Abstract
Bacterial surface colonization is a universal adaptation strategy in aquatic environments. However, neither the identities of early colonizers nor the temporal changes in surface assemblages are well understood. To determine the identities of the most common bacterial primary colonizers and to assess the succession process, if any, of the bacterial assemblages during early stages of surface colonization in coastal water of the West Pacific Ocean, nonnutritive inert materials (glass, Plexiglas, and polyvinyl chloride) were employed as test surfaces and incubated in seawater off the Qingdao coast in the spring of 2005 for 24 and 72 h. Phylogenetic analysis of the 16S rRNA gene sequences amplified from the recovered surface-colonizing microbiota indicated that diverse bacteria colonized the submerged surfaces. Multivariate statistical cluster analyses indicated that the succession of early surface-colonizing bacterial assemblages followed sequential steps on all types of test surfaces. The Rhodobacterales, especially the marine Roseobacter clade members, formed the most common and dominant primary surface-colonizing bacterial group. Our current data, along with previous studies of the Atlantic coast, indicate that the Rhodobacterales bacteria are the dominant and ubiquitous primary surface colonizers in temperate coastal waters of the world and that microbial surface colonization follows a succession sequence. A conceptual model is proposed based on these findings, which may have important implications for understanding the structure, dynamics, and function of marine biofilms and for developing strategies to harness or control surface-associated microbial communities.
Submerged surfaces in aquatic environments are rapidly colonized by bacteria (14). Surface colonization may provide many advantages to the colonizers, such as (i) protection from adverse environments and predation, (ii) increasing nutrient availability and metabolic cooperativity, (iii) facilitating new genetic trait exchange and acquisition, (iv) maintaining extracellular enzyme activities, and (v) formation of a higher level of microbial organization (15, 23, 29, 38). Surface-colonizing bacteria play important roles in environmental and geomicrobiological processes, including biomineralization of organic matter, biodegradation of xenobiotic compounds, and biogeochemical cycling of nutrients and other earth elements (7, 18, 32, 40). Surface bacteria may have critical effects on larval settlement in benthic invertebrates (51), which is important in marine fisheries and ecosystems. They also participate in biocorrosion and biofouling (6), causing substantial damage and economic losses in marine engineering and industries. Surface colonization may provide a reservoir of harmful or pathogenic microbes as well (9, 43), posing problems and difficulties in public health and management of the environment. Despite this importance, very little is known about the structure and dynamics of the surface-colonizing microbiota in natural aquatic environments (15, 25).
Microbial surface colonization may proceed via ordered steps (15, 25, 31, 46, 52, 59), starting with initial sensing and interaction of the primary colonizers with surfaces (30, 42, 51). Prokaryotes are usually the major pioneer colonizers (34). Bacteria contribute the most to the initial surface microbiota, while Archaea are seldom detected (16, 60). Bacterial primary colonizers play critical roles in modifying the surface physicochemical properties by multiplication and production of extracellular polymeric substances (EPS) or other metabolites, making the surface suitable or unsuitable for recruitment of successive colonizers (10, 23). Primary colonizers may also influence the community function, as identified in dental biofilm formation (27). They are responsible for the initiation of biofouling as well and are important in the success or failure of microbiological engineering applications (62). These facts indicate the paramount importance of the bacterial primary colonizers in surface community formation, dynamics, and function.
The Rhodobacterales group bacteria have been found to be rapid primary surface colonizers in temperate Atlantic coastal waters, and surface community succession (the temporal change in microbial community composition and structure) has been identified (15, 16, 25). However, little is known about the identities of the primary colonizers and their succession pattern in other coastal waters of the world. The importance of surface-colonizing communities in medical, engineering, and natural environments has been recognized for a long time (13, 27, 43, 62). However, research progress in marine environments has been slow due to the lack of research effort and comprehensive understanding. Most research has focused on bacterial isolates as simplified models to reduce the complexity inherent in multispecies natural biofilms. It is difficult to extrapolate the results of such research to the real world in order to develop strategies to exploit or prevent surface microbial communities in natural environments (62). In the past, the complexity, technical difficulty, and lack of conceptual formulation hindered us from determining the identities and processes of the surface-colonizing bacteria in nature.
Cultivation-independent 16S rRNA gene clone library analyses were employed in the current study to determine the identities and dynamics of the early surface-colonizing bacteria in coastal water of the temperate West Pacific Ocean. Our goals were to (i) determine the phylogenetic affiliations of the most common primary colonizers on nonnutritive surfaces, (ii) identify the diversity and succession pattern, if any, of early bacterial surface colonizers, and (iii) compare the identities and colonization dynamics of the currently studied Pacific coast and the previously studied Atlantic coast (15, 16, 25) in order to identify any generic properties of bacterial surface colonization and succession in temperate coastal waters. Due to the global distribution of the Rhodobacterales group bacteria, especially the marine Roseobacter clade members (12, 57), and their importance in the establishment and succession of the surface-colonizing microbial assemblages discovered in the Atlantic coastal waters (15, 16, 25), we hypothesized that members of this bacterial group could be common primary surface colonizers that are ubiquitous and dominant in coastal environments of the temperate oceans.
MATERIALS AND METHODS
Surface incubation and microbe collection.
Glass, Plexiglas (polymethylmethacrylate), and polyvinyl chloride (PVC) plates (30 by 30 cm), with different degrees of surface hydrophilicity (glass > Plexiglas > PVC) according to water contact angle measurements obtained previously (26, 50), were used as test surfaces. So that data could be compared, a construct similar to that used in a previous experiment (15) was employed to deploy the surfaces in seawater at a depth of 1 m below the water surface near Wheat Island off the Qingdao coast (36.31°N, 120.25°E) on 23 May 2005. Three replicate test surfaces were used for each surface type. The average seawater salinity, temperature, and pH during the incubation period were 31.0‰, 16.0°C, and 8.0, respectively.
The original field surface incubation time frame of Dang and Lovell (15) was used to detect the early surface colonizers so that experiments performed in the two different world oceans could be compared. Using the procedures of Dang and Lovell (15), surface-colonizing microbes were collected after test surfaces were incubated in seawater for 24 or 72 h and concentrated for genomic DNA extraction.
Extraction of genomic DNA.
DNA was extracted using a modification of the method of Rivera et al. (47). The differences were as follows: the cell pellet was resuspended in 360 μl SET buffer (20% [wt/vol] sucrose, 50 mM Tris-HCl [pH 8.0], 50 mM EDTA), and the volumes of all solutions added thereafter were one-fifth the original volumes described (47). The quality and quantity of the DNA extracted from the surface-colonizing microbes were determined by electrophoresis on 0.8% (wt/vol) agarose gels in 0.5× Tris-borate-EDTA.
Amplification of 16S rRNA gene.
Bacteria-specific primers 27F and 1387R were used for 16S rRNA gene amplification (15). The PCR mixtures (final volume, 25 μl) contained each deoxynucleoside triphosphate at a concentration of 100 μM, 1.5 μM MgCl2, 1× Taq buffer, each primer at a concentration of 0.1 μM, and 1.5 U of Taq DNA polymerase (MBI, United States). PCR amplification was performed with a PTC-200 thermal controller (Bio-Rad, United States), and the following program was used: initial denaturation at 94°C for 3 min, followed by 25 cycles of denaturation at 94°C for 45 s, annealing at 55°C for 60 s, and extension at 72°C for 90 s and then a final extension step consisting of 72°C for 10 min. PCR products were examined by electrophoresis on a 1% agarose gel.
Construction of clone libraries.
Products from five PCRs were pooled, purified using a Gel Spin DNA purification kit (Novagen, United States), and ligated into pMD19-T simple vectors (Takara, Japan). The hybrid vectors were used to transform Escherichia coli TOP10 competent cells prepared by using the calcium chloride protocol (49). Recombinants were selected using LB indicator plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and isopropyl-β-d-thiogalactopyranoside (IPTG) with 100 μg/ml ampicillin. Clone libraries were constructed by random selection of approximate 100 white colonies from a single plate for each library. All clone libraries were named based on the surface type and incubation time; GL1 was the library constructed for a glass surface incubated for 24 h, GL2 was the library constructed for a glass surface incubated for 72 h, PM1 was the library constructed for a Plexiglas surface incubated for 24 h, PM2 was the library constructed for a Plexiglas surface incubated for 72 h, PV1 was the library constructed for a PVC surface incubated for 24 h, and PV2 was the library constructed for a PVC surface incubated for 72 h.
Amplified rRNA gene restriction analyses (ARDRA).
A plasmid miniprep method was used for recombinant plasmid preparation (15). Cloned 16S rRNA gene fragments were reamplified by using primers RV-M (5′-GAGCGGATAACAATTTCACACAGG-3′) and M13-D (5′-AGGGTTTTCCCAGTCACGACG-3′) flanking the insertion site of the cloning vector. The PCR mixtures (final volume, 25 μl) contained 1 μl of a plasmid preparation, each deoxynucleoside triphosphate at a concentration of 100 μM, 1.5 μM MgCl2, 1× Taq buffer, each primer at a concentration of 0.1 μM, and 1.5 U of Taq DNA polymerase (MBI, United States). The following thermal cycling program was used: initial denaturation at 94°C for 3 min, followed by 30 cycles consisting of 94°C for 45 s, 58°C for 60 s, and 72°C for 90 s and then a final extension step of 72°C for 10 min. PCR products were screened for the correct size and purity by 1% agarose gel electrophoresis.
Amplimers that were the correct size were restriction digested in separate reactions using MspI and HhaI (MBI, United States). The restriction fragments were resolved by electrophoresis on 4% agarose gels (Akeaibao, Spain) in 0.5× Tris-borate-EDTA and digitally photographed with an ImageMaster VDS imaging system (Pharmacia Biotech, United States). The band patterns were compared to identify redundant clones and to define operational taxonomic units (OTUs).
Statistical analyses.
The 16S rRNA gene library coverage was calculated as follows: [1 − (n1/N)] × 100, where n1 is the number of unique OTUs (frequency, 1) detected in the library and N is the total number of clones in the same library (39). The value obtained approximated the probability that all species present in a given sample were represented at least once in the library.
The sequence phylotype diversity and sampling efficiency of the libraries were analyzed by the rarefaction curve method using the aRarefactWin program (Steven Holland, Stratigraphy Lab, University of Georgia [http://www.uga.edu/strata/software/Software.html]).
Similarities of the surface-colonizing microbial assemblages on different test surfaces with different incubation periods were determined by performing cluster analyses of the 16S rRNA gene clone libraries with the software package SPSS11.5 (SPSS, United States). As studies have shown that biased PCR amplification occurs with heterogeneous DNA samples (56), log-transformed frequency data for the OTUs were used to minimize the influence of preferential PCR amplification (15). The frequency (F) was calculated as follows: F = (m/N) × 100, where m is the number of clones of an OTU in a library and N is the total number of clones in the same library. Two cluster analysis methods, hierarchical and k-means clustering, were employed to test the robustness of the analysis methods and the consistency of the analysis results.
16S rRNA gene sequence analyses.
A total of 236 unique OTUs were identified, and 126 of them were selected for sequencing and sequence analysis, including all the most common or predominant OTUs of the six clone libraries and all the OTUs obtained from the PM2 library. Two primers, V-27 (5′-GGATCTTCCAGAGATTAGRGTTTGATC-3′) and V-1387 (5′-CTTCCAGAGATTGGGCGGWGTG-3′), were designed to span both the pMD19-T simple cloning vector RV-M annealing end (boldface type indicates the vector end) and one of the 16S rRNA gene insert ends (underlined in primer V-27 for the 27F end and underlined in primer V-1387 for the 1387R end). The primers were used separately with primer M13-D for PCR determination of the orientations of the inserts. Primer M13-D or RV-M was chosen based on insert orientation, and sequencing was performed with an ABI 3770 automatic sequencer (Applied Biosystems, United States). The resultant sequences were usually about 800 bp long, covering at least the V1 to V3 hypervariable regions of the 16S rRNA gene.
Standard methods were used for bioinformatic determination of sequence affiliations (15). All sequences were submitted to the online RDP-II CHECK_CHIMERA program to check the sequence integrity and to check for chimeras (33). For each nonchimera sequence, the GenBank BLAST program was used to determine the nearest phylogenetic neighbor sequence (4). Sequences were aligned with the ClustalX program (55), and phylogenetic trees were constructed using the DNADIST and NEIGHBOR programs in the PHYLIP package (version 3.65c) (20).
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences determined have been deposited in the GenBank database under accession numbers EF215704 to EF215824.
RESULTS
ARDRA and surface assemblage succession.
Six 16S rRNA gene libraries were constructed for three surface types (glass, Plexiglas, and PVC) and two incubation periods (24 and 72 h). A total of 584 clones were screened, and 544 of them contained a proper insert. A total of 236 OTUs (unique ARDRA patterns or phylotypes) were identified, and 44 (18.6%) of them occurred in at least two libraries obtained with different surface types or incubation periods. The library coverage ranged from 50.0 to 80.0% (Table 1). Rarefaction analysis further confirmed the OTU phylotype diversity of the surface-colonizing microbiota (see Fig. S1 in the supplemental material).
TABLE 1.
Analysis of the clone libraries constructed for three surface types and two incubation periods (24 and 72 h)
| Library | No. of clones | No. of OTUs | % Coveragea | No. of OTUs shared by libraries from the same surface type |
|---|---|---|---|---|
| GL1 | 94 | 56 | 62.4 | 7 |
| GL2 | 90 | 65 | 50.0 | |
| PM1 | 91 | 51 | 69.2 | 11 |
| PM2 | 85 | 35 | 80.0 | |
| PV1 | 88 | 54 | 69.3 | 14 |
| PV2 | 91 | 50 | 74.7 |
The coverage values were calculated as described in Materials and Methods.
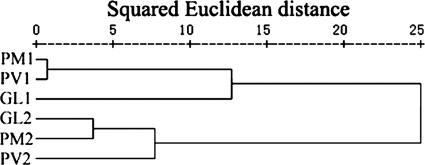
Clone libraries obtained from the same surface type but with different incubation periods typically shared only 7 to 14 common OTUs (Table 1), accounting for 10.8 to 31.4% of the OTUs in each of the libraries. Surface microbiota succession was further confirmed by statistical analyses. The hierarchical (Fig. 1) and k-means (data not shown) clustering results were consistent and showed that the libraries obtained from different surface types but with the same incubation period were grouped together. All the clone libraries obtained from the 24-h incubation (GL1, PM1, and PV1) were grouped into a single class, and the three other libraries, obtained from the 72-h incubation (GL2, PM2, and PV2), were grouped into another single class, demonstrating that the early colonization process was highly dynamic and that succession changes occurred on all test surfaces, regardless of the surface hydrophilicity characteristics.
FIG. 1.
Dendrogram based on a hierarchical clustering analysis of the 16S rRNA gene clone libraries, constructed using the squared Euclidean distance similarity and Ward linkage procedures.
16S rRNA gene sequence analyses.
Of the 126 OTUs selected for sequence analysis, which accounted for 431 (79.2%) of the 544 clones with the proper insert size, only 5 (each from a single clone) were probably chimeras based on CHECK_CHIMERA results. Slightly more than one-half (54.5%) of the 121 valid OTUs had high levels of similarity (≥97% identity) with known sequences in the GenBank database, and 18.2% were only moderately related (<94% identity) to known sequences. Phylogenetic analysis showed that 106 OTUs were affiliated with the domain Bacteria, and the remaining 15 OTUs were related to mitochondrion or chloroplast 16S rRNA genes from algae. Besides the predominant prokaryotes, microalgae and algal spores also contributed to the colonization of freshly incubated surfaces in Qingdao coastal water.
Seven bacterial groups were obtained from the clone libraries, including the following divisions or phyla: Actinobacteria, Bacteroidetes, Cyanobacteria, Firmicutes, Planctomycetes, Proteobacteria, and Spirochaetes (see Fig. S2 in the supplemental material). Four Proteobacteria subdivisions were present (α-, β-, γ-, and δ-Proteobacteria). The α- and γ-Proteobacteria sequences were highly diverse in our libraries and accounted for 24.8 and 34.7%, respectively, of the 121 OTUs characterized; they each accounted for 33.1% of the 426 clones analyzed (excluding the 5 chimeric clones). Bacteroidetes sequences were diverse as well, accounting for 18.2% of the OTUs characterized and 8.9% of the clones analyzed.
In order to obtain a better understanding of the surface colonizer diversity, all 36 OTUs in the PM2 library were sequenced, which resulted in one chimera and 35 valid sequences. The nonchimera sequences were affiliated with the α-, β-, γ-, and δ-Proteobacteria, Bacteroidetes, Cyanobacteria, and plastids of some marine algae. OTUs affiliated with the α- and γ-Proteobacteria showed the greatest diversity (40.0 and 31.4% of the 35 OTUs), and OTUs belonging to Bacteroidetes were diverse (14.3%) as well. Sequences affiliated with Loktanella, Ochrobactrum, Pseudoalteromonas, and the β-Proteobacteria were found only in the PM2 library, the least diverse library in the current study (Table 1; see Fig. S1 in the supplemental material), indicating that new bacterial groups might be found among the remaining unsequenced OTUs of the other libraries and that greater phylotype diversity should be expected. The 121 OTUs characterized might have covered the diversity of the surface colonizers in the coastal environment studied only partially. However, these OTUs might be the most important OTUs in terms of occurrence and dominance in the libraries.
(i) Sequences of the α-Proteobacteria.
The α-Proteobacteria clones dominated the libraries, accounting for 33.1% of the 426 clones analyzed (11.1% of the 24-h library clones and 53.9% of the 72-h library clones). The significant increase in abundance with time was probably caused by both growth and recruitment on the surfaces.
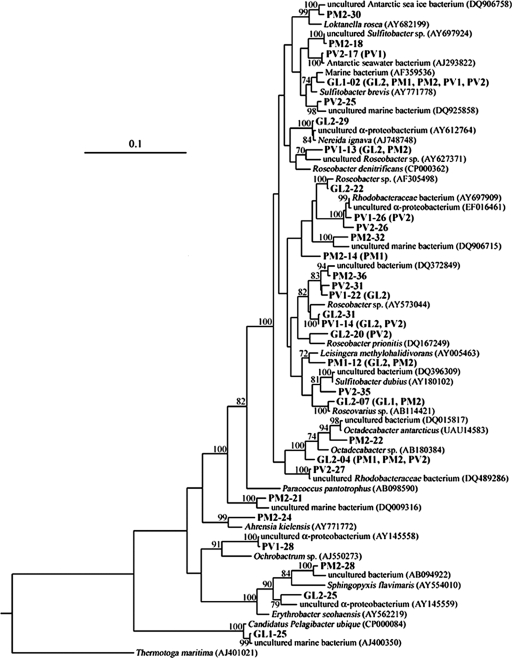
Thirty sequenced OTUs (141 clones) were affiliated with the α-Proteobacteria and were related to the genera Ahrensia, Erythrobacter, Leisingera, Loktanella, Nereida, Octadecabacter, Ochrobactrum, Paracoccus, Roseobacter, Roseovarius, Sphingopyxis, Sulfitobacter, and “Candidatus Pelagibacter” (Fig. 2). Twenty-six OTUs (137 clones) were affiliated with the Rhodobacterales. All of them belonged to the Rhodobacteraceae subgroup, and they included 24 OTUs (135 clones) belonging to the marine Roseobacter clade, including the genera Leisingera, Loktanella, Nereida, Octadecabacter, Roseobacter, Roseovarius, and Sulfitobacter. Besides being quite diverse, the clones of the Roseobacter clade bacteria also were dominant (31.7%) in the libraries and had high levels of sequence similarity (96 to 99%) to known GenBank sequences. There was a significant increase in clone abundance with the increase in surface incubation time. The clones related to Rhodobacterales and especially the Roseobacter clade accounted for 10.1% of the clones in the 24-h libraries and for 53.0 and 52.1% of the clones, respectively, in the 72-h libraries.
FIG. 2.
Phylogenetic tree of the α-Proteobacteria sequences recovered from the six 16S rRNA gene clone libraries, constructed using the neighbor-joining method with the Kimura two-parameter model for nucleotide change. The branch distances indicate nucleotide substitution rates. Scale bar = 0.1 expected change per homologous nucleotide position. Bootstrap values greater than 70% (based on 100 bootstrap resamplings) are indicated at the nodes. OTUs that occurred in two or more libraries are shown, and the additional library names are indicated in parentheses.
The most common Roseobacter clade OTUs belonged to Sulfitobacter, which occurred in all six libraries and accounted for 35.6% of the 135 Roseobacter clade clones detected in the libraries. OTU GL1-02, which was affiliated with Sulfitobacter brevis, was present in all clone libraries. In addition, clones related to Octadecabacter and Roseobacter were also common and abundant, accounting for 18.5 and 17.0%, respectively, of the 135 Roseobacter clade clones. Three OTUs were related to Octadecabacter, and OTU GL2-04 occurred in four libraries. Several OTUs affiliated with Roseobacter occurred in at least two libraries, and sequences of this genus were found in five libraries.
Comparison of the 24- and 72-h libraries indicated that the α-Proteobacteria OTUs exhibited obvious succession during early stages of bacterial surface colonization in the coastal environment studied. These OTUs accounted for 10.3, 12.9, and 19.4% of the sequenced OTUs in libraries GL1, PM1, and PV1, respectively, and for 36.4, 40.0, and 29.7% of the sequenced OTUs in libraries GL2, PM2, and PV2, respectively (see Fig. S3 in the supplemental material). The clone abundance increased even more dramatically (see Fig. S4 in the supplemental material). As the major group of the α-Proteobacteria detected, the Rhodobacterales (especially the Roseobacter clade) OTUs also showed obvious succession with the increase in surface incubation time; they accounted for 6.9% (6.9%), 12.9% (12.9%), and 16.7% (16.7%) of the OTUs in the 24-h libraries and for 33.3% (33.3%), 37.1% (31.4%), and 29.7% (29.7%) of the OTUs in the 72-h libraries. Most of the 24-h α-Proteobacteria OTUs were also present in the 72-h libraries; the only exceptions were the two sequences that did not belong to the Rhodobacterales group (Fig. 2). Eighteen α-Proteobacteria OTUs were present only in the 72-h libraries; none of them was found in the 24-h libraries.
The Rhodobacterales bacteria, especially those in the marine Roseobacter clade, might represent a successful primary surface-colonizing bacterial group with the potential to colonize, grow, and recruit on submerged surfaces during early stages of surface community development in the Qingdao coastal water studied.
(ii) Sequences of the γ-Proteobacteria.
The γ-Proteobacteria OTUs were abundant as well, with 42 OTUs accounting for 33.1% of the 426 clones analyzed. They were quite diverse and were related to Alteromonas, Glaciecola, Halomonas, Methylococcus, Oceanospirillum, Oleispira, Pseudoalteromonas, Psychrobacter, Vibrio, some unidentified γ-Proteobacteria, and marine invertebrate bacterial symbionts (see Fig. S5 in the supplemental material). Oceanospirillales sequences were the most dominant γ-Proteobacteria phylotypes in the libraries, accounting for 42.9% of the 42 OTUs and 69.5% of the 141 related clones. These sequences had 89 to 99% similarity to known GenBank sequences, including sequences of Halomonas, Oleispira, Oceanospirillum, and some uncultured bacteria. Five sequenced OTUs were affiliated with Alteromonadales, with 94 to 99% similarity to known sequences, including sequences of Alteromonas, Glaciecola, and Pseudoalteromonas.
The Oceanospirillales OTUs were found in all the libraries with high frequency. OTUs GL-03, GL1-04, and GL1-05 occurred in five libraries. Several other OTUs were found in two to four clone libraries. In addition, the sequences that were related to some marine invertebrate bacterial symbionts and some uncultured γ-Proteobacteria also occurred frequently in the libraries. OTU PM1-06 was found in four libraries, GL2-08 was found in three libraries, and GL1-17 and PV2-21 were found in two libraries.
The surface-colonizing γ-Proteobacteria assemblage was quite dynamic based on comparison analyses of the 24- and 72-h libraries. Sixteen OTUs were present only in the 24-h libraries, and 17 OTUs were present only in the 72-h libraries. For the Oceanospirillales, eight OTUs were present only in the 24-h libraries and three OTUs were present only in the 72-h libraries. The γ-Proteobacteria OTUs were dominant in both the 24- and 72-h libraries, accounting for 27.6, 54.8, and 36.1% of the sequenced OTUs in libraries GL1, PM1, and PV1, respectively, and 36.4, 31.4, and 32.4% of the sequenced OTUs in libraries GL2, PM2, and PV2, respectively (see Fig. S3 in the supplemental material). However, the relative abundance of the γ-Proteobacteria clones decreased from 40.1% in the 24-h libraries to 26.5% in the 72-h libraries (see Fig. S4 in the supplemental material). The abundance of the Oceanospirillales clones, the major group of the surface-colonizing γ-Proteobacteria in the Qingdao coastal environment studied, decreased as well. The Oceanospirillales clones accounted for 10.4, 60.0, and 27.1% of the clones analyzed in libraries GL1, PM1, and PV1, respectively, and 8.6, 18.8, and 11.8% of the clones analyzed in libraries GL2, PM2, and PV2, respectively. The Oceanospirillales bacteria might represent a fast-surface-colonizing bacterial group, while the contribution of the other γ-Proteobacteria to development of the surface microbiota might be less important. Except for the Oceanospirillales species, many of the γ-Proteobacteria might leave the colonized surface early, and the strains that remain might not establish stable populations very well on the submerged surfaces.
(iii) Sequences of the Bacteroidetes.
Twenty-two sequenced OTUs (38 clones) were affiliated with the Bacteroidetes, including the groups Bacteroidales, Flavobacteriales, and Sphingobacteriales (see Fig. S6 in the supplemental material). The Flavobacteriales OTUs had 91 to 99% similarity to known GenBank sequences. They were the major members of the Bacteroidetes detected in the libraries, accounting for 81.8% of the related OTUs. These bacteria were highly diverse and were related to the genera Dokdonia, Fluviicola, Formosa, Maribacter, Mesonia, Pibocella, Polaribacter, Psychroserpens, Tenacibaculum, and Ulvibacter.
The Bacteroidetes OTUs accounted for 10.3, 19.4, and 16.7% of the sequenced OTUs in the GL1, PM1, and PV1 libraries, respectively, and 12.1, 14.3, and 24.3% of the sequenced OTUs in the GL2, PM2, and PV2 libraries, respectively (see Fig. S3 in the supplemental material). Eight OTUs occurred only in the 24-h libraries, and 10 OTUs occurred only in the 72-h libraries. The Bacteroidetes and Flavobacteriales clones accounted for 9.2 and 6.8% of the OTUs in the 24-h libraries, respectively, and 8.7 and 8.2% of the OTUs in the 72-h libraries, respectively (see Fig. S4 in the supplemental material).
DISCUSSION
In the current study, PCR-based bacterial community analyses were employed to study the primary colonizers and succession process in early stages of surface colonization at the Qingdoo coast of the temperate West Pacific Ocean. So that the results could be compared with the results of previous studies of the Atlantic coast (15), the same experimental and analysis procedures were used, except for the DNA extraction method and PCR conditions. Our PCR procedure included 25 cycles, and the annealing temperature was 55 °C in order to obtain exponential amplification of the diverse 16S rRNA gene targets and to minimize the occurrence of artifacts and bias. This was a significant improvement over the previous PCR procedures (15, 25). Sequences of the common and dominant surface-colonizing bacteria were probably amplified and recovered using the improved PCR conditions. The molecular technique used provided a means to probe the underappreciated issue of microbial surface colonization in a natural environment.
Cluster analyses indicated that the microbial assemblages obtained with the same surface incubation time were similar regardless of the surface type, and the early colonizing process was highly dynamic, consistent with the early findings obtained for the Atlantic coast (15, 16, 25). These findings indicate that successional change in microbial communities in early stages of surface colonization is a common feature in temperate coastal waters. However, it is also important to recognize that OTUs identified in environmental libraries reflect only the number of different sequences and not the number of different species originally in the samples (15, 19). Fluorescence in situ hybridization (FISH) will be carried out to further test our findings (16).
Submerged surfaces in aquatic environments may be rapidly covered by a conditioning film of organic compounds, which affects the surface physicochemical properties prior to bacterial adhesion (21). The exposure to seawater might result in convergence of surface hydrophobicity, roughness, and nutrient conditions due to the formation of a conditioning film (5). Thus, the initial surface-colonizing communities may be similar due to the masking effect of the conditioning film on the surface chemistry and to some common rules that may govern the microbial cell-surface interactions (25), resulting in similar initial surface-colonizing bacterial communities on distinct surface types in coastal waters in different regions of the world (15, 16, 25).
Our data indicated that clones of the surface-colonizing Rhodobacterales, especially the marine Roseobacter clade, became more diverse and dominant with an increase in the surface incubation time. Analyses based on clone libraries and FISH quantification consistently demonstrated that the Rhodobacterales bacteria were the ubiquitous dominant early surface colonizers in the Atlantic coastal waters (15, 16, 25), and they were also dominant on marine particles (17). Similar conclusions can be drawn across half of the globe from the Atlantic Ocean to the Pacific Ocean. The marine Roseobacter clade bacteria have also frequently been isolated from surfaces or biofilms in marine waters (28). All these findings indicate that the Rhodobacterales group bacteria are the primary surface colonizers in temperate coastal waters of the world.
Besides being abundant, ubiquitous, and frequently surface associated (57), the Roseobacter clade bacteria are physiologically active as well, and they are the fastest utilizers of nutrient enrichments in coastal environments (1-3). These bacteria play important roles in marine ecosystems, such as carbon and sulfur cycling and trace metal reduction (12, 57). To adapt to surface life, bacteria may also exploit intercellular communication and interaction (36, 37), facilitating diverse collective behaviors within the colonizing community, such as cooperation and/or competition (10, 44, 61). Many Roseobacter clade bacteria produce acylated homoserine lactones (22, 35, 58), the quorum-sensing signals involved in various microbial processes, such as biofilm formation and development. Thus, these bacteria may have the ability to shape the subsequent biofilm community structure and development via cell-cell communication and interaction. Our current data, along with previous studies (15, 16, 25), indicated the global importance of the Roseobacter clade bacteria in early surface colonization and community succession.
The surface Roseobacter clade OTUs from the Atlantic coastal waters were related to Roseobacter, Roseovarius, Ruegeria, and Sagittula (15, 16, 25), mainly in the Antarctobacter-Sagittula, Octadecabacter-Ruegeria, Roseobacter, and Silicibacter-Ruegeria superlineages, while some of the Qingdao coast sequences were mainly affiliated with the Octadecabacter-Ruegeria, Roseobacter, and Sulfitobacter-Staleya-Oceanibulbus superlineages as defined previously by Buchan et al. (12). Both similar and distinct bacteria lineages within the Roseobacter clade could be identified as primary surface colonizers in different coastal waters of the world. The local microbial composition and environmental factors might have important effects on the primary surface-colonizing microbiota.
The γ-Proteobacteria, especially the Oceanospirillales, were also very diverse and common on the submerged surfaces at the Qingdao coast. However, Oceanospirillales sequences were only occasionally found in the surface 16S rRNA gene libraries from the Atlantic coast (15, 16, 25), where the most important surface-colonizing γ-Proteobacteria might be the Alteromonadales species (15, 25). Thus, for γ-Proteobacteria, the early surface assemblages might be quite different in distinct coastal waters of the world.
FISH quantification indicated that the γ-Proteobacteria contributed less to the surface-colonizing microbiota on submerged glass, stainless steel, and polycarbonate surfaces in the Atlantic coastal waters than the Rhodobacterales bacteria contributed (16, 25). Our current study indicated that the relative abundance of the γ-Proteobacteria clones decreased with surface incubation time. The γ-Proteobacteria might be fast-surface-colonizing species. However, most of them, with the possible exception of the Oceanospirillales species, could not establish stable colonizing populations in the early steps of surface community development, in contrast to the surface-colonizing Rhodobacterales species. It has been found that certain Roseobacter clade bacteria produce antibacterial compounds when they are growing on a surface, preventing other bacteria, including γ-Proteobacteria, from colonizing the surface and forming a biofilm (8, 10, 11, 24, 44, 48). A strain was even found to invade and disperse preestablished biofilms formed by other bacteria (45). Genes encoding nonribosomal peptide synthetases and polyketide synthases, key enzymes involved in the synthesis of various pharmaceutically important natural products, have been identified in many Roseobacter clade bacteria (35). The ability to produce antibacterial compounds and the enhancement of biofilm formation via cell aggregate formation may give the Roseobacter clade bacteria selective advantages that help explain their prevalence and dominance in marine surface microbiota (11, 35, 46, 57). Whether the decrease in the γ-Proteobacteria relative abundance on the submerged surfaces in Qingdao coast water was related to the potential inhibitory effect of the surface-colonizing Roseobacter clade bacteria during early stages of surface microbiota development is worth further investigation.
The Bacteroidetes are often abundant in riverine and estuarine biofilms and particles (25, 31, 41). However, the success of Bacteroidetes in particle assemblages may be due more to their use of detritus as a carbon source rather than to their preference for surfaces (25). Although diverse Bacteroidetes sequences, especially those of the Flavobacteriales group, were found in our clone libraries, most of these sequences were not abundant. However, the importance of Bacteroidetes could be underestimated, as their sequences were usually underrepresented in 16S rRNA gene clone libraries constructed from environmental samples (25).
Based on our current knowledge and understanding, a conceptual model sequence of surface-colonizing bacterial community dynamics during early stages of development is proposed, with the Rhodobacterales, especially the marine Roseobacter clade bacteria, as the keystone primary surface colonizers. The major steps are described below.
The first step is the initial deposition and attachment of free-living bacteria on a conditioning film-covered submerged surface. The Roseobacter clade bacteria may be among the fastest colonizers on freshly submerged surfaces (15, 16, 25). These bacteria not only are abundant and ubiquitous in coastal waters (57) but also react to a low level of nutrient enrichment faster than other bacteria, such as the γ-Proteobacteria and Bacteroidetes (1-3). The conditioning film could be a significant source of nutrients for the Roseobacter bacteria (5, 21). The initial attachment may be an active and selective process, and surface sensing via two-component regulatory systems may be important (30). Furthermore, the formation of a conditioning film may result in convergence of surface physicochemical properties, masking the original differences of the surfaces (5) and encouraging selection of specific bacterial groups to develop and harness specific physiological and genetic adaptive advantages.
The second step is competition for surface resources via active growth and recruitment. This step involves quick growth of the Roseobacter clade bacteria to form microcolonies on surfaces, stimulated by surface-accumulated nutrients (1-3, 15, 16, 25). The Roseobacter and other bacteria may also recruit cells on the surfaces via formation of aggregates consisting of surface-attached cells and aquatic free-living cells (46, 57). During this step, due to quick utilization of the nutrients that were originally accumulated on the submerged surfaces by the Roseobacter clade bacteria, some of the initially attached bacteria belonging to other groups, such as the γ-Proteobacteria, may actually leave the surfaces. The increase in the number of key primary surface colonizers, such as the Roseobacter clade bacteria, and the decrease in the number of other bacteria are the main dynamic characteristics at this stage.
The third step is competition for surface resources via quorum-sensing-regulated processes, such as EPS and antibiotic production. Once the Roseobacter clade bacterial populations reach a certain density, they may actively expel or take advantage of the other surface-attaching bacteria via quorum-sensing-regulated antibiotic production and other mechanisms (8, 10, 11, 24, 35, 44, 45, 48, 57). Quorum sensing may also be important for multispecies biofilm formation (22, 35, 58). Microbial competition and cooperation may happen at the same time on a surface via cell-cell communication and interaction.
The fourth step is a shift in the surface-colonizing microbial community. Once the Roseobacter clade bacterial climax population is reached, some of the cells may leave the surface to avoid overcrowding, or they may become less active due to limited availability of nutrients and other resources or to aging and death of the bacterial cells because of phage infection, protozoan grazing, or other microbial interactions (23, 42, 52, 54). The accumulation of EPS and other biopolymers may make the surface physicochemical properties significantly different from those of the initial conditioning film-covered surface, thus allowing other microbes to grow or colonize (63). One of the options may be the Bacteroidetes bacteria, which are active biopolymer utilizers and are abundant in natural aquatic biofilms and particles (25, 41, 60). The biofilm matrix structure may vary depending on the microbes present, their physiological status, the nutrients available, and the prevailing physicochemical conditions (53). A change in the biofilm matrix composition may occur with the change in the bacterial community. At this stage, the surface-colonizing bacterial community and biofilm matrix composition shift from the early primary structure to the secondary structure usually seen in nature.
In this simplified conceptual model, the early surface colonizers, such as the Roseobacter clade bacteria (15, 16, 25), are the initial active utilizers of surface resources during surface microbiota formation and development (1-3). These bacteria convert the initial surface-accumulated nutrients and energy (whose levels may be very low in nature) into biomass and extracellular products, which may be exploited by the late-stage microbes for nutrients or microhabitats (63). As the keystone primary colonizers, the Roseobacter bacteria provide the important transition of the submerged surfaces from low-nutrient status (covered with conditioning film) to high-nutrient status (covered with abundant EPS and other biopolymers) for the succession and maturation of the surface-colonizing microbial community. The Roseobacter bacteria may also influence the surface community structure, dynamics, and function via cell-cell communication and interaction (8, 10, 11, 22, 24, 35, 44-46, 48, 57, 58). Decoding the cellular and ecological processes of the keystone primary colonizers should significantly increase our knowledge of marine biofilm formation, development, and function.
The current study, along with previous studies, demonstrated the global importance of the Rhodobacterales group bacteria in microbial surface colonization and community succession in coastal environments. The conclusion that the Rhodobacterales group bacteria are the keystone primary surface colonizers is based on several lines of evidence, including their abundance, omnipresence, cellular physiology involving quick reactions to low-level nutrient enrichment (such as the formation of conditioning films on submerged surfaces), genetic potential for cell-cell communication and interaction, and ecological and environmental functions, as shown in the various studies cited in this paper. However, these key primary surface colonizers have seldom been studied to determine their roles in surface microbiota structure, dynamics, and function. Further investigation of how bacteria interact with surfaces and with one another and what common rules govern the initial colonization and succession process should help workers decode biofilm ecology in marine environments and develop strategies to harness or control the surface-associated microbial communities.
Supplementary Material
Acknowledgments
This work was financially supported by the Pilot Projects of Knowledge Innovation Project (CAS grants KZCX2-YW-211-03 and KZCX3-SW-233), the National Natural Science Foundation of China (grants 40476058 and 40576069), and China Ocean Mineral Resources R&D Association grants (COMRA acceptances 020 and 126).
Footnotes
Published ahead of print on 26 October 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alonso, C., and J. Pernthaler. 2006. Concentration-dependent patterns of leucine incorporation by coastal picoplankton. Appl. Environ. Microbiol. 72:2141-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso, C., and J. Pernthaler. 2006. Roseobacter and SAR11 dominate microbial glucose uptake in coastal North Sea waters. Environ. Microbiol. 8:2022-2030. [DOI] [PubMed] [Google Scholar]
- 3.Alonso-Saez, L., and J. M. Gasol. 2007. Seasonal variations in the contributions of different bacterial groups to the uptake of low-molecular-weight compounds in northwestern Mediterranean coastal waters. Appl. Environ. Microbiol. 73:3528-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakker, D. P., H. J. Busscher, J. van Zanten, J. de Vries, J. W. Klijnstra, and H. C. van der Mei. 2004. Multiple linear regression analysis of bacterial deposition to polyurethane coatings after conditioning film formation in the marine environment. Microbiology 150:1779-1784. [DOI] [PubMed] [Google Scholar]
- 6.Beech, I. B., J. A. Sunner, and K. Hiraoka. 2005. Microbe-surface interactions in biofouling and biocorrosion processes. Int. Microbiol. 8:157-168. [PubMed] [Google Scholar]
- 7.Bhaskar, P. V., and N. B. Bhosle. 2005. Microbial extracellular polymeric substances in marine biogeochemical processes. Curr. Sci. 88:45-53. [Google Scholar]
- 8.Brinkhoff, T., G. Bach, T. Heidorn, L. Liang, A. Schlingloff, and M. Simon. 2004. Antibiotic production by a Roseobacter clade-affiliated species from the German Wadden Sea and its antagonistic effects on indigenous isolates. Appl. Environ. Microbiol. 70:2560-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, M. R. W., and J. Barker. 1999. Unexplored reservoirs of pathogenic bacteria: protozoa and biofilms. Trends Microbiol. 7:46-50. [DOI] [PubMed] [Google Scholar]
- 10.Bruhn, J. B., K. F. Nielsen, M. Hjelm, M. Hansen, J. Bresciani, S. Schulz, and L. Gram. 2005. Ecology, inhibitory activity, and morphogenesis of a marine antagonistic bacterium belonging to the Roseobacter clade. Appl. Environ. Microbiol. 71:7263-7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruhn, J. B., L. Gram, and R. Belas. 2007. Production of antibacterial compounds and biofilm formation by Roseobacter species are influenced by culture conditions. Appl. Environ. Microbiol. 73:442-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchan, A., J. M. Gonzalez, and M. A. Moran. 2005. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71:5665-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costerton, J. W., K.-I. Cheng, G. G. Geesey, T. I. Ladd, N. C. Nickel, M. Dosgupton, and I. J. Marine. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41:435-464. [DOI] [PubMed] [Google Scholar]
- 14.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 15.Dang, H. Y., and C. R. Lovell. 2000. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 66:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dang, H. Y., and C. R. Lovell. 2002. Numerical dominance and phylotype diversity of marine Rhodobacter species during early colonization of submerged surfaces in coastal marine waters as determined by 16S ribosomal DNA sequence analysis and fluorescence in situ hybridization. Appl. Environ. Microbiol. 68:496-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dang, H. Y., and C. R. Lovell. 2002. Seasonal dynamics of particle-associated and free-living marine Proteobacteria in a salt marsh tidal creek as determined using fluorescence in situ hybridization. Environ. Microbiol. 4:287-295. [DOI] [PubMed] [Google Scholar]
- 18.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farrelly, V., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 21.Fletcher, M. 1996. Bacterial attachment in aquatic environments: a diversity of surfaces and adhesion strategies, p. 1-24. In M. Fletcher (ed.), Bacterial adhesion: molecular and ecological diversity. Wiley-Liss, New York, NY.
- 22.Gram, L., H.-P. Grossart, A. Schlingloff, and T. Kiorboe. 2002. Possible quorum sensing in marine snow bacteria: production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl. Environ. Microbiol. 68:4111-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 24.Hjelm, M., A. Riaza, F. Formoso, J. Melchiorsen, and L. Gram. 2004. Seasonal incidence of autochthonous antagonistic Roseobacter spp. and Vibrionaceae strains in a turbot larva (Scophthalmus maximus) rearing system. Appl. Environ. Microbiol. 70:7288-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, P. R., M. T. Cottrell, D. L. Kirchman, and S. C. Dexter. 2007. Bacterial community structure of biofilms on artificial surfaces in an estuary. Microb. Ecol. 53:153-162. [DOI] [PubMed] [Google Scholar]
- 26.Katsikogianni, M., I. Spiliopoulou, D. P. Dowling, and Y. F. Missirlis. 2006. Adhesion of slime producing Staphylococcus epidermidis strains to PVC and diamond-like carbon/silver/fluorinated coatings. J. Mater. Sci. Mater. Med. 17:679-689. [DOI] [PubMed] [Google Scholar]
- 27.Kolenbrander, P. E., R. N. Andersen, D. S. Blehert, P. G. Egland, J. S. Foster, and R. J. Palmer, Jr. 2002. Communication among oral bacteria. Microbiol. Mol. Biol. Rev. 66:486-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwon, K. K., H. S. Lee, S.-Y. Jung, J.-H. Yim, J.-H. Lee, and H. K. Lee. 2002. Isolation and identification of biofilm-forming marine bacteria on glass surfaces in Dae-Ho Dike, Korea. J. Microbiol. 40:260-266. [Google Scholar]
- 29.Lehman, R. M., and S. P. O'Connell. 2002. Comparison of extracellular enzyme activities and community composition of attached and free-living bacteria in porous medium columns. Appl. Environ. Microbiol. 68:1569-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lejeune, P. 2003. Contamination of abiotic surfaces: what a colonizing bacterium sees and how to blur it. Trends Microbiol. 11:179-184. [DOI] [PubMed] [Google Scholar]
- 31.Lyautey, E., C. R. Jackson, J. Cayrou, J. L. Rols, and F. Garabetian. 2005. Bacterial community succession in natural river biofilm assemblages. Microb. Ecol. 50:589-601. [DOI] [PubMed] [Google Scholar]
- 32.Magalhaes, C. M., A. A. Bordalo, and W. J. Wiebe. 2003. Intertidal biofilms on rocky substratum can play a major role in estuarine carbon and nutrient dynamics. Mar. Ecol. Prog. Ser. 258:275-281. [Google Scholar]
- 33.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marszalek, D. S., S. M. Gerchakov, and L. R. Udey. 1979. Influence of substrate composition on marine microfouling. Appl. Environ. Microbiol. 38:987-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martens, T., L. Gram, H. P. Grossart, D. Kessler, R. Muller, M. Simon, S. C. Wenzel, and T. Brinkhoff. 2007. Bacteria of the Roseobacter clade show potential for secondary metabolite production. Microb. Ecol. 54:31-42. [DOI] [PubMed] [Google Scholar]
- 36.McLean, R. J., M. B. Barnes, M. K. Windham, M. Merchant, M. R. Forstner, and C. Fuqua. 2005. Cell-cell influences on bacterial community development in aquatic biofilms. Appl. Environ. Microbiol. 71:8987-8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLean, R. J., M. Whiteley, D. J. Stickler, and W. C. Fuqua. 1997. Evidence of autoinducer activity in naturally occurring biofilms. FEMS Microbiol. Lett. 154:259-263. [DOI] [PubMed] [Google Scholar]
- 38.Molin, S., and T. Tolker-Nielsen. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14:255-261. [DOI] [PubMed] [Google Scholar]
- 39.Mullins, T. D., T. B. Britschgi, R. L. Krest, and S. J. Giovannoni. 1995. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol. Oceanogr. 40:148-158. [Google Scholar]
- 40.Newman, D. K., and J. F. Banfield. 2002. Geomicrobiology: how molecular-scale interactions underpin biogeochemical systems. Science 296:1071-1077. [DOI] [PubMed] [Google Scholar]
- 41.Nocker, A., J. E. Lepo, and R. A. Snyder. 2004. Influence of an oyster reef on development of the microbial heterotrophic community of an estuarine biofilm. Appl. Environ. Microbiol. 70:6834-6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 43.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 44.Rao, D., J. S. Webb, and S. Kjelleberg. 2005. Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 71:1729-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rao, D., J. S. Webb, and S. Kjelleberg. 2006. Microbial colonization and competition on the marine alga Ulva australis. Appl. Environ. Microbiol. 72:5547-5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rickard, A. H., P. Gilbert, N. J. High, P. E. Kolenbrander, and P. S. Handley. 2003. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 11:94-100. [DOI] [PubMed] [Google Scholar]
- 47.Rivera, I. N. G., E. K. Lipp, A. Gil, N. Choopun, A. Huq, and R. R. Colwell. 2003. Method of DNA extraction and application of multiplex polymerase chain reaction to detect toxigenic Vibrio cholerae O1 and O139 from aquatic ecosystems. Environ. Microbiol. 5:599-606. [DOI] [PubMed] [Google Scholar]
- 48.Ruiz-Ponte, C., J. F. Samain, J. L. Sanchez, and J. L. Nicolas. 1999. The benefit of a Roseobacter species on the survival of scallop larvae. Mar. Biotechnol. 1:52-59. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 50.Shadpour, H., H. Musyimi, J. Chen, and S. A. Soper. 2006. Physiochemical properties of various polymer substrates and their effects on microchip electrophoresis performance. J. Chromatogr. A 1111:238-251. [DOI] [PubMed] [Google Scholar]
- 51.Steinberg, P. D., R. De Nys, and S. Kjelleberg. 2002. Chemical cues for surface colonization. J. Chem. Ecol. 28:1935-1951. [DOI] [PubMed] [Google Scholar]
- 52.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 53.Sutherland, I. W. 2001. The biofilm matrix—an immobilized but dynamic microbial environment. Trends Microbiol. 9:222-227. [DOI] [PubMed] [Google Scholar]
- 54.Sutherland, I. W., K. A. Hughes, L. C. Skillman, and K. Tait. 2004. The interaction of phage and biofilms. FEMS Microbiol. Lett. 232:1-6. [DOI] [PubMed] [Google Scholar]
- 55.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 57.Wagner-Dobler, I., and H. Biebl. 2006. Environmental biology of the marine Roseobacter lineage. Annu. Rev. Microbiol. 60:255-280. [DOI] [PubMed] [Google Scholar]
- 58.Wagner-Dobler, I., V. Thiel, L. Eberl, M. Allgaier, A. Bodor, S. Meyer, S. Ebner, A. Hennig, R. Pukall, and S. Schulz. 2005. Discovery of complex mixtures of novel long-chain quorum sensing signals in free-living and host-associated marine Alphaproteobacteria. Chembiochem 6:2195-2206. [DOI] [PubMed] [Google Scholar]
- 59.Watnick, P., and R. Kolter. 2000. Biofilm, city of microbes. J. Bacteriol. 182:2675-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Webster, N. S., and A. P. Negri. 2006. Site-specific variation in Antarctic marine biofilms established on artificial surfaces. Environ. Microbiol. 8:1177-1190. [DOI] [PubMed] [Google Scholar]
- 61.Xavier, J. B., and K. R. Foster. 2007. Cooperation and conflict in microbial biofilms. Proc. Natl. Acad. Sci. USA 104:876-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, K., H. Choi, D. D. Dionysiou, G. A. Sorial, and D. B. Oerther. 2006. Identifying pioneer bacterial species responsible for biofouling membrane bioreactors. Environ. Microbiol. 8:433-440. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, X., and P. L. Bishop. 2003. Biodegradability of biofilm extracellular polymeric substances. Chemosphere 50:63-69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.