection (30, 44, 71). As an L. monocytogenes ΔsigB null mutant also showed attenuated virulence in a guinea pig model beyond that caused by reduced transcription of the σB-dependent inlA (27), it is possible that interactions between DegU, MogR, and σB are important for transcription of virulence genes; experiments using appropriate sigB, degU, and mogR null mutants will be required to further test this hypothesis. In B. subtilis, the DegS/DegU two-component system has been shown to sense salt stress (16, 46, 64) and phosphorylated DegU is thought to repress transcription of motility genes through repression of sigD, which encodes the flagellum-specific sigma factor σD in B. subtilis (1, 67), which is absent in L. monocytogenes. These data support the idea that interactions among regulators contribute to regulation of flagellar proteins in different bacteria. Our results extend the list of possible regulators of flagellar protein expression in L. monocytogenes to include σB.
The L. monocytogenes and L. innocua σB regulons show considerable conservation with a core of at least 49 genes that appear to be σB dependent in both species.
Initial HMM analysis using the L. innocua CLIP 11262 genome (29) identified 232 putative L. innocua σB promoter sequences with e-values of ≤0.01 located between 20 and 350 nt upstream of a start codon of a predicted ORF (see Table S4 in the supplemental material); 123 of these ORFs had homologues in both L. monocytogenes and L. innocua and were preceded by a putative σB-dependent promoter in both species. Interestingly, a total of 29 L. innocua ORFs with no homologue in L. monocytogenes (as determined by the ListiList server [http://genolist.pasteur.fr/ListiList/]) were predicted to have upstream σB-dependent promoters. Similarly, a total of 26 L. monocytogenes ORFs with no homologue in L. innocua (as determined by the ListiList server) were predicted to have upstream σB-dependent promoters (e.g., prfA, sepA, and bsh), providing preliminary evidence of diversification of the σB regulons between the two species. To enable sufficient hybridization between microarray probes and the transcripts from putative σB-dependent L. innocua genes, additional L. innocua-specific probes were designed to target genes that were predicted to be σB dependent by HMM but either (i) did not have a homologue in L. monocytogenes (29 genes) or (ii) were not expected to hybridize with the probe targeting the L. monocytogenes homologue due to sequence divergence, as the L. monocytogenes-specific probe showed a CHI of <90 with the L. innocua target gene based on the genome sequence for L. innocua CLIP 11262 (28 genes; see Materials and Methods for further explanation).
Application of our modified L. monocytogenes whole-genome array identified 64 genes that displayed significantly, and at least 2.0-fold-, higher expression in the L. innocua parent than in the isogenic ΔsigB null mutant under at least one of the two stress conditions (LIN-NaCl or LIN-STAT; see Table S6 in the supplemental material). Among the 64 genes positively regulated by σB in L. innocua, 28 genes showed σB-dependent transcription under both experimental conditions; 28 and 8 genes had higher transcript levels in the parent strain only in salt-stressed cells (e.g., ctc and fri) or only in stationary-phase cells (e.g., pdhA and pdp), respectively (see Fig. S2 and Table S6 in the supplemental material). Among these 64 genes, 42 were preceded by a putative σB-dependent promoter as identified by HMM (e-value of ≤0.01 with distance of 20 to 350 nt [see Table S6 in the supplemental material]). Reevaluation of the upstream regions of the remaining 22 genes with relaxed HMM criteria for σB promoter identification (i.e., higher e-values and less stringent distance requirement; see Table S3 in the supplemental material) identified an additional 13 genes with putative upstream σB-dependent promoters. Visual evaluation identified putative σB-dependent promoters upstream of an additional two genes to yield a total of at least 57 genes that appear to be directly positively regulated by σB in L. innocua (see Table S6 in the supplemental material).
Our approach of applying L. monocytogenes microarrays to L. innocua only partially identified the L. innocua σB regulon, since transcripts for some σB-dependent L. innocua genes were unlikely to have been detected due to low CHI of a given L. innocua gene for the L. monocytogenes-specific microarray probe. Of the 1,030 L. innocua genes targeted with a probe with a CHI of ≥95, a total of 6% were identified as σB dependent by microarray analyses, while only 0.3% of L. innocua genes targeted with a probe with a CHI of <95 were identified as σB dependent. Genes targeted with a highly homologous probe (CHI of ≥95) were significantly more likely to be identified as σB dependent (P < 0.001; χ2 test of independence). While our approach thus may have led to a number of false negatives in L. innocua (i.e., σB-dependent L. innocua genes were not identified as such), it likely generated very few false positives (i.e., non-σB-dependent L. innocua genes identified as σB dependent). Consequently, the σB-dependent L. innocua genes identified here should be considered a partial σB regulon with additional σB-dependent L. innocua genes likely to be identified in future experiments with L. innocua-specific microarrays.
The 64 L. innocua genes positively regulated by σB included nine operons with more than one gene identified as σB dependent; eight of these operons also showed clear evidence of σB dependence in L. monocytogenes (see Fig. S4 in the supplemental material). While the remaining operon (lin2550 to lin2552) was σB dependent in L. innocua, only one gene from the operon (pgm, homologous to lin2550) showed σB dependence in L. monocytogenes. This operon is situated in a six-gene locus (lin2549 to lin2554) that encodes proteins related to glucose metabolism (see Fig. S4 in the supplemental material). The first gene in the locus (lin2554 or lmo2460) is preceded by a predicted σB promoter in both species, supporting the idea that this operon is transcribed by σB in both species.
By comparing the 64 σB-dependent L. innocua genes with the L. monocytogenes σB regulon, we were able to identify a total of 49 σB-dependent genes common to both L. monocytogenes and L. innocua (representing genes that show higher transcript levels in the parent strain than in the ΔsigB strain under at least one stress condition in both species) (Table 2). A total of 20 genes (e.g., hfq and ldh) were found to be positively regulated by σB under both stress conditions in both L. monocytogenes and L. innocua (see Fig. S2 in the supplemental material). While the 49 conserved σB-dependent genes include a number of genes encoding hypothetical proteins, which we propose are important candidates for functionally important σB-dependent genes, they also include a number of genes encoding stress response proteins, transporters, and metabolic enzymes. For example, the homologues lin0674 and lmo0669 (which both encode an oxidoreductase [see Fig. S4 in the supplemental material]) as well as lin2332 and lmo2230 (which both encode an arsenate reductase) were σB dependent in both species and showed considerably lower transcript levels in the ΔsigB mutant in both species (Table 2). Some of the genes found to be σB dependent in both L. monocytogenes and L. innocua also have homologues previously found to be σB dependent in B. subtilis (e.g., ctc and yvyD) (57), S. aureus (e.g., N315-SAS0202, encoding a phophoglycerate mutase; N315-SA0163, encoding a cation efflux transport protein) (4), or both (e.g., ywmG/csbD; yhxD/ydaD, encoding a short-chain dehydrogenase or oxidoreductase) (4, 34, 57), indicating conservation of certain σB-dependent stress response mechanisms across gram-positive genera. Interestingly though, early-stationary-phase cells for both L. monocytogenes and L. innocua ΔsigB strains did not show reduced ability to survive ethanol stress (compared to the respective parent strains [see Fig. S7 in the supplemental material]); while bacterial numbers for all strains were reduced by ∼5 logs, there were no significant (P > 0.05) differences among the four strains. While this is consistent with previous observations that ethanol resistance in stationary-phase L. monocytogenes cells is largely σB independent (21), ethanol resistance in B. subtilis has been shown to be σB dependent (36), supporting some diversification of σB-dependent stress response functions between the closely related genera Bacillus and Listeria.
TABLE 2.
Genes identified by microarray analyses as positively regulated by σB in both L. monocytogenes and L. innocua
| Function and L. monocytogenesa/L. innocuab name (gene name) | Fold change (parent/ΔsigB) forc:
|
σB promoter ford:
|
EGD-e probe CHI fore:
|
Description of proteinf | |||||
|---|---|---|---|---|---|---|---|---|---|
| LMO-NaCl | LMO-STAT | LIN-NaCl | LIN-STAT | L. monocytogenes | L. innocua | L. monocytogenes 10403S | L. innocua CLIP 11262 | ||
| Stress | |||||||||
| lmo0211/lin0243 (ctc) | 1.4 | 2.1 | 2.4 | 1.4 (NS) | + | + | 97 | 100 | Similar to B. subtilis general stress protein |
| lmo0232/lin0264 (clpC) | 2.5 | −1.2 (NS) | 2.2 | 1.1 (NS) | + | + | 100 | 98 | Endopeptidase Clp ATP-binding chain C |
| lmo0669/lin0674 | 34.4 | 34.4 | 19.1 | 15.1 | + | + | 98 | 98 | Similar to oxidoreductase |
| lmo1433/lin1472g | 2.7 | 2.6 | 7.3 | 3.4 (NS) | + | + | 100 | 100 (LI) | Similar to glutathione reductase |
| lmo1601/lin1642 | 2.5 | 4.2 | 4.4 | 9.7 | + | + | 100 | 98 | Similar to general stress protein |
| lmo2230/lin2332 | 33.9 | 21.8 | 11.6 | 37.4 | + | + | 100 | 98 | Similar to arsenate reductase |
| Virulence and virulence associated | |||||||||
| lmo1295/lin1333 (hfq) | 2.5 | 3.9 | 4.2 | 8.8 | + | + | 100 | 98 | Similar to host factor 1 protein |
| Transcriptional regulation | |||||||||
| lmo0893/lin0892 (rsbV) | 2.2 | 1.5 | 4.2 | 3.5 | + | + | 100 | 98 | Anti-anti-sigma factor (antagonist of RsbW) |
| lmo0894/lin0893 (rsbW) | 2.4 | 1.3 | 4.3 | 3.4 (NS) | + | + | 100 | 100 | σB activity negative regulator RsbW |
| lmo0958/lin0957 | 2.0 | 1.5 (NS) | 2.3 | 5.5 (NS) | + | + | 100 | 98 | Similar to transcription regulator (GntR family) |
| Transporter | |||||||||
| lmo0169/lin0212 | 2.8 | 2.1 | 2.1 | 1.2 (NS) | + | + | 100 | 98 | Similar to a glucose uptake protein |
| lmo0654/lin0657g | 2.2 | 2.1 | 15.4 | 39.7 | + | + | 100 | 100 (LI) | Similar to ABC transporter, ATP-binding protein |
| lmo0782/lin0775 | 10.3 | 12.6 | 5.2 | 8.5 | + | + | 100 | 98 | Similar to mannose-specific PTS component IIC |
| lmo1425/lin1464 (opuCD) | 7.0 | 1.8 | 5.6 | 1.6 (NS) | + | + | 100 | 97 | Similar to betaine/carnitine/choline ABC transporter (membrane protein) |
| lmo1428/lin1467 (opuCA) | 10.1 | 2.9 | 13.6 | 2.4 (NS) | + | + | 98 | 98 | Similar to glycine betaine/carnitine/choline ABC transporter (ATP-binding protein) |
| Metabolism | |||||||||
| lmo0210/lin0242 (ldh) | 2.0 | 3.1 | 2.0 | 3.7 | + | + | 100 | 98 | Similar to l-lactate dehydrogenase |
| lmo0231/lin0263 (mcsB) | 2.0 | 1.1 (NS) | 2.4 | 1.3 (NS) | + | + | 100 | 98 | Similar to arginine kinase |
| lmo0539/lin0543 | 18.8 | 13.7 | 9.6 | 14.3 | + | + | 98 | 98 | Similar to tagatose-1,6-diphosphate aldolase |
| lmo0913/lin0913 | 9.7 | 5.8 | 3.7 | 5.2 | − | − | 100 | 91 | Similar to succinate semialdehyde dehydrogenase |
| lmo1883/lin1996 | 2.5 | 12.0 | 1.9 | 10.2 | + | + | 100 | 100 | Similar to chitinases |
| lmo2456/lin2550 (pgm) | 2.5 | −1.1 (NS) | 2.0 | 2.4 (NS) | + | + | 98 | 98 | Phosphoglycerate mutase |
| lmo2571/lin2716 | 6.2 | 4.0 | 4.3 | 8.4 | + | + | 100 | 100 | Similar to nicotinamidase |
| lmo2674/lin2821 | 1.8 | 2.7 | 1.6 | 4.3 | + | + | 92 | 92 | Similar to ribose 5-phosphate epimerase |
| lmo2695/lin2843 | 7.0 | 1.3 (NS) | 8.0 | 1.9 (NS) | + | + | 98 | 98 | Similar to dihydroxyacetone kinase |
| lmo2696/lin2844 | 6.5 | 1.5 | 6.9 | 2.0 (NS) | + | + | 92 | 91 | Similar to hypothetical dihydroxyacetone kinase |
| lmo2724/lin2872 | 3.7 | 1.8 | 3.3 | 4.8 | + | + | 100 | 100 | Similar to unknown proteins |
| DNA metabolism and transport | |||||||||
| lmo1606/lin1647g | 1.2 (NS) | 2.8 | 1.9 | 5.9 | + | + | 97 | 100 (LI) | Similar to DNA translocase |
| Protein synthesis and modification | |||||||||
| lmo2511/lin2655 | 1.3 (NS) | 3.2 | 3.1 | 1.3 (NS) | + | + | 100 | 98 | Similar to conserved hypothetical proteins similar to B. subtilis YvyD protein |
| Cell envelope and cellular processes | |||||||||
| lmo0880/lin0879g | 12.3 | 13.0 | 9.0 | 23.7 | + | + | 100 | 100 (LI) | Similar to wall-associated protein precursor (LPXTG motif) |
| lmo1694/lin1802 | 3.4 | 2.6 | 2.1 | 3.6 | + | + | 100 | 98 | Similar to CDP-abequose synthase |
| Unknown | |||||||||
| lmo0133/lin0180g | 2.4 | 1.2 (NS) | 11.0 | 10.2 | + | + | 94 | 100 (LI) | Similar to Escherichia coli YjdI protein |
| lmo0170/lin0213 | 4.0 | 1.9 (NS) | 4.7 | 3.0 | + | + | 100 | 98 | Hypothetical protein |
| lmo0291/lin0319 | 2.0 | 2.1 | 1.4 (BS) | 2.8 | + | − | 98 | 94 | Conserved hypothetical protein similar to B. subtilis YycJ protein |
| lmo0321/lin0346 | 3.0 | 4.3 | 2.2 | 1.5 (NS) | + | + | 100 | 98 | Similar to unknown proteins |
| lmo0589/lin0598 | 3.9 | 1.8 | 3.7 | 1.3 (NS) | + | + | 100 | 92 | Hypothetical protein |
| lmo0592/lin0601 | 2.0 | 1.5 | 2.0 | 1.1 (NS) | + | + | 100 | 95 | Hypothetical protein |
| lmo0596/lin0605 | 17.7 | 12.3 | 14.5 | 13.2 | + | + | 98 | 98 | Similar to unknown proteins |
| lmo0628/lin0637 | 2.2 | 3.8 | 2.3 | 3.9 | + | + | 100 | 95 | Hypothetical protein |
| lmo0647/lin0650 | 4.8 | 3.6 | 8.9 | −2.1 (BS) | + | + | 100 | 98 | Hypothetical protein |
| lmo0670/lin0675 | 12.8 | 8.3 | 4.0 | 3.1 (NS) | + | + | 98 | 95 | Hypothetical protein |
| lmo0953/lin0952g | 3.0 | 3.0 | 9.9 | 19.5 | + | + | 100 | 100 (LI) | Hypothetical protein |
| lmo0995/lin0994 | 3.9 | 2.8 | 5.9 | 3.8 | + | − | 95 | 95 | Similar to B. subtilis YkrP protein |
| lmo1241/lin1205 | 2.3 | 2.6 | 3.2 | 3.2 | + | + | 100 | 98 | Hypothetical protein |
| lmo1602/lin1643 | 3.6 | 5.1 | 4.2 | 12.4 (BS) | + | + | 100 | 98 | Similar to unknown proteins |
| lmo2158/lin2261 | 17.8 | 15.6 | 5.1 | 6.8 | + | + | 100 | 94 | Similar to B. subtilis YwmG protein |
| lmo2191/lin2295 | 3.5 | 2.7 | 2.1 | 2.2 (NS) | + | + | 100 | 98 | Similar to unknown proteins |
| lmo2232/lin2334 | 4.9 | 1.6 | 1.5 | 3.3 | + | + | 100 | 98 | Similar to unknown proteins |
| lmo2269/lin2370 | 2.5 | 5.7 | 6.4 | 3.1 | + | + | 100 | 100 | Hypothetical protein |
| lmo2391/lin2490 | 5.7 | 7.5 | 2.0 | 8.5 | + | + | 100 | 91 | Conserved hypothetical protein similar to B. subtilis YhfK protein |
Probe name based on L. monocytogenes EGD-e gene.
L. innocua homologue as given by ListiList server (http://genolist.pasteur.fr/Listilist).
Positive changes represent higher transcript levels in the parent strain than in the ΔsigB strain. Negative changes represent higher transcript levels in the ΔsigB mutant than in the parent strain. Bold indicates σB-dependent genes (adjusted P < 0.05; change, ≥2.0-fold). NS, not significant (adjusted P > 0.05); BS, borderline significant (adjusted P between 0.05 and 0.1).
A plus sign indicates the presence of a σB-dependent promoter upstream of the ORF or the first gene of an operon. A minus sign denotes that no σB-dependent promoter was identified. See Tables S3 and S6 in the supplemental material for promoter descriptions and classifications of σB-dependent promoters, respectively.
L. monocytogenes EGD-e probe CHI to L. monocytogenes 10403S and L. innocua CLIP 11262. LI denotes L. innocua-specific probe (CHI, 100) designed by ArrayOligoSelector.
Description of protein as given by ListiList server (http://genolist.pasteur.fr/ListiList/).
Denotes L. innocua-specific probe. Changes in LIN-NaCl and LIN-STAT reflect transcript levels from this probe.
L. monocytogenes and L. innocua include species-specific σB-dependent genes.
Identification of L. monocytogenes and L. innocua species-specific σB-dependent genes is likely to provide further insight into gene regulation during infection and adaptation of stress response systems to pathogenic and saprophytic lifestyles. Specifically, σB-dependent genes present in L. monocytogenes and absent in L. innocua are likely candidates for virulence genes, as σB has been shown to contribute to L. monocytogenes virulence (27). Overall, 10 of the σB-dependent L. monocytogenes genes identified do not have homologues in L. innocua, including inlA, inlB, inlD, sepA, lmo2085, bsh, lmo0445, lmo2671, lmo2290, and lmo2387. Five of these genes (inlA, inlB, sepA, lmo2085, and lmo0445) have previously been shown to be upregulated in intracellular bacteria (12), consistent with the well-defined virulence contributions of InlA and InlB (19, 26, 51). While no clear contributions to virulence have been ascribed to lmo2085, sepA, and lmo0445 or to the other five L. monocytogenes-specific σB-dependent genes, we hypothesize that these genes include candidate virulence genes. In particular, sepA, which encodes a protein of the metallo-beta-lactamase family (41), is likely to contribute to virulence as its transcription is regulated both by σB and by the virulence gene regulator PrfA (12, 52), similar to the σB-PrfA coregulation reported for two internalins, inlA and inlB (50). Interestingly, the only σB-dependent L. innocua gene with no clear homologue in L. monocytogenes (lin0372 [see Fig. S3 in the supplemental material]) also encodes a putative internalin, indicating that σB-dependent transcription of internalins is evolutionarily conserved among Listeria spp. This observation suggests that internalin expression may contribute to Listeria survival in nonhost environments, consistent with recent evidence that σB-dependent L. monocytogenes internalins are highly transcribed at temperatures below mammalian body temperatures (49).
While gene homologues that were identified as σB dependent in L. monocytogenes, but not in L. innocua, are likely to include a number of false-negative L. innocua genes targeted by microarray probes with relatively low CHI values, the 14 genes that were initially identified as σB dependent in L. innocua but not identified as such in L. monocytogenes represent candidates for genes that have evolved to be regulated by different mechanisms in these two species. Further evaluation of these 14 genes showed that one L. monocytogenes gene (lmo0265, encoding a succinyldiaminopimelate desuccinylase) was targeted by a microarray probe with a low CHI (CHI of 67), as the DNA sequence of the lmo0265 allele in EGD-e differs from that in 10403S in the probe binding site. qRT-PCR clearly showed σB-dependent transcription of this gene in both L. monocytogenes and L. innocua (Fig. 1). An additional 10 L. monocytogenes genes (which showed σB-dependent transcription in L. innocua) showed lower transcript levels in the L. monocytogenes ΔsigB mutant, although at levels below the 2.0-fold criterion (adjusted P < 0.05 for eight genes; adjusted P between 0.05 and 0.1 for two genes), indicating either that these genes are σB dependent in both species or that quantitative differences in σB-dependent transcription for some of these genes are subtle. As these data suggested that few of the genes with homologues in both L. monocytogenes and L. innocua may have species-specific patterns of σB-dependent transcription, qRT-PCR was used to further evaluate σB-dependent transcription of an additional four genes with homologues in both L. monocytogenes and L. innocua. lmo2434/lin2528 (encoding GadA, a glutamate decarboxylase) and lmo2748/lin2891 (encoding a stress protein similar to B. subtilis YdaG) were included in the qRT-PCR experiments as microarray data showed positive σB-dependent regulation for the L. monocytogenes, but not the L. innocua, homologues, possibly because of the relatively low CHI of the probe for the L. innocua genes (CHIs of 94 and 91, respectively). qRT-PCR clearly showed σB-dependent transcription for both of these genes in both L. monocytogenes and L. innocua (Fig. 1), supporting the idea that probes with low CHIs may yield false-negative results. lmo1879/lin1992 (encoding CspD, a cold shock protein) was included in qRT-PCR experiments as the microarray data (based on a probe with CHIs of 100 and 98 for L. monocytogenes and L. innocua, respectively) showed lower transcript levels in the salt-stressed L. monocytogenes parent strain (compared to the ΔsigB strain), while showing higher transcript levels in the stationary-phase L. innocua parent strain (compared to the ΔsigB strain) (see Table S6 in the supplemental material). Initial analysis of qRT-PCR data for cells exposed to salt showed lower transcript levels in both the L. monocytogenes and L. innocua parent strain than in the respective sigB null mutants with no difference in transcript levels for stationary-phase cells (Fig. 1). When transcript levels for L. innocua cspD were normalized to rpoB only (rather than to the geometric mean of rpoB and gap, an approach chosen as L. innocua gap showed some σB-dependent transcription), L. innocua cspD showed higher transcript levels in the parent strain in stationary phase and no difference between parent and ΔsigB strain for salt-stressed cells, representing the same trends observed in the microarray data for L. innocua. While these findings illustrate the effect of different normalization procedures on data interpretation, including the possibility that normalization to a large number of genes (as performed in a microarray) may provide for more robust data, the biological relevance of these findings remains to be determined. Importantly though, for all other genes where transcript levels were evaluated by qRT-PCR, normalization of L. innocua transcript levels to rpoB alone (rather than both rpoB and gap) did not change the conclusions (Fig. 1 legend). Finally, lmo1428/lin1467 (encoding OpuCA, a component of the carnitine transporter) were included in the qRT-PCR experiments, as opuCA has previously been shown to be σB dependent in L. monocytogenes (40, 66). qRT-PCR showed that L. monocytogenes opuCA transcription is σB dependent in both stationary-phase and salt-stressed cells, while L. innocua opuCA was σB dependent only in salt-stressed cells, consistent with microarray data (Fig. 1). These data support the idea that σB-dependent regulation of transcription may differ between L. monocytogenes and L. innocua, even for genes found in both species.
Phenotypic characterization reveals different stress survival patterns for L. monocytogenes and L. innocua sigB null mutants.
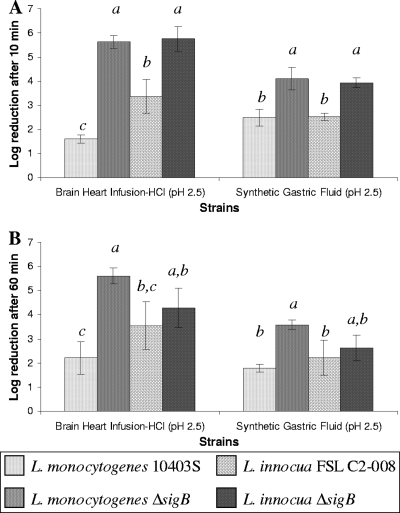
Acid stress survival assays showed that exposure of log-phase cells to BHI broth acidified to pH 2.5 or SGF at pH 2.5 leads to a clear reduction of L. monocytogenes and L. innocua bacterial numbers, with the ΔsigB strains showing significantly (P < 0.05) (Fig. 2A) greater reduction after acid exposure than the respective parent strains, indicating the importance of σB for acid stress survival in log-phase L. monocytogenes and L. innocua. Exposure of stationary-phase cells to BHI-HCl or SGF for 60 min also led to a clear reduction of L. monocytogenes and L. innocua bacterial numbers; while the L. monocytogenes ΔsigB strain showed significantly (P < 0.05) (Fig. 2B) higher reduction than the parent strain after acid exposure, stationary-phase L. innocua ΔsigB and parent strains did not differ in their susceptibility to either BHI-HCl or SGF, suggesting that σB contributes less to stationary-phase acid stress survival in L. innocua than in L. monocytogenes, suggesting that the relative contributions of σB to acid stress survival differ between these species. The observed difference in acid resistance phenotype may relate to the fact that the L. monocytogenes σB stress response system has adapted to suit a pathogenic lifestyle (as also supported by σB-dependent virulence gene transcription), including the need to rapidly adapt to and subsequently survive in the low-pH environment of the mammalian stomach. This hypothesis is consistent with the emerging picture that L. monocytogenes σB is particularly critical during the gastrointestinal stages of infection, by regulating transcription of genes encoding bile-associated proteins (3, 65, 66), internalins important for attachment to intestinal epithelial cells and other cells (42, 43), and other proteins important for gastrointestinal infection (e.g., OpuC [60]). L. innocua, on the other hand, may have evolved a more constitutively expressed mechanism of stationary-phase acid resistance, which relies less on σB.
FIG. 2.
Log reduction of L. monocytogenes and L. innocua parent and ΔsigB strains for log-phase cells (A) and stationary-phase cells (B) after exposure to acid stress. Strains with higher log reduction showed greater sensitivity to acid stress (e.g., L. monocytogenes ΔsigB showed a 5.5-log reduction while L. monocytogenes parent strain 10403S showed a 2-log reduction; the ΔsigB strain is thus more sensitive to acid stress). Experiments were performed in three independent trials, and data shown represent the averages of these trials; error bars show the standard deviations. Bacterial numbers after acid stress were analyzed using the general linear model with Tukey's multiple-comparisons procedure; bars labeled with different letters indicate bacterial numbers that differed significantly (P < 0.05), while bars labeled with identical letters indicate bacterial numbers that did not differ significantly.
Conclusions.
Overall, our data clearly support the importance of σB as a transcriptional regulator in both pathogenic and nonpathogenic Listeria species, as indicated by the considerable number of genes positively regulated as well as indirectly repressed by σB. Our data also suggest the involvement of σB in regulatory networks and interactions that appear to be critical to ensure appropriate gene expression in Listeria species under different environmental stress conditions as well as during host infections. In addition to the well-supported interactions between PrfA and σB, which appear to be critical to virulence and virulence gene regulation (26, 40), σB also appears to contribute to transcriptional regulation of genes encoding other regulators (e.g., hrcA) and appears to affect transcription of a number of genes, including flagellar genes, that are also regulated by the motility gene repressor MogR and the two-component response regulator DegU. The role of σB in gene regulation, including in regulatory networks, also appears to have adapted to the differing requirements for gene regulation in a pathogen versus a nonpathogen, as supported by pathogen-specific regulatory networks (i.e., the σB-PrfA network [40, 50]) and σB-dependent transcription of genes unique to the pathogen L. monocytogenes (e.g., inlAB and bsh) and of at least one internalin-like gene unique to the nonpathogen L. innocua.
Supplementary Material
Acknowledgments
We thank R. Ivanek for assistance with the statistical analyses and helpful discussions.
This work was supported in part by National Institutes of Health award no. RO1-AI052151-01A1 (to K.J.B.). S. Raengpradub was supported by USDA National Needs Fellowship grant 2002-38420-11738.
Footnotes
Published ahead of print on 16 November 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amati, G., P. Bisicchia, and A. Galizzi. 2004. DegU-P represses expression of the motility fla-che operon in Bacillus subtilis. J. Bacteriol. 186:6003-6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, L. A., M. S. Cetin, R. W. Hutkins, and A. K. Benson. 1998. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Begley, M., R. D. Sleator, C. G. M. Gahan, and C. Hill. 2005. Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect. Immun. 73:894-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bachi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop, D., and D. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 139:2005-2009. [PubMed] [Google Scholar]
- 6.Bozdech, Z., J. Zhu, M. Joachimiak, F. Cohen, B. Pulliam, and J. DeRisi. 2003. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 4:R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brody, M. S., and C. W. Price. 1998. Bacillus licheniformis sigB operon encoding the general stress transcription factor σB. Gene 212:111-118. [DOI] [PubMed] [Google Scholar]
- 8.Brody, M. S., K. Vijay, and C. W. Price. 2001. Catalytic function of an α/β hydrolase is required for energy stress activation of the σB transcription factor in Bacillus subtilis. J. Bacteriol. 183:6422-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabanes, D., O. Dussurget, P. Dehoux, and P. Cossart. 2004. Auto, a surface associated autolysin of Listeria monocytogenes required for entry into eukaryotic cells and virulence. Mol. Microbiol. 51:1601-1614. [DOI] [PubMed] [Google Scholar]
- 10.Chan, Y. C., K. J. Boor, and M. Wiedmann. 2007. σB-dependent and σB-independent mechanisms contribute to transcription of Listeria monocytogenes cold stress genes during cold shock and cold growth. Appl. Environ. Microbiol. 73:6019-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan, Y. C., S. Raengpradub, K. J. Boor, and M. Wiedmann. 2007. Microarray-based characterization of the Listeria monocytogenes cold regulon in log- and stationary-phase cells. Appl. Environ. Microbiol. 73:6484-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatterjee, S. S., H. Hossain, S. Otten, C. Kuenne, K. Kuchmina, S. Machata, E. Domann, T. Chakraborty, and T. Hain. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74:1323-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaturongakul, S., and K. J. Boor. 2004. RsbT and RsbV contribute to σB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 70:5349-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conte, M. P., G. Petrone, A. M. Di Biase, C. Longhi, M. Penta, A. Tinari, F. Superti, G. Fabozzi, P. Visca, and L. Seganti. 2002. Effect of acid adaptation on the fate of Listeria monocytogenes in THP-1 human macrophages activated by gamma interferon. Infect. Immun. 70:4369-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotter, P. D., C. G. M. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 16.Dartois, V., M. Debarbouille, F. Kunst, and G. Rapoport. 1998. Characterization of a novel member of the DegS-DegU regulon affected by salt stress in Bacillus subtilis. J. Bacteriol. 180:1855-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Hoon, M. J. L., Y. Makita, K. Nakai, and S. Miyano. 2005. Prediction of transcriptional terminators in Bacillus subtilis and related species. PLoS Comput. Biol. 1:e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doan, T., and S. Aymerich. 2003. Regulation of the central glycolytic genes in Bacillus subtilis: binding of the repressor CggR to its single DNA target sequence is modulated by fructose-1,6-bisphophate. Mol. Microbiol. 47:1709-1721. [DOI] [PubMed] [Google Scholar]
- 19.Dramsi, S., I. Biswas, E. Maguin, L. Braun, P. Mastroeni, and P. Cossart. 1995. Entry of Listeria monocytogenes into hepatocytes requires expression of inlB, a surface protein of the internalin multigene family. Mol. Microbiol. 16:251-261. [DOI] [PubMed] [Google Scholar]
- 20.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferreira, A., C. P. O'Byrne, and K. J. Boor. 2001. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67:4454-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferreira, A., D. Sue, C. P. O'Byrne, and K. J. Boor. 2003. Role of Listeria monocytogenes σB in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl. Environ. Microbiol. 69:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flamm, R. K., D. J. Hinrichs, and M. F. Thomashow. 1984. Introduction of pAMβ1 into Listeria monocytogenes by conjugation and homology between native L. monocytogenes plasmids. Infect. Immun. 44:157-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fouet, A., O. Namy, and G. Lambert. 2000. Characterization of the operon encoding the alternative σB factor from Bacillus anthracis and its role in virulence. J. Bacteriol. 182:5036-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gahan, C. G. M., J. O'Mahony, and C. Hill. 2001. Characterization of the groESL operon in Listeria monocytogenes: utilization of two reporter systems (gfp and hly) for evaluating in vivo expression. Infect. Immun. 69:3924-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaillard, J. L., P. Berche, C. Frehel, E. Gouin, and P. Cossart. 1991. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from Gram-positive cocci. Cell 65:1127-1141. [DOI] [PubMed] [Google Scholar]
- 27.Garner, M. R., B. L. Njaa, M. Wiedmann, and K. J. Boor. 2006. Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect. Immun. 74:876-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gentleman, R., V. Carey, D. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Yang, and J. Zhang. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K.-D. Entian, H. Fsihi, F. G.-D. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L.-M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J.-C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J.-A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 30.Grundling, A., L. S. Burrack, H. G. A. Bouwer, and D. E. Higgins. 2004. Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc. Natl. Acad. Sci. USA 101:12318-12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haldenwang, W., and R. Losick. 1979. A modified RNA polymerase transcribes a cloned gene under sporulation control in Bacillus subtilis. Nature 282:256-260. [DOI] [PubMed] [Google Scholar]
- 32.Hanawa, T., M. Kai, S. Kamiya, and T. Yamamoto. 2000. Cloning, sequencing, and transcriptional analysis of the dnaK heat shock operon of Listeria monocytogenes. Cell Stress Chaperones 5:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hecker, M., and U. Volker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35-91. [DOI] [PubMed] [Google Scholar]
- 34.Helmann, J. D., M. F. W. Wu, P. A. Kobel, F.-J. Gamo, M. Wilson, M. M. Morshedi, M. Navre, and C. Paddon. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183:7318-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffman, A. D., K. L. Gall, D. M. Norton, and M. Wiedmann. 2003. Listeria monocytogenes contamination patterns for the smoked fish processing environment and for raw fish. J. Food Prot. 66:52-60. [DOI] [PubMed] [Google Scholar]
- 36.Hoper, D., U. Volker, and M. Hecker. 2005. Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis. J. Bacteriol. 187:2810-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu, Y., H. F. Oliver, S. Raengpradub, R. H. Orsi, M. Wiedmann, and K. J. Boor. 2007. Transcriptomic and phenotypic analyses suggest a network between the transcriptional regulators HrcA and σB in Listeria monocytogenes. Appl. Environ. Microbiol. 73:7981-7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joseph, B., K. Przybilla, C. Stuhler, K. Schauer, J. Slaghuis, T. M. Fuchs, and W. Goebel. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188:556-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karatzas, K. A., J. A. Wouters, C. G. M. Gahan, C. Hill, T. Abee, and M. H. Behnik. 2003. The CtsR regulator of Listeria monocytogenes contains a variant glycine repeat region that affects piezotolerance, stress resistance, motility and virulence. Mol. Microbiol. 49:1227-1238. [DOI] [PubMed] [Google Scholar]
- 40.Kazmierczak, M. J., K. J. Boor, and M. Wiedmann. 2006. Contributions of Listeria monocytogenes σB and PrfA to expression of virulence and stress response genes during extra- and intracellular growth. Microbiology 152:1827-1838. [DOI] [PubMed] [Google Scholar]
- 41.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes σB regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim, H., K. J. Boor, and H. Marquis. 2004. Listeria monocytogenes σB contributes to invasion of human intestinal epithelial cells. Infect. Immun. 72:7374-7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim, H., H. Marquis, and K. J. Boor. 2005. σB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology 151:3215-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knudsen, G. M., J. E. Olsen, and L. Dons. 2004. Characterization of DegU, a response regulator in Listeria monocytogenes, involved in regulation of motility and contributes to virulence. FEMS Microbiol. Lett. 240:171-179. [DOI] [PubMed] [Google Scholar]
- 45.Kreft, J., and J. A. Vazquez-Boland. 2001. Regulation of virulence genes in Listeria. Int. J. Med. Microbiol. 291:145-157. [DOI] [PubMed] [Google Scholar]
- 46.Kunst, F., and G. Rapoport. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larisch, C., D. Nakunst, A. Huser, A. Tauch, and J. Kalinowski. 2007. The alternative sigma factor SigB of Corynebacterium glutamicum modulates global gene expression during transition from exponential growth to stationary phase. BMC Genomics 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lou, Y., and A. Yousef. 1997. Adaptation to sublethal environmental stresses protects Listeria monocytogenes against lethal preservation factors. Appl. Environ. Microbiol. 63:1252-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGann, P., R. Ivanek, M. Wiedmann, and K. J. Boor. 2007. Temperature-dependent expression of Listeria monocytogenes internalin and internalin-like genes suggests functional diversity for these proteins among the listeriae. Appl. Environ. Microbiol. 73:2806-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGann, P., M. Wiedmann, and K. J. Boor. 2007. The alternative sigma factor σB and the virulence gene regulator PrfA both regulate transcription of Listeria monocytogenes internalins. Appl. Environ. Microbiol. 73:2919-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mengaud, J., H. Ohayon, P. Gounon, R. M. Mege, and P. Cossart. 1996. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84:923-932. [DOI] [PubMed] [Google Scholar]
- 52.Milohanic, E., P. Glaser, J.-Y. Coppee, L. Frangeul, Y. Vega, J. A. Vazquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 53.Nadon, C. A., B. M. Bowen, M. Wiedmann, and K. J. Boor. 2002. Sigma B contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect. Immun. 70:3948-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nair, S., I. Derre, T. Msadek, O. Gaillot, and P. Berche. 2000. CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol. Microbiol. 35:800-811. [DOI] [PubMed] [Google Scholar]
- 55.O'Neil, H. S., and H. Marquis. 2006. Listeria monocytogenes flagella are used for motility, not as adhesins, to increase host cell invasion. Infect. Immun. 74:6675-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paillard, D., V. Dubois, R. Duran, F. Nathier, C. Guittet, P. Caumette, and C. Quentin. 2003. Rapid identification of Listeria species by using restriction fragment length polymorphism of PCR-amplified 23S rRNA gene fragments. Appl. Environ. Microbiol. 69:6386-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 58.Rauch, M., Q. Luo, S. Muller-Altrock, and W. Goebel. 2005. SigB-dependent in vitro transcription of prfA and some newly identified genes of Listeria monocytogenes whose expression is affected by PrfA in vivo. J. Bacteriol. 187:800-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwab, U., B. Bowen, C. Nadon, M. Wiedmann, and K. J. Boor. 2005. The Listeria monocytogenes prfAP2 promoter is regulated by sigma B in a growth phase dependent manner. FEMS Microbiol. Lett. 245:329-336. [DOI] [PubMed] [Google Scholar]
- 60.Sleator, R. D., J. Wouters, C. G. M. Gahan, T. Abee, and C. Hill. 2001. Analysis of the role of OpuC, an osmolyte transport system, in salt tolerance and virulence potential of Listeria monocytogenes. Appl. Environ. Microbiol. 67:2692-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smyth, G. K. 2005. Limma: linear models for microarray data, p. 397-420. In R. Gentleman, V. Carey, S. Dudoit, R. Irizarry, and W. Huber (ed.), Bioinformatics and computational biology solutions using R and bioconductor. Springer, New York, NY.
- 62.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:1-25. [DOI] [PubMed] [Google Scholar]
- 63.Smyth, G. K., J. Michaud, and H. S. Scott. 2005. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21:2067-2075. [DOI] [PubMed] [Google Scholar]
- 64.Steil, L., T. Hoffmann, I. Budde, U. Volker, and E. Bremer. 2003. Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J. Bacteriol. 185:6358-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sue, D., K. J. Boor, and M. Wiedmann. 2003. σB-dependent expression patterns of compatible solute transporter genes opuCA and lmo1421 and the conjugated bile salt hydrolase gene bsh in Listeria monocytogenes. Microbiology 149:3247-3256. [DOI] [PubMed] [Google Scholar]
- 66.Sue, D., D. Fink, M. Wiedmann, and K. J. Boor. 2004. σB-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology 150:3843-3855. [DOI] [PubMed] [Google Scholar]
- 67.Tokunaga, T., M. H. Rashid, A. Kuroda, and J. Sekiguchi. 1994. Effect of degS-degU mutations on the expression of sigD, encoding an alternative sigma factor, and autolysin operon in Bacillus subtilis. J. Bacteriol. 176:5177-5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Schaik, W., M. H. Tempelaars, J. A. Wouters, W. M. de Vos, and T. Abee. 2004. The alternative sigma factor σB of Bacillus cereus: response to stress and role in heat adaptation. J. Bacteriol. 186:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180-188. [DOI] [PubMed] [Google Scholar]
- 70.Wiedmann, M., T. J. Arvik, R. J. Hurley, and K. J. Boor. 1998. General stress transcription factor σB and its role in acid tolerance and virulence of Listeria monocytogenes. J. Bacteriol. 180:3650-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williams, T., S. Bauer, D. Beier, and M. Kuhn. 2005. Construction and characterization of Listeria monocytogenes mutants with in-frame deletions in the response regulator genes identified in the genome sequence. Infect. Immun. 73:3152-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Williams, T., B. Joseph, D. Beier, W. Goebel, and M. Kuhn. 2005. Response regulator DegU of Listeria monocytogenes regulates the expression of flagella-specific genes. FEMS Microbiol. Lett. 252:287-298. [DOI] [PubMed] [Google Scholar]
- 73.Woodling, S. E., and C. I. Moraru. 2005. Influence of surface topography on the effectiveness of pulsed light treatment for the inactivation of Listeria innocua on stainless-steel surfaces. J. Food Sci. 70:345-351. [Google Scholar]
- 74.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu, S., H. de Lencastre, and A. Tomasz. 1996. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J. Bacteriol. 178:6036-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng, W., and S. Kathariou. 1995. Differentiation of epidemic-associated strains of Listeria monocytogenes by restriction fragment length polymorphism in a gene region essential for growth at low temperatures (4°C). Appl. Environ. Microbiol. 61:4310-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zheng, W., and S. Kathariou. 1994. Transposon-induced mutants of Listeria monocytogenes incapable of growth at low temperature (4°C). FEMS Microbiol. Lett. 121:287-291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.