Abstract
A microbial community analysis using 16S rRNA gene sequencing was performed on borehole water and a granite rock core from Henderson Mine, a >1,000-meter-deep molybdenum mine near Empire, CO. Chemical analysis of borehole water at two separate depths (1,044 m and 1,004 m below the mine entrance) suggests that a sharp chemical gradient exists, likely from the mixing of two distinct subsurface fluids, one metal rich and one relatively dilute; this has created unique niches for microorganisms. The microbial community analyzed from filtered, oxic borehole water indicated an abundance of sequences from iron-oxidizing bacteria (Gallionella spp.) and was compared to the community from the same borehole after 2 weeks of being plugged with an expandable packer. Statistical analyses with UniFrac revealed a significant shift in community structure following the addition of the packer. Phospholipid fatty acid (PLFA) analysis suggested that Nitrosomonadales dominated the oxic borehole, while PLFAs indicative of anaerobic bacteria were most abundant in the samples from the plugged borehole. Microbial sequences were represented primarily by Firmicutes, Proteobacteria, and a lineage of sequences which did not group with any identified bacterial division; phylogenetic analyses confirmed the presence of a novel candidate division. This “Henderson candidate division” dominated the clone libraries from the dilute anoxic fluids. Sequences obtained from the granitic rock core (1,740 m below the surface) were represented by the divisions Proteobacteria (primarily the family Ralstoniaceae) and Firmicutes. Sequences grouping within Ralstoniaceae were also found in the clone libraries from metal-rich fluids yet were absent in more dilute fluids. Lineage-specific comparisons, combined with phylogenetic statistical analyses, show that geochemical variance has an important effect on microbial community structure in deep, subsurface systems.
The deep terrestrial subsurface biosphere represents an emerging frontier for studies of biodiversity, the physiological limits to life, microbial mechanisms of adaptation, and potentially analogous environments for extraterrestrial life (18). Intensive studies of deep boreholes in South Africa (19, 37), Finland (22), Sweden (49, 50), the United States (14, 26), and Japan (23), using a variety of molecular techniques, have shown an active biosphere composed of diverse groups of microorganisms. The microbial communities reported from different subsurface communities vary widely; such differences are due to different host rock types and varied water origins and chemistry, as well as geography.
The study of the deep subsurface biosphere has yielded valuable information about the ecology of microbes, the chemical and geological factors driving microbial community composition, and the energetics of microbial metabolisms in systems that are deficient in or devoid of photosynthetically derived carbon. Knowledge is gradually emerging that in the deep subsurface, rock-water interactions (49, 59) and radiolysis of water (35, 36) can generate abundant reductants (e.g., H2) along with biologically relevant oxidants such as sulfate (35, 36).
The Henderson Mine near Empire, CO, is a >1,000-meter-deep, active molybdenum mine. Molybdenum is mined from a 27- to 30-megaannum orebody intrusion surrounded by Precambrian Silverplume granite. Preexisting boreholes within the Henderson Mine, as well as a new borehole drilled and cored into pristine granite, provided an opportunity to sample and characterize the geochemistry and microbiology of deep-fracture waters and granite at this site. Geochemical analyses revealed that some fracture fluids are rich in dissolved metals (mM concentrations of Fe and Mn), carbon dioxide (CO2), and sulfate and that steep geochemical gradients exist over small spatial scales, where these fluids mix with more dilute waters. Correspondingly, the diversity of microbial communities varies widely across these interfaces, which expands what is known about microbial ecology in granitic fractures. In particular, we report a new bacterial candidate division that appears to be hosted primarily in anoxic fluids within the mine.
MATERIALS AND METHODS
Site and sampling methods.
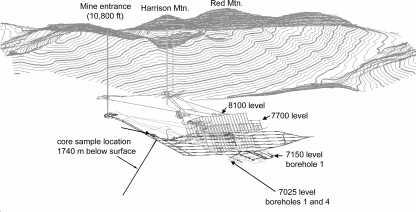
Groundwater samples were collected from existing, flowing boreholes in the Henderson Mine on 1 December 2005, 9 March 2006, and 23 March 2006. The boreholes were drilled in March 2005, approximately horizontal, into mine tunnel walls using a tri-cone bit (∼8-cm diameter) to distances of approximately 13 m. At the time of sampling, long, soft streamers of secondary minerals [predominantly Fe(III) hydroxides and gypsum/anhydrite (CaSO4)] precipitated from the fluids surrounded the mouth of each borehole, and dissolved gas (presumably CO2) vigorously degassed during discharge. Boreholes sampled in December 2005 were on the 7150 level (2,180 m above sea level) of the mine (approximately 1,004 m deeper than the mine entrance and approximately 1,450 m below the elevation of Red Mountain peak), and boreholes sampled in March 2006 were located at the 7025 level (2,140 m above sea level and 1,044 m below the mine entrance) (Fig. 1). In all, four boreholes were sampled for fluid chemistry at two different elevations: boreholes 1 and 4 at the 7025 level and borehole 1 at the 7150 level. The 7025 level is the “drain” level, where deep-seated fluids are released from depth through a long series of boreholes and pumped out of the mine to keep the overlying mining operations dry.
FIG. 1.
Schematic diagram of the Henderson Mine, showing the two levels where water samples were taken (7150 and 7025) as well as the location of the rock core sample.
Fluids were collected directly from the flowing boreholes into sterile, sealed, anaerobic, acid-washed vials with no residual headspace. After initial water sampling at the 7025 level, autoclaved borehole packers (constructed from PVC) were inserted into the boreholes to eliminate aeration of the groundwater as it emerged from the borehole, which allowed the water to completely fill the borehole, thus better representing water in the fracture(s) intersected by the borehole. Autoclaved Tygon tubing (Saint-Gobain, Akron, OH) was attached to the packers to allow continuous partial flow of water from the boreholes. The packers were left in place for 2 weeks, after which the water was again sampled into sterile sample bottles on 23 March 2006. Additionally, borehole water was filtered (0.2 μm; Millipore Durapore GVWP) using a stainless steel, autoclaved, high-pressure filter manifold (Millipore, Billerica, MA) immediately following installation of the packers and also 2 weeks after the packers were installed. Twenty liters of water was filtered for phospholipid fatty acid (PLFA) analysis, while an additional 2 liters was filtered for DNA extraction and microbial characterization. The filters were transported on dry ice to the surface and frozen (−80°C) within 3 h after collection. Clone libraries constructed from postpacker filters are designated with the letter P, while prepacker libraries are designated with the letter D. For example, a postpacker sample obtained from borehole 1 at the 7025 level is labeled 7025-P1; a prepacker sample from the same location is labeled 7025-D1.
Rock cores were collected from a borehole that was drilled from an elevation of 2,292 m at a −45° angle to an elevation of 1,477 m (above sea level) beneath Harrison Mountain (Fig. 1). Drilling was by a diamond core rig drilling method using wireline and an inner tube core barrel, with a borehole diameter of 96 mm and a recovered core diameter of 63.5 mm. Core samples were collected between 635.5 m and 635.8 m down hole (∼50°C) at an elevation of 1,840 m above sea level and 1,740 m below the surface. Fluorescently labeled (yellow-green), carboxylated, 1.0-μm-diameter, polycarbonate microspheres (Polysciences, Warrington, PA) were used as particulate tracers during collection of cores intended for microbiological analyses. The microspheres (1010 microspheres/ml in approximately 50 ml water) were placed in a Whirl-Pak bag (Nasco, Fort Atkinson, WI) that was attached inside the shoe of the core collector such that it would break on contact with the rock to be collected in the borehole (28, 52). Cores were aseptically collected and processed in a sterile laminar flow hood within 3 h of collection. Cores were subcored using a flame-sterilized hammer and chisel to break the core into 1- to 2-cm slices and then to remove the perimeters of the slices (28, 52). The subcores were crushed using a flame-sterilized ore crusher and then frozen at −80°C. The subcores were tested for fluorescent microspheres by suspending the crushed rock in 0.1% (wt/vol sodium pyrophosphate (pH 7.0), shaking the suspension for 10 min, allowing it to settle for 2 min, and then examining the supernatant for fluorescent microspheres by epifluorescence microscopy.
Chemical analysis.
Chemical analysis of major element, trace element, and anion concentrations in the borehole fluids was conducted at the Laboratory for Environmental and Geological Studies at the University of Colorado, Boulder. All water samples were prefiltered with a 0.45-μm filter. Water samples for anion analysis were kept at 9°C until measured on a Dionex 4500i AS14 anion chromatography column according to U.S. Environmental Protection Agency (EPA) method 300.0. Water samples for major cations and trace metals were acidified with concentrated trace metal grade nitric acid (0.01%) to pH 3 and kept at 9°C until analysis. Major elements were analyzed by inductively coupled plasma atomic emission spectroscopy on a 3410 Applied Research Laboratories instrument, following EPA method 6010 C. Separate aliquots were analyzed for trace element concentrations by inductively coupled plasma mass spectrometry on a Perkin-Elmer Elan DRC-E mass spectrometer using EPA method 6020. Water samples for determination total organic carbon, total inorganic carbon, and total nitrogen were stored at 9°C, aliquoted into 40-ml vials, and acidified with two drops of concentrated hydrochloric acid (0.1%). The samples were analyzed on a Shimadzu TOC-V equipped with a TNM-1. Dissolved inorganic carbon (DIC) was determined by the difference between dissolved organic carbon values obtained before and after acidification.
Nitrate, nitrite, and ammonia were measured at the U.S. Geological Survey in Boulder, CO, by ion chromatography (55). Samples were filtered (0.45 μm) into 40-ml amber glass vials and kept on ice until analysis. Dissolved nitrous oxide and methane were measured using a headspace equilibration technique. Briefly, 60-ml water samples were collected with a syringe and injected into a stoppered, helium-flushed, evacuated serum bottle that contained 0.7 ml of 12.5 N KOH. The samples were stored at room temperature and analyzed by gas chromatography using electron capture (N2O) (6) and flame ionization (CH4) (2) detectors.
Determination of stable isotope, tritium, and noble gas levels and time since recharge of the borehole water.
One liter of borehole water from a postpacker borehole (7025-P4) was filtered through a 0.45-μm filter and analyzed by Beta Analytic Inc. using the benzene method for 14C and δ13C. Oxygen and hydrogen isotopes (δ18O and δ2H) were analyzed at the Colorado School of Mines Stable Isotope Laboratory. Results are reported as per mille differences from the Vienna Standard Mean Ocean Water (VSMOW) standard. Precision from replicate analyses of laboratory standards and blind duplicate samples was 0.08‰ for O2 and 0.4‰ for H2.
The age of the water was determined by the common decay equation using the half-life of 14C. The maximum age of the sampled water was determined by assuming the initial 14C activity to be 100% modern carbon. To estimate the initial activity of 14C and the subsequent residence time of DIC in the system, two methods were used, those of Pearson et al. (48) and of Fontes and Garnier (17). Measured input values included temperature of the sample (35.4°C), pH (5.95), and DIC content (20.3 mM); a δ13C value for the soil gas was assumed to be −30‰ (9). A 500-ml sample was collected for tritium analysis.
DNA extraction, cloning, and sequencing.
Portions of borehole filter papers were torn using flame-sterilized forceps and placed directly into the PowerSoil extraction kit from Mobio, Inc. (Carlsbad, CA). For the rock core (taken 1,740 m below the ground surface), a 10-g fragment of the pared core was placed into an ethanol-washed and flame-sterilized ore crusher and pulverized in a laminar flow hood. Approximately 0.5 g of crushed rock was then immediately placed into the PowerSoil DNA extraction kit. Concentrations of extracted DNA were quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop, Wilmington, DE). PCRs were then conducted in duplicate using protocols and materials described elsewhere (57). The PCR primers 515F (5′-GTG CCA GCM GCC GCG GTA A-3′) and 1391R (5′-GAC GGG CGG TGW GTR CA-3′) were selected to amplify small-subunit rRNA gene fragments from all three domains of life. After this initial screen of diversity, forward primers specific for Bacteria (8F [5′-AGA GTT TGA TCC TGG CTC AG-3′]) and Archaea (21Fa [5′-TTC CGG TTG ATC CYG CCG GA-3′]) were amplified with reverse primer 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′) to obtain longer sequences. Duplicate PCR products were pooled, gel purified with Montage (Millipore, Billerica, MA), TOPO TA (Invitrogen, Carlsbad, CA) cloned using electrocompetent Escherichia coli TOP10 cells, and sequenced on a MegaBACE 1000 dye-terminating sequencer.
Sequence analysis.
Successful sequence reads were called by PHRED and assembled with PHRAP in conjunction with XplorSeq (Daniel Frank, unpublished). Contiguous sequences were aligned with the NAST aligner (11), screened for chimeras with Mallard (3), analyzed for nearest neighbors by the basic local alignment search tool (BLAST) (1), and manually corrected using the ARB editor (40). Once sequences were aligned, a Phylip (16) distance matrix was generated, and operational taxonomic units (OTUs) were determined by 97% raw sequence similarity by the furthest-neighbor method in DOTUR (53). Nonparametric species richness estimates were made through DOTUR using the abundance-based coverage estimator (ACE) (7) at a distance of 0.03. Phylogenetic relationships of sequenced clones were determined by inserting sequences into an ARB tree composed of the most recent Greengenes database release (greengenes.lbl.gov) using the Lanemask filter. Phylogenetic relationships of sequences that failed to group with any known divisions of bacteria were further determined by building maximum-likelihood trees with RAxML-VI-HPC (58) from 1,000 inferences on the original alignment. Tree topology and posterior probability values were verified using Bayesian inference coupled with Markov chain Monte Carlo methods with MrBayes 3.1 (24). Representative sequences from the ARB database were included in the tree-building analysis to verify the unique relationship of ungrouped sequences.
Weighted (using OTU data), pairwise statistical comparisons were made with UniFrac (38) by analyzing 1,000 samples. A UniFrac significance test (39) and the P test (43) were performed on all samples.
Clone names.
Clones were named according to the depth or type of the sample taken (7025, 7150, or core), the primer pair used (U, 515F and 1391R; B, 8F and 1492R; A, 21Fa and 1492R), and the clone number (1 to 96).
Lipid analysis.
Total lipids were extracted from filters by a modified Bligh-Dyer method and were separated into three fractions on solid-phase extraction columns (0.5 g silica; Burdick and Jackson) with 5 ml chloroform (nonpolar lipids), 10 ml acetone (lipids of intermediate polarity), and 10 ml methanol (polar lipids, including phospholipids) (61, 62). Fatty acid methyl esters (FAMEs) were released from phosphoester lipids by mild alkaline methanolysis (64, 65). FAMEs were then separated from any archaeal phosphoether lipids present in the samples on the same solid-phase extraction columns described above. FAMEs were eluted with 5 ml chloroform and phosphoether lipids with 10 ml methanol (65). Isoprenoidal hydrocarbon chains were released from phosphoethers by acid hydrolysis and subsequent reduction with LiAlH4 (29). An aliquot of each FAME sample was derivatized with dimethyl disulfide to determine the double bond position of monounsaturated FAMEs (13, 65).
FAMEs were analyzed on a Hewlett-Packard HP6890 series gas chromatograph coupled to a JEOL GC-mate mass spectrometer equipped with a Zebron ZB-1 column (60 m by 0.32 mm by 0.25 μm). FAME concentrations were determined by comparing total ion chromatogram peak areas with that of a 13:0 internal standard. Identification was made by comparing retention times and mass spectra with those of known standards. Ether lipid hydrocarbons were analyzed by gas chromatography-mass spectrometry as described above.
Nucleotide sequence accession numbers.
A total of 605 sequences were submitted to GenBank under accession numbers EF561677 to EF562281.
RESULTS
Groundwater geochemistry.
The chemistry of the Henderson molybdenum mine was analyzed in deep granitic fractures to understand the biologically relevant chemical constituents of the fluids and the corresponding effect on microbial community composition. Henderson Mine fluid temperatures ranged from 35 to 40°C, and the pH varied between 5.8 and 6.3. Reacted fluids, sampled at 7150-D1, were highly enriched in sulfate (48 mM), fluorine (12 mM) and dissolved metals (Fe, Mn, and Al at 1 to 20 mM) (Table 1). Dissolved O2 concentrations of as high as 119 μM were measured in free-flowing fluids. The boreholes were only approximately 25% saturated with fluid, which would explain the presence of oxygen in prepacker boreholes. However, within 1 h of packer insertion, the dissolved O2 concentrations dropped to <5 μM, and water was anoxic when sampled 2 weeks after packer insertion.
TABLE 1.
Groundwater chemistry of fracture fluids collected in boreholes on the 7150 and 7025 levels at Henderson Mine
| Parameter or species (unit) | Valuea for sample (mo/yr):
|
||||
|---|---|---|---|---|---|
| 7150-D1 (11/05) | 7025-D1 (3/06) | 7025-D4 (3/06) | 7025-P1 (3/06) | 7025-P4 (3/06) | |
| O2 (%) | NM | 3.75 | 55 | 1.4 | 0.4 |
| O2 (μM) | NM | 8.13 | 118.13 | 4.38 | 1.25 |
| Temp (°C) | 40 | 38.4 | 35.3 | 39.6 | 35.4 |
| pH | 5.85 | 5.9 | 6.28 | 5.82 | 5.95 |
| DIC (mM) | NM | 28.9 | 32.0 | 15.7 | 20.3 |
| DOC (mM) | NM | 0.05 | 0.05 | 0.03 | 0.03 |
| Total N (mM) | NM | 0.118 | 0.42 | 0.05 | 0.22 |
| Organic N (mM) | NM | 0.055 | 0.18 | 0.005 | 0.011 |
| NH4+ (mM) | NM | 0.009 | 0.113 | 0.005 | 0.112 |
| Anions (mM) | |||||
| Cl− | 0.15 | 0.11 | 0.14 | 0.1 | 0.14 |
| F− | 12.4 | 0.71 | 1.49 | 0.7 | 1.3 |
| SO42− | 47.5 | 4.33 | 10.4 | 4.0 | 9.8 |
| PO42− | BD | BD | BD | BD | BD |
| NO3− | NM | 0.021 | 0.072 | 0.017 | 0.047 |
| NO2− | NM | 0.032 | 0.048 | 0.027 | 0.046 |
| Major cations (mM) | |||||
| Si | 0.05 | 0.89 | 0.8 | 0.77 | 0.65 |
| Mn | 21.02 | 0.62 | 2.72 | 0.66 | 2.84 |
| Fe | 1.46 | 0.01 | 0.001 | 0.13 | 0.075 |
| Mg | 1.29 | 0.4 | 0.73 | 0.39 | 0.71 |
| Ca | 8.67 | 3 | 5.21 | 2.97 | 4.93 |
| Al | 4.78 | 0.06 | 0.26 | 0.66 | 0.27 |
| Sr | 0.04 | 0.014 | 0.03 | 0.014 | 0.03 |
| Na | 7.93 | 5.89 | 9.3 | 5.9 | 8.6 |
| Li | NM | 0.05 | 0.06 | NM | NM |
| K | 1.06 | 1.02 | 1.3 | 0.93 | 1.23 |
| Trace elements (μM) | |||||
| V | BD | 0.01 | BD | BD | BD |
| Ni | 1.45 | 0.17 | 0.5 | 0.21 | 0.51 |
| Cu | 0.26 | 0.04 | 0.05 | 0.17 | 0.25 |
| Zn | 2859 | 31.87 | 153.52 | 35.44 | 154.3 |
| As | 0.04 | 0.06 | 0.02 | 0.1 | 0.03 |
| Rb | 1.5 | 2.96 | 3.96 | 3.3 | 4.56 |
| Sr | 18.7 | 12.8 | 25.03 | 14.17 | 25.05 |
| Mo | 0.02 | 0.07 | 0.05 | 0.02 | 0.054 |
| W | 0.005 | 0.01 | BD | 0.001 | 0.001 |
| Stable isotopes | |||||
| δ2HVSMOW (‰) | NM | NM | NM | NM | −133.55 |
| δ18OVSMOW (‰) | NM | NM | NM | NM | −16.812 |
| Carbon | |||||
| 14C (%) | NM | NM | NM | NM | 3.54 ± 0.07 |
| δ13C (‰) | NM | NM | NM | NM | −6.0 |
NM, not measured; BD, below detection.
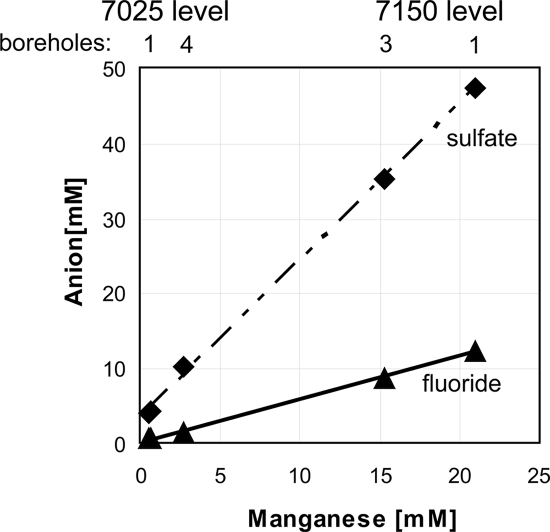
All of the water samples analyzed appear to be mixtures of the rock-dominated fluid composition (7150-D1 in Table 1) with more dilute waters. There is a clear linear relationship between the dissolved Mn (II) concentration in the fluids and both the sulfate and fluoride concentrations (Fig. 2). Fluid mixing shifts the water chemistry over short spatial scales, as indicated by the fluid compositions measured between two boreholes (7025-D1 and -D4) separated by only a few meters on opposite sides of the same drift.
FIG. 2.
Concentrations of manganese, fluoride, and sulfate in water sampled from boreholes at the 7150 and 7025 levels in the Henderson mine.
The mouths of the boreholes were often surrounded by white precipitated minerals that were likely calcium sulfates, since solubility calculations show that gypsum (CaSO4·2H2O) and anhydrite (CaSO4) were supersaturated in the fluids. Red precipitated minerals that formed were amorphous Fe(III) oxides [for example, Fe(OH)3] that precipitated as the Fe(II) in the fluids was rapidly oxidized at the oxic/anoxic interface. In addition, colorimetric assays using Leucoberbelin blue (31) were used to demonstrate that black Mn(III)/Mn(IV) oxide minerals coated the tunnel walls immediately adjacent to the boreholes. None of the minerals could be fully characterized, since they were relatively amorphous and produced only broad X-ray diffraction peaks when analyzed by both conventional and synchrotron-based X-ray diffraction techniques (data not shown).
The carbon dissolved in the fluids was dominated by inorganic rather than organic carbon. DIC values averaged 31 mM, whereas dissolved organic carbon (DOC) values in the fluids averaged 0.04 mM. Inorganic phosphate was below detection; however, inorganic N species were detectable, with dissolved nitrate, nitrite, and ammonium concentrations all in the micromolar range (Table 1). Nitrous oxide was present in the fluids after installation of the packers (12 to 34 μM as N), but methane concentrations were low (<0.2 μM). Zn was the most abundant trace element, with fluid concentrations ranging from 30 μM to 3 mM in the most reacted fluids.
Groundwater ages.
A groundwater age could be estimated for only one of the relatively dilute water samples, 7025-D4. The results from the carbon analyses were 14C = 3.54% ± 0.07% modern carbon and δ13C = −6‰ for 7025-D4. These results suggest that the average time for some of the DIC to travel from the surface to the collection point was 13,000 years. In deep subsurface systems the distribution of travel times to the discharge point can be quite large, i.e., from a decade to greater than the estimated ages (51). The age derived in this study reflects the average residence time of the youngest, least reacted fluids but cannot be used to constrain the “rock-dominated,” metal-rich fluids, which are likely significantly older.
The stable isotope composition of the water was δ2HVSMOW = −133.55‰ and δ18OVSMOW = −16.81‰. These values fall on the cold-region portion of the global meteoric water line (δ2H = 8.13‰; δ18O = 10.8‰), which indicates that the water is of meteoric origin (snowmelt).
The tritium content of the sample was 0.38 ± 0.2 tritium units (TU). Modern water in Colorado likely contains 10 to 20 TU (41). A value of 0.4 TU indicates a mixture of water that is predominantly pre-1950s (pre-atomic bomb) with potentially a small amount of modern (or post-atomic bomb) water. This result is consistent with most of the sampled water being thousands of years old.
Microbial community composition.
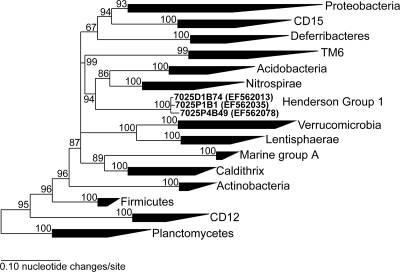
16S rRNA gene sequencing was performed on the filterable fraction of borehole water to determine the microbial community composition in each of the samples. The 16S rRNA gene sequences, or “phylotypes,” obtained from four borehole water samples revealed relatively low microbial diversity. 7025-D1 contained a majority of sequences (90%) belonging to one phylotype that grouped within the genus Gallionella yet also contained sequences grouping with the genus Siderooxidans (1%) and the divisions Chloroflexi and Firmicutes. A group of sequences, referred to as the Henderson group 1 candidate division, appeared to be independent of all known divisions of bacteria. Maximum-likelihood, Bayesian inference, and neighbor-joining tree analyses confirmed that sequences grouping with the Henderson group 1 did not group with closely related divisions of bacteria (Fig. 3). The lack of phylogenetic similarity with other groups on a phylogenetic tree containing closely related divisions (Fig. 3) suggests that these sequences represent a novel division-level bacterial lineage. The division of ungrouped bacteria was defined by the criteria that there must be three or more sequences from independent PCR products, the sequences must be greater than 1,000 nucleotides in length, and there must be high support levels in phylogenetic analyses (25). Sequences from Henderson group 1 were present in oxic waters from 7025-D1 but represented a very low percentage of the clone library.
FIG. 3.
Phylogenetic tree showing representative bacterial divisions constructed with Bayesian inference. Sequences that form the Henderson group 1 are shown in bold. Numbers at bifurcations indicate posterior probabilities calculated by MrBayes v3.1. The rooting archaeal outgroup (a sequence from Halobacteria) has been removed for clarity.
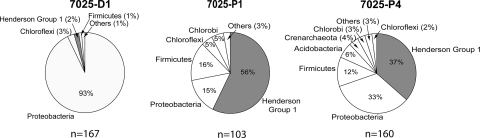
The clone library from 7025-P1 constructed from filters collected after the addition of the packer showed a substantial community shift (Fig. 4). The majority of sequences grouped with Henderson group 1, while the Gallionella sequences were mostly absent from the library (<1%). Sequences from four divisions which were not present in the library created from samples collected prior to the packer insertion were observed in the postpacker sample.
FIG. 4.
Pie charts showing microbial community compositions of drill hole 1 (pre- and postpacker) and drill hole 4 (postpacker), all at the 7025 level. “Others” refers to pooled divisions representing <1% of each respective library.
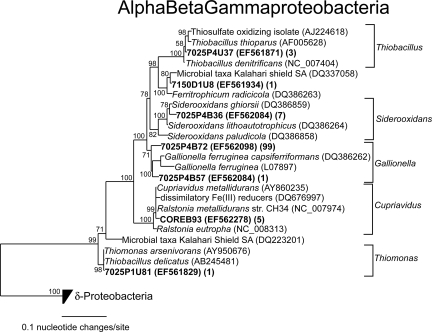
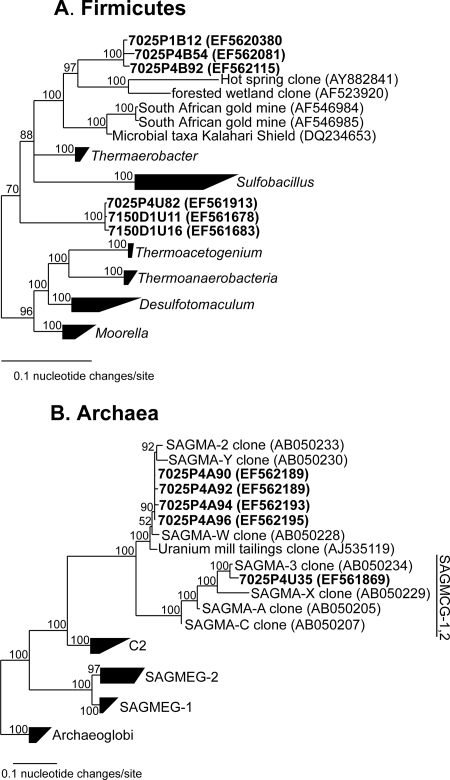
The clone library generated from 7025-P4 was similar to the library from 7025-P1, showing small differences in phylotype distributions and community compositions (Fig. 4). However, differences were observed in the analysis of specific lineages, such as the presence of the family Ralstoniaceae and the division Acidobacteria, which were present in the metal-rich fluid at borehole 4 and absent in the more dilute fluid at borehole 1. Sequences were represented primarily by Proteobacteria and Henderson group 1 but also included sequences grouping with Chlorobi, Chloroflexi, Acidobacteria, and Firmicutes. Within Proteobacteria, sequences were found that showed >99% BLAST identity and related phylogeny with Thiobacillus denitrificans (Fig. 5), a phylotype also isolated from deep mines in South Africa (19). In addition, two radiations of sequences were formed within Firmicutes that showed low relatedness to sequences in public databases (87% BLAST identity) and may represent novel genus-level lineages (Fig. 6A).
FIG. 5.
Phylogenetic tree of the division Proteobacteria. Numbers at nodes represent posterior probabilities. Numbers following accession numbers indicate how many sequences grouped with each phylotype.
FIG. 6.
Phylogenetic trees of the division Firmicutes (A) and the domain Archaea (B). Sequences obtained in this study are in bold. Numbers at branching nodes indicate posterior probabilities. The rooting outgroup for the Firmicutes has been removed for clarity.
The clone library created from 7150-D1 contained a number of phylotypes not present in samples from the 7025 level. The library was comprised primarily of sequences from Proteobacteria (52%) and Firmicutes (34%) but also contained one sequence that grouped with the Henderson group 1. Two sequences showed >99% BLAST identity with Ralstonia metallidurans, a microbe that tolerates millimolar concentrations of heavy metals, including Zn(II), Cd(II), and Co(II) (45). Zn concentrations were high in 7150-D1 (Table 1), which supports the presence of this particular phylotype. Other sequences grouping with the family Ralstoniaceae (15% of library) showed 99% BLAST identity with sequences obtained from spent nuclear fuel pools (8).
A clone library was also obtained from the pared subcore. Although no microspheres were observed in the pared core, DNA was successfully extracted and amplified. The amount of DNA extracted from the core was small (112 ng in 20 μl), but enough product was amplified through PCR to clone. The phylotype distribution was similar to that in the library from 7150-D1. The library contained very low division-level diversity; only Proteobacteria (85%), and Firmicutes (15%) were represented. Within the division Proteobacteria, the majority of sequences (46%) were represented by the family Ralstoniaceae. Other sequences grouping with Proteobacteria included members of the Alphaproteobacteria and Deltaproteobacteria.
Archaeal primers amplified gene products from sample 7025-P4 only; no PCR product was observed from other samples analyzed by gel electrophoresis. Although the diversity of archaeal sequences was low and limited to the main archaeal group Crenarchaeota, many sequences were phylogenetically similar to other sequences obtained from deep underground systems (Fig. 6B).
Coverage estimates.
ACE coverage estimates were made to determine how well the sequencing effort covered the anticipated diversity of each sample. Sequences were binned through DOTUR into OTUs showing ≥97% sequence similarity. ACE coverage estimates, as calculated by the observed OTUs/predicted OTUs at 97% shared sequence identity, were 34% (7025-D1), 36% (7025-P1), 67% (7025-P4), 57% (7150-D1), and 86% (CORE). The calculated sequence coverage in each sample allowed us to make statistical comparisons of microbial communities between samples.
UniFrac analyses.
Statistical analyses were performed with UniFrac to determine if microbial communities from samples analyzed in this study were statistically similar or different by utilizing phylogenetic comparative methods incorporating branch length differences. A UniFrac significance P value threshold of 0.05 was selected for all pairwise comparisons. Weighted, pairwise comparisons of microbial communities isolated from different sources revealed statistically similar UniFrac P values (P > 0.05) between 7025-P1 and 7025-P4 and between 7150-D1 and the rock core. UniFrac P values also revealed significant differences (P ≤ 0.05) in the microbial communities from 7025-D1 and 7025-P1.
Total PLFA concentration and profiles.
PLFA analysis was used to verify the results of the gene sequencing analysis as well as to estimate viable biomass from each borehole. As PLFAs degrade quickly after cell death (65), they can provide a measure of viable biomass in an environmental sample. Total PLFA concentrations were 103.6, 127.7, and 393.1 pmol PLFA liter−1 at sites 7025-P4, 7025-P1, and 7025-D1, respectively. A general conversion factor of 2.5 × 104 cells pmol−1 PLFA was used to convert this concentration into viable biomass (65), with values ranging from of 2,590, 3,190, and 9,830 cells ml−1 for sites 7025-P4, 7025-P1, and 7025-D1, respectively.
Similar to what was observed from the clone libraries, the fluids from 7025-P1 and 7025-P4 also showed similar PLFA profiles. In both samples, 16:0 was the major straight-chain PLFA and br15:0 and 10me16:0 were the major branched saturated PLFAs (Table 2). The PLFA profile of 7025-D1, collected from the same borehole prior to packer insertion, was conspicuously different. This sample was dominated by 16:0 and 16:1ω7, with mole percentages of 41.9 and 42.2, respectively. No isoprenoid, ether-bound archaeal lipids were detected in any of the boreholes.
TABLE 2.
PLFA profiles of cells filtered from groundwater in borehole fluids collected from the 7025 level
| PLFAa | mol% in sample:
|
||
|---|---|---|---|
| 7025-D1 | 7025-P1 | 7025-P4 | |
| 14:0 | 1.3 | 10.3 | 8.9 |
| br15:0b | 0.0 | 15.7 | 15.7 |
| i15:0 | 0.0 | 1.9 | 2.4 |
| ai15:0 | 0.2 | 6.0 | 4.1 |
| a16:0c | 0.0 | 1.0 | 1.0 |
| 16:1ω7 | 41.9 | 3.2 | 8.0 |
| 16:0 | 42.2 | 26.5 | 30.6 |
| 10me16:0 | 1.6 | 15.2 | 9.6 |
| i17:0c | 0.0 | 5.9 | 0.9 |
| ai17:0c | 0.0 | 5.1 | 2.9 |
| cyc17d | 0.3 | 3.7 | 3.0 |
| 18:1ω7 | 0.0 | 0.0 | 1.4 |
| 18:1ω9 | 4.2 | 0.3 | 0.0 |
| 18:1ω11 | 2.5 | 2.4 | 3.2 |
| 18:0 | 2.1 | 2.7 | 8.4 |
| Total | |||
| Saturated | 45.6 | 39.5 | 47.8 |
| Branched | 1.8 | 50.9 | 36.6 |
| Unsaturated/cyclic | 48.9 | 9.6 | 15.6 |
14:0, 14-carbon chain and no double bonds; 16:1ω7, 16-carbon chain with one double bond at seven carbons from the aliphatic end; i, iso branching at the aliphatic end; ai, anteiso branching; 10me, methyl group at 10 carbons from the carbonyl carbon; cyc17, one cyclopropyl group in the carbon chain.
Methyl branch position not determined.
Identified by mass spectrum and expected retention time, with no comparison to authentic standard.
Peak area estimated, as peak was overlain by a contamination peak.
DISCUSSION
Groundwater geochemistry.
The majority of carbon present in the Henderson system was DIC, but low levels of organic carbon were also detected. The concentrations of organic carbon detected in boreholes were lower than those reported from mines in Sweden (49) and South Africa (19, 47). Total nitrogen levels were highly variable, but all N species measured (NH4+, NO2−, and NO3−) were above detection limits (Table 1), suggesting that this is not an N-limited system. However, the source of nitrogen and potential for the existence of a subsurface microbial N cycle must be explored further. CO2 levels were high in all of the waters sampled, and CO2 likely plays an important role in buffering the pH of the fluids to near-neutral values rather than the acidic values expected due to mineral dissolution. Furthermore, CO2 likely provides the carbon for autotrophic iron-oxidizing bacteria that thrive in oxic borehole water.
Chemical differences existed between boreholes on the same mine level, which suggests that small-scale heterogeneities in fluid flow paths were present. For example, borehole 4 at the 7025 level, although only meters away from borehole 1 at the 7025 level, exhibited a fluid chemistry that is clearly intermediate between those of the dilute 7025 fluids at borehole 1 and the metal- and sulfate-rich fluids detected at the 7150 level (Table 1). Correspondingly, the microbial community structure changed across the fluid gradient, with the percentage of sequences grouping with the family Ralstoniaceae increasing from the dilute fluids (0%) to the intermediate fluids (5%) to the most reacted fluids (17%). Statistically significant shifts in community structure were also seen before and after the addition of the borehole packer. This is similar to the findings of Moser et al. (47), who characterized the chemistry and microbiology of groundwater from a borehole in a deep South African gold mine before and after sealing with a packer. However, our study used quantitative, statistical analyses of sequence data, incorporating phylogeny through weighted UniFrac, instead of terminal restriction fragment length polymorphism, to analyze the shift in microbial community structure.
Prepacker microbial community analysis.
The dominance of Gallionella spp. in the prepacker sample (7025-D1) likely reflects the presence of O2 and iron species, as well as the low concentrations of DOC. Consistent with the 16S rRNA gene analysis, PLFA profiles suggested similar trends in microbial community structure in 7025-D1. The PLFAs were dominated (>84%) by 16:0 and 16:1ω7; these have been shown to be the dominant PLFAs in microbes within Nitrosomonadales (5).
Whether the Gallionella sequences are from indigenous microbes or from airborne contamination is unknown. When analyzing microbial communities from mine samples, the potential for contamination from mine air (Henderson Mine has an extensive ventilation network using a high volume of surface air), humans, drilling fluids, and machinery cannot be ignored. For example, one sequence closely related to Propionobacter acnes, a species normally found on human skin (44), was observed in 7150-D1 yet showed ≥98% raw sequence similarity with sequences from an underground laboratory in Sweden (50) and a borehole in South Africa (19). Whether this and other sequences are from indigenous microbes or are a product of mining and/or sampling contamination is not known, but the potential certainly exists for the contamination of subsurface mines by normal drilling/mining operations (18).
After Gallionella, the second-most-represented group of sequences from 7025-D1 was Chloroflexi, whose major PLFAs have been reported to be 16:0, 18:0, and 18:1 (27). Correspondingly, 18:0 and 18:1 make up the remainder of the 7025-D1 PLFAs (8.8 mol% total) and are consistent with the presence of Chloroflexi (Table 2). In addition, sequences grouping with Chloroflexi were also observed in postpacker samples, with similar lipids (18:0 and 18:1) attributed to Chloroflexi. Many of the Chloroflexi phylotypes showed >98% BLAST identity with sequences from an alpine tundra environment in Niwot Ridge, CO (10), located approximately 25 miles from the Henderson Mine. Although the ecology of these microbes in Henderson Mine fluids is unclear, their presence may indicate transport through the subsurface due to snowmelt.
Postpacker microbial community analysis.
UniFrac significance values showed a clear community shift before and after the addition of the expandable packer in drill hole 1 at the 7025 level. However, the postpacker sample represents a single point in time, and the precise dynamics of community shift are unclear. PLFA profiles support the observed shift in community structure between pre- and postpacker samples. In general, the postpacker samples (7025-P1 and 7025-P4) showed a greater diversity of PLFAs, suggesting a more diverse microbial community. The microbial communities from the two postpacker samples (7025-P1 and 7025-P4) were statistically similar when compared by using UniFrac, which suggests that despite differences in geochemistry, the microbial community structures are statistically similar.
Clone libraries from each borehole showed the presence of a novel bacterial lineage, Henderson group 1. Although 123 sequences which grouped with Henderson group 1 were amplified in this study, only three distinct OTUs were formed (Fig. 3). The fact that the Henderson group 1 sequences were nearly absent from water samples collected before installation of the packer but dominated after the packer had been in place for 2 weeks suggests that these are indigenous groundwater bacteria with anaerobic metabolism. Although it is difficult to speculate on the bioenergetics of microbes associated with these sequences, the low concentrations of organic carbon combined with available electron acceptors (e.g., sulfate and nitrate) and low concentrations of dissolved oxygen in postpacker borehole water suggest that these microbes are anaerobic chemoautotrophs that fix their carbon from high concentrations of dissolved CO2, as has been observed in other deep subsurface systems (37).
The predominance of branched saturated PLFAs in the postpacker bore holes is suggestive of anaerobic gram negative bacteria (32). Iso- and anteiso-branched PLFAs (i15:0, ai15:0, ai16:0, i17:0, and ai17:0) were all detected in 7025-P1 and 7025-P4 and have been associated with several genera of sulfate-reducing bacteria (12, 30, 62). Furthermore, the PLFA 10me16:0 was detected in 7025-P1 and 7025-P4 and has been associated with Actinobacteria (61), Planctomycetes (54, 60), and Desulfobacter (12, 30); however, no sequences that are associated with these groups were retrieved from our study. Given that the novel division of bacteria detected by 16S rRNA gene analysis represented a significant percentage of each postpacker clone library and that these iso- and anteiso-branched PLFAs composed a significant mole percentage of the total PLFAs, we speculate that members of Henderson group 1 produce the various branched saturated PLFAs that are commonly observed in these other genera of Bacteria.
Rock core microbial community analysis.
Sequences from two divisions of Bacteria were recovered from the pared core of pristine granite. Although other studies have looked at microbes that live in pristine rock cores (19, 33, 34), the sequences from those studies were not phylogenetically similar to those observed in our library. Forty-five percent of the library was composed of sequences grouping with the family Ralstoniaceae. Sequences grouping with Ralstoniaceae have been associated with some deep subsurface environments (42, 63) but are absent from others; such differences are likely due to different chemical and geological conditions (18). UniFrac analysis revealed that the microbial community isolated from the rock core was significantly similar to the library constructed from the 7150 level. The high concentrations of metals and anions in the fluid from this level suggest a higher contact time of the fluids with rock than for the fluids sampled at the 7025 level. The metal-rich fluid could potentially be filling microfractures in the granite matrix as the fluid moves through the subsurface; potential microfractures (∼3 μm in width) were observed in the rock core by scanning electron microscopy (not shown).
Microbial metabolisms inferred from phylogeny.
Microbial metabolisms were inferred from the sequence identity of certain phylotypes based on high BLAST identity to cultivated microbes (≥98%), closely related phylogeny (Fig. 5), and the geochemistry of the mine, as done elsewhere (56). For example, one lineage of sequences found in Henderson fluids showed 100% BLAST identity and identical phylogeny with Thiomonas arsenivorans, a chemoautotrophic microbe that grows on As(III), sulfur, and thiosulfate (4). Another lineage of sequences showed >97% BLAST identity with Siderooxidans lithoautotrophicus, an iron-oxidizing lithoauthotroph (15). Additionally, sequences that showed >99% BLAST identity with Ralstonia metallidurans (also known as Cupriavidus metallidurans) were observed in metal-rich fluids and the rock core. R. metallidurans is a hydrogen-oxidizing lithoautotroph that is capable of subsisting on H2 and CO2 as its primary sources of energy and carbon (46) while using inorganic electron acceptors such as nitrate (21), a theme possibly common to subsurface life, as originally speculated by Gold (20).
Conclusion.
Overall, the microbial diversity found in pristine, anoxic granitic-fracture water at the Henderson Mine was low, although several novel lineages of Bacteria were observed. Extensive geochemistry data, combined with phylogenetic comparative analyses, showed how the microbial community structure shifts with dynamic fluid chemistries across relatively small spatial scales. Additionally, the related phylogeny of sequences obtained in this study to known chemoautotrophs, combined with low concentrations of DOC, suggests that the fixation of inorganic carbon may be an important microbial metabolism in this system, a result that is consistent with other studies of deep subsurface microbial ecology. Future work looking at potential inorganic electron donors, combined with cultivation and subsequent characterization of native microbes, will attempt to determine the dominant modes of microbial metabolism in this deep biosphere system.
Acknowledgments
We thank the Climax Molybdenum Company for access to the Henderson Mine and Mark Ramirez, Chip deWolfe, Brad Nelson, and Dick Propernick for assistance in sampling. We also thank Chris Mills for his helpful editorial comments on an early draft of the manuscript and Leah Feazel at the University of Colorado, Boulder, for assistance in operating the MegaBACE 1000.
Support for the research was provided by the NSF S2 grant for the DUSEL project to the Colorado School of Mines and by the Department of Local Affairs and the Office of Economic Development and International Trade in the State of Colorado.
The use of trade or product names in this report is for identification purposes only and does not constitute endorsement by the U.S. Geological Survey.
Footnotes
Published ahead of print on 2 November 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Antweiler, R. C., R. L. Smith, M. A. Voytek, J. K. Bohlke, and K. D. Richards. 2004. Water-quality data from two agricultural drainage basins in the northwestern Indiana and northeastern Illinois. I. Lagrangian and synoptic data, 1999-2002. U.S. Geological Survey Open-File Report 2004-1317. U.S. Geological Survey, Reston, VA.
- 3.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battaglia-Brunet, F., C. Joulian, F. Garrido, M.-C. Dictor, D. Morin, K. Coupland, D. B. Johnson, K. B. Hallberg, and P. Baranger. 2005. Oxidation of arsenite by Thiomonas strains and characterization of Thiomonas arsenivorans sp. nov. Antoine Leeuwenhoek 89:99-108. [DOI] [PubMed] [Google Scholar]
- 5.Blumer, M., T. Chase, and S. W. Watson. 1969. Fatty acids in the lipids of marine and terrestrial nitrifying bacteria. J. Bacteriol. 99:366-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks, M. H., R. L. Smith, and D. L. Macalady. 1992. Inhibition of existing denitrification enzyme activity by chloramphenicol. Appl. Environ. Microbiol. 58:1746-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chao, A., and S. M. Lee. 1992. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 87:210-217. [Google Scholar]
- 8.Chicote, E., A. M. Garcia, D. A. Moreno, M. I. Sarro, P. I. Lorenzo, and F. Montero. 2005. Isolation and identification of bacteria from spent nuclear fuel pools. J. Ind. Microbiol. Biotechnol. 32:155-162. [DOI] [PubMed] [Google Scholar]
- 9.Clark, I. D., and P. Fritz. 1997. Environmental isotopes in hydrogeology. CRC Press, Boca Raton, FL.
- 10.Costello, E. K., and S. K. Schmidt. 2006. Microbial diversity in alpine tundra wet meadow soil: novel Chloroflexi from a cold, water-saturated environment. Environ. Microbiol. 8:1471-1486. [DOI] [PubMed] [Google Scholar]
- 11.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowling, N., J. E. Widdel, and D. C. White. 1986. Phospholipid ester-linked fatty acid biomarkers of acetate-oxidizing sulfate-reducers and other sulfide-forming bacteria. J. Gen. Microbiol. 132:1815-1825. [Google Scholar]
- 13.Dunkelblum, E., S. H. Tan, and P. J. Silk. 1985. Double-bond location in monounsaturated fatty acids by dimethyl disulfide derivatization and mass spectrometry. J. Chem. Ecol. 11:265-277. [DOI] [PubMed] [Google Scholar]
- 14.Edwards, R. A., B. Rodriguez-Brito, L. Wegley, M. Haynes, M. Breitbart, D. M. Peterson, M. O. Saar, S. Alexander, E. C. Alexander, Jr., and F. Rohwer. 2006. Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics 7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emerson, D., and C. L. Moyer. 1997. Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl. Environ. Microbiol. 63:4784-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenstein, J. 1989. Phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 17.Fontes, J.-C., and J. M. Garnier. 1979. Determination of the initial 14C activity of the total dissolved inorganic carbon: a review of the existing models and a new approach. Water Resour. Res. 15:399-413. [Google Scholar]
- 18.Fredrickson, J. K., and D. Balkwill. 2006. Geomicrobial processes and biodiversity in the deep terrestrial subsurface. Geomicrobiology J. 23:345-356. [Google Scholar]
- 19.Gihring, T. M., D. P. Moser, L.-H. Lin, M. Davidson, T. C. Onstott, L. Morgan, M. Milleson, T. L. Kieft, E. Trimarco, D. L. Balkwill, and M. E. Dollhopf. 2006. The distribution of microbial taxa in the subsurface water of the Kalahari Shield, South Africa. Geomicrobiol. J. 23:415-430. [Google Scholar]
- 20.Gold, T. 1992. The deep, hot biosphere. Proc. Natl. Acad. Sci. USA 89:6045-6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goris, J., P. De Vos, T. Coenye, B. Hoste, D. Janssens, H. Brim, L. Diels, M. Mergeay, K. Kersters, and P. Vandamme. 2001. Classification of metal-resistant bacteria from industrial biotopes as Ralstonia campinensis sp. nov., Ralstonia metallidurans sp. nov. and Ralstonia basilensis Steinle et al. 1998 emend. Int. J. Syst. Evol. Microbiol. 51:1773-1782. [DOI] [PubMed] [Google Scholar]
- 22.Haveman, S. A., and K. Pedersen. 1999. Distribution and metabolic diversity of microorganisms in deep igneous rock aquifers of Finland. Geomicrobiol. J. 16:277-294. [Google Scholar]
- 23.Hirayama, H., K. Takai, F. Inagaki, Y. Yamato, M. Suzuki, K. H. Nealson, and K. Horikoshi. 2005. Bacterial community shift along a subsurface geothermal water stream in a Japanese gold mine. Extremophiles 9:169-184. [DOI] [PubMed] [Google Scholar]
- 24.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 25.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiménez, L. 1990. Molecular analysis of deep-subsurface bacteria. Appl. Environ. Microbiol. 56:2108-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenyon, C. N., and A. M. Gray. 1974. Preliminary analysis of lipids and fatty acids of green bacteria and Chloroflexus aurantiacus. J. Bacteriol. 120:131-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kieft, T. L., T. J. Phelps, and J. K. Fredrickson. 2007. Drilling, coring, and sampling subsurface environments, p. 799-818. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, M. J. McInerney, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 3rd ed. ASM Press, Washington, DC.
- 29.Koga, Y., and H. Morii. 2006. Special methods for the analysis of ether lipid structure and metabolism in archaea. Anal. Biochem. 348:1-14. [DOI] [PubMed] [Google Scholar]
- 30.Kohring, L. L., D. B. Ringelberg, R. Devereux, D. A. Sahl, M. W. Mittelman, and D. C. White. 1994. Comparison of phylogenetic relationships based on phospholipid fatty acid profiles and ribosomal RNA sequence similarities among dissimilatory sulfate-reducing bacteria. FEMS Microbiol. Lett. 303-308. [DOI] [PubMed]
- 31.Krumbein, W. E., and H. J. Altmann. 1973. A new method for the detection and enumeration of manganese oxidizing and reducing microorganisms. Helgo. Wiss. Meeresunters. 25:347-356. [Google Scholar]
- 32.Lechevalier, M. P. 1977. Lipids in bacterial taxonomy: a taxonimists view. Crit. Rev. Microbiol. 5:109-210. [DOI] [PubMed] [Google Scholar]
- 33.Lehman, R. M., S. P. O'Connell, A. Banta, J. K. Fredrickson, A.-L. Reysenbach, T. L. Kieft, and F. S. Colwell. 2004. Microbiological comparison of core and groundwater samples collected from a fractured basalt aquifer with that of dialysis chambers incubated in situ. Geomicrobiol. J. 21:169-182. [Google Scholar]
- 34.Lehman, R. M., F. F. Roberto, D. Earley, D. F. Bruhn, S. E. Brink, S. P. O'Connell, M. E. Delwiche, and F. S. Colwell. 2001. Attached and unattached bacterial communities in a 120-meter corehole in an acidic, crystalline rock aquifer. Appl. Environ. Microbiol. 67:2095-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, L.-H., J. Hall, J. Lippmann-Pipke, J. A. Ward, B. S. Lollar, M. DeFlaun, R. Rothmel, D. P. Moser, T. M. Gihring, B. Mislowack, and T. C. Onstott. 2005. Radiolytic H2 in continental crust: nuclear power for deep subsurface microbial communities. Geochem. Geophys. Geosyst. 6:1-13. [Google Scholar]
- 36.Lin, L.-H., G. F. Slater, B. S. Lollar, G. Lacrampe-Couloume, and T. C. Onstott. 2005. The yield and isotopic composition of radiolytic H2, a potential energy source for the deep subsurface biosphere. Geochim. Cosmochim. Acta 69:893-903. [Google Scholar]
- 37.Lin, L.-H., P.-L. Wang, D. Rumble, J. Lippmann-Pipke, E. Boice, L. M. Pratt, B. S. Lollar, E. L. Brodie, T. C. Hazen, G. L. Andersen, T. Z. DeSantis, D. P. Moser, D. Kershaw, and T. C. Onstott. 2006. Long-term sustainability of a high-energy, low-diversity crustal biome. Science 314:479-482. [DOI] [PubMed] [Google Scholar]
- 38.Lozupone, C., M. Hamady, and R. Knight. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. Bioinformatics 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lozupone, C., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manning, A. H., and J. S. Caine. 4 April 2007. Groundwater noble gas, age, and temperature signatures in an alpine watershed: valuable tools in conceptual model development. Water Resour. Res. 43:W04404. doi: 10.1029/2006WR005349. [DOI]
- 42.Marchesi, J. R., A. J. Weightman, B. A. Cragg, R. J. Parkes, and J. C. Fry. 2001. Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadia Margin as revealed by 16S rRNA molecular analysis. FEMS Microbiol. Ecol. 34:221-228. [DOI] [PubMed] [Google Scholar]
- 43.Martin, A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGinley, K. J., G. F. Webster, and J. J. Leyden. 1978. Regional variations of cutaneous propionibacteria. Appl. Environ. Microbiol. 35:62-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mergeay, M., S. Monchy, T. Vallaeys, V. Auquier, A. Benotmane, P. Bertin, S. Taghavi, J. Dunn, D. van der Lelie, and R. Wattiez. 2003. Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes. FEMS Microbiol. Rev. 27:385-410. [DOI] [PubMed] [Google Scholar]
- 46.Mergeay, M., D. Nies, H. G. Schlegel, J. Gerits, P. Charles, and F. Van Gijsegem. 1985. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 162:328-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moser, D. P., T. C. Onstott, J. K. Fredrickson, F. J. Brockman, D. L. Balkwill, G. R. Drake, S. M. Pfiffner, and D. C. White. 2003. Temporal shifts in the geochemistry and microbial community structure of an ultradeep mine borehold following isolation. Geomicrobiol. J. 20:517-548. [Google Scholar]
- 48.Pearson, F. J., C. J. Noronha, and R. W. Andrews. 1983. Mathematical-modeling of the distribution of natural C-14, U-234, and U-238 in a regional groundwater system. Radiocarbon 25:291-300. [Google Scholar]
- 49.Pedersen, K. 1997. Microbial life in deep granitic rock. FEMS Microbiol. Rev. 20:399-414. [Google Scholar]
- 50.Pedersen, K., L. Hallbeck, J. Arlinger, A.-C. Erlandson, and N. Jahromi. 1997. Investigation of the potential for microbial contamination of deep granitic aquifers during drilling using 16S rRNA gene sequencing and cultureing methods. J. Microbiol. Methods 30:179-192. [Google Scholar]
- 51.Plummer, L. N., M. Bexfield, S. K. Anderson, and W. E. Sanford. 2004. Hydrochemical tracers in the middle Rio Grande Basin, USA. 1. Conceptualization of groundwater flow. Hydrogeol. J. 12:359-388. [Google Scholar]
- 52.Russell, B. F., T. J. Phelps, W. T. Griffin, and K. A. Sargent. 1992. Procedures for sampling deep subsurface microbial communities in unconsolidated sediments. Ground Water Monitor Rev. 12:96-104. [Google Scholar]
- 53.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmid, M., K. Walsh, R. Webb, W. I. Rijpstra, K. van de Pas-Schoonen, M. J. Verbruggen, T. Hill, B. Moffett, J. Fuerst, S. Schouten, J. S. Damste, J. Harris, P. Shaw, M. Jetten, and M. Strous. 2003. Candidatus “Scalindua brodae”, sp. nov., Candidatus “Scalindua wagneri”, sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol. 26:529-538. [DOI] [PubMed] [Google Scholar]
- 55.Smith, R. L., L. K. Baumgartner, D. N. Miller, D. A. Repert, and J. K. Bohlke. 2006. Assessment of nitrification potential in ground water using short term, single-well injection experiments. Microb. Ecol. 51:22-35. [DOI] [PubMed] [Google Scholar]
- 56.Spear, J. R., J. J. Walker, T. M. McCollom, and N. R. Pace. 2005. Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc. Natl. Acad. Sci. USA 102:2555-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spear, J. R., J. J. Walker, and N. R. Pace. 2005. Hydrogen and primary productivity: inference of biogeochemistry from phylogeny in a geothermal ecosystem, p. 113-128. In W. P. Inskeep and T. R. McDermott (ed.), Gemothermal biology and geochemistry in Yellowstone National Park. Thermal Biology Institute, Montana State University, Bozeman.
- 58.Stamatakis, A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688-2690. [DOI] [PubMed] [Google Scholar]
- 59.Stevens, T. O., and J. P. McKinley. 1995. Lithoautotrophic microbia, ecosystems in deep basalt aquifers. Science 270:450-454. [Google Scholar]
- 60.Strous, M., J. A. Fuerst, E. H. M. Kramer, S. Logemann, G. Muyzer, K. T. van de Pas-Schoonen, R. Webb, J. G. Kuenen, and M. S. M. Jetten. 1999. Missing lithotroph identified as a new planctomycete. Nature 400:446-449. [DOI] [PubMed] [Google Scholar]
- 61.Sundh, I., M. Nilsson, and P. Borga. 1997. Variation in microbial community structure in two boreal peatlands as determined by analysis of phospholipid fatty acid profiles. Appl. Environ. Microbiol. 63:1476-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taylor, J., and R. J. Parkes. 1983. The cellular fatty acids of sulphate-reducing bacteria, Desulfobacter sp., Desulfobulbus sp. and Desulfovibrio desulfuricans. J. Gen. Microbiol. 129:3303-3309. [Google Scholar]
- 63.Trimarco, E., D. Balkwill, M. Davidson, and T. C. Onstott. 2006. In situ enrichment of a diverse community of bacteria from a 4-5 km deep fault zone in South Africa. Geomicrobiol. J. 23:463-473. [Google Scholar]
- 64.White, D. C., W. M. Davis, J. S. Jickels, J. D. King, and R. J. Bobbie. 1979. Determination of the sedimentary microbial biomass by extractable lipid phosphate. Oeologia 40:51-62. [DOI] [PubMed] [Google Scholar]
- 65.White, D. C., and D. B. Ringelberg. 1998. Signature lipid biomarker analysis. Oxford University Press, Oxford, United Kingdom.